Background: The SALMs are neuronal cell adhesion molecules.
Results: Deletion of the SALM1 PDZ-binding motif or mutation of a dileucine motif affects ER retention and surface expression.
Conclusion: Enhanced SALM1 surface causes formation of elongated processes.
Significance: SALMs can regulate neuronal morphology and may be involved in developmental disorders like autism.
Keywords: Cell Adhesion, Glycosylation, Neurite Outgrowth, Neurodevelopment, Protein Targeting, ER Retention, Cell Adhesion Molecules, Dileucine Motif, Synapse Formation
Abstract
Synaptic adhesion-like molecules (SALMs) are a family of cell adhesion molecules involved in neurite outgrowth and synapse formation. Of the five family members, only SALM1, -2, and -3 contain a cytoplasmic C-terminal PDZ-binding motif. We have found that SALM1 is unique among the SALMs because deletion of its PDZ-binding motif (SALM1ΔPDZ) blocks its surface expression in heterologous cells. When expressed in hippocampal neurons, SALM1ΔPDZ had decreased surface expression in dendrites and the cell soma but not in axons, suggesting that the PDZ-binding domain may influence cellular trafficking of SALMs to specific neuronal locations. Endoglycosidase H digestion assays indicated that SALM1ΔPDZ is retained in the endoplasmic reticulum (ER) in heterologous cells. However, when the entire C-terminal tail of SALM1 was deleted, SALM1 was detected on the cell surface. Using serial deletions, we identified a region of SALM1 that contains a putative dileucine ER retention motif, which is not present in the other SALMs. Mutation of this DXXXLL motif allowed SALM1 to leave the ER and enhanced its surface expression in heterologous cells and neurons. An increase in the number of protrusions at the dendrites and cell body was observed when this SALM1 mutant was expressed in hippocampal neurons. With electron microscopy, these protrusions appeared to be irregular, enlarged spines and filopodia. Thus, enrichment of SALM1 on the cell surface affects dendritic arborization, and intracellular motifs regulate its dendritic versus axonal localization.
Introduction
Cell adhesion molecules (CAMs)2 are membrane-anchored molecules that directly interact through their extracellular domains to hold the membranes of two neurons together, at synapses or other points of contact (1–4). SALMs, also known as Lrfn (leucine-rich repeat and fibronectin III domain-containing), are a recently identified family of adhesion molecules that are largely, but not exclusively, restricted to neurons (5–8). They are enriched in growth cones and then later at synapses, and changes in SALM expression alter neurite outgrowth and synapse formation (5, 6, 9, 10). The SALMs have recently been associated with autism spectrum disorders (11–14), suggesting that changes in their expression have a significant biological impact, possibly by altering neuronal morphology.
The SALMs contain an extracellular leucine-rich region, an immunoglobulin C2-like domain, a fibronectin type III domain, a transmembrane domain, and an intracellular C-terminal domain. Although very similar in their extracellular N-terminal domains, their C termini are highly divergent (supplemental Fig. 1A). SALM1 to -3 have PDZ-binding domains (PDZ-BDs), whereas SALM4 and -5 do not. Overexpression of each SALM enhances neurite outgrowth in hippocampal neurons in culture, although the patterns of outgrowth vary with individual SALMs (9). SALMs also differ in their associational properties. SALM4 and -5 form homophilic intercellular interactions, whereas SALM1 to -3 do not, and binding partners for SALM1 to -3 remain unidentified (15). Co-immunoprecipitation studies showed that SALMs can form both homomeric and heteromeric complexes in neurons and heterologous cells, but individual SALMs show specific preferences (15). These studies suggest that individual members of the SALM family have different functions and perhaps different localizations in neurons.
In our initial study of SALMs, we found that deletion of the PDZ-BD at the C terminus of SALM1 drastically reduces its surface expression in heterologous cells and in neurons, showing that a PDZ interaction is required for normal trafficking of SALM1 (5). In the present study, we studied the surface expression of SALM2 and -3 and asked if deletion of their PDZ-BDs also led to a reduction of surface expression; SALM4 and -5, which naturally lack PDZ-BDs, are abundantly expressed on the cell surface in heterologous cells (15). Only SALM1 had reduced surface expression when its PDZ-BD was removed. Therefore, we investigated the mechanisms underlying the role of the PDZ-BD in the surface expression of SALM1. Our results indicate that deletion of the PDZ-BD results in retention of SALM1 in the ER due to the presence of a second motif in the cytoplasmic C terminus. Mutation of this dileucine motif, in the absence of the PDZ-BD, allows the release of SALM1 from the ER in heterologous cells. In neurons, removal of the SALM1 PDZ-BD mainly localizes SALM1 to the surface of axons. Mutation of the dileucine motif increases somatodendritic localization and also enhances the neuronal morphological complexity. Therefore, our results highlight the different trafficking mechanisms of SALM1 that may underlie its unique functional properties among the SALMs.
EXPERIMENTAL PROCEDURES
SALM cDNA Constructs
Cloning of the SALM family members was described previously (5). The SALM deletion constructs were made using the QuikChange site-directed mutagenesis method (Stratagene, La Jolla, CA). The C-terminal deletion of SALM1 (Myc-SALM1ΔCT) was constructed by inserting a stop codon after the transmembrane domain, resulting in termination at Lys-558. Myc-SALM1 was also truncated by inserting a stop codon at Glu-608 (608 stop), Pro-680 (680 stop), Gly-691 (691 stop), Pro-698 (698 stop), Gly-701 (701 stop), Ala-704 (704 stop), Glu-716 (716 stop), Tyr-741 (741 stop), or Val-788 (Δ1). The Myc-SALM1ΔPDZ, Myc-SALM2ΔPDZ, and SALM3ΔPDZ constructs were generated by inserting a stop codon before the PDZ-binding motif as described previously (9). Mutagenesis was used to mutate Leu-700, Pro-703, or Asp-695 to alanine in Myc-SALM1ΔPDZ (L700AΔPDZ, P703AΔPDZ, and D695AΔPDZ, respectively).
Antibodies
Production of SALM polyclonal antiserum was described previously (15). Briefly, a SALM1 N-terminal antibody was generated in rabbits using a synthetic peptide directed toward amino acids 384–397 (Covance, Denver, PA). A 1:250 dilution was used for surface staining of HeLa cells and hippocampal neurons. A SALM2 C-terminal antibody was generated using amino acids 651–666 and used at a 1:500 dilution for total staining in HeLa cells and Western blotting. Anti-SALM3 antiserum was made using a peptide targeted to N-terminal amino acids 377–389 and used at a 1:250 dilution for Western blotting. Anti-Myc monoclonal antibody (1:500, clone 9E10, hybridoma purchased from ATCC, Manassas, VA), PSD-95 (1:500, Affinity BioReagents/Thermo Fisher Scientific), and guinea pig VGLUT1 (1:2000, Millipore) were used for immunofluorescence.
Biotinylation and Western Blotting
HeLa cells were plated at 20–30% confluence and maintained at 50–90% confluence in DMEM (Invitrogen) containing heat-inactivated fetal bovine serum (FBS) (Invitrogen), 2 mm l-glutamine, and 1 mm sodium pyruvate. Forty-eight hours after cells were transfected with SALM cDNA using calcium phosphate, surface proteins were biotinylated with N-hydroxysulfosuccininimide-biotin (NHS-SS-biotin) (1 mg/ml) (Pierce) for 20 min at 4 °C. Cells were washed three times with cold glycine buffer (25 mm) to terminate the biotinylation reaction. Proteins were solubilized with 1% SDS buffer (1% SDS, 150 mm NaCl, 50 mm Tris, pH 7.4, 5 mm EDTA, and a protease inhibitor mixture) (Complete, EDTA-free pills, Roche Applied Science) and heated at 37 °C for 20 min. After high speed centrifugation (110,000 × g for 30 min at 4 °C), the protein concentration was determined using the BCA assay (Pierce). NeutrAvidin-agarose beads (Pierce) were added to 500 μg of cell lysate for precipitation of biotinylated proteins and incubated for 2 h at 4 °C. The beads were washed with 0.1% Triton X-100 in TBS three times and resuspended in 2× SDS loading buffer. Samples were heated at 95 °C for 5 min and resolved by SDS-PAGE on a 10% Tris-glycine gel (Invitrogen). Immunoblotting was performed using SALM antibodies, peroxidase-coupled secondary antibodies (GE Healthcare), and chemiluminescence (Amersham Biosciences ECL Plus, GE Healthcare).
Endoglycosidase H (Endo H) Digestion and Deglycosylation
HeLa cells, expressing the SALM1 constructs, were harvested, and pellets were resuspended in denaturing buffer (10 mm NaH2PO4, pH 6, 0.5% SDS, and 2% glycerol), incubated for 3 min at 95 °C, and diluted with 1% Nonidet P-40 in 10 mm NaH2PO4, pH 6, with protease inhibitors. Lysate was incubated with N-glycosidase F (3 units) (Prozyme, Hayward, CA) or Endo H (15 milliunits) (Prozyme) for 6 h at 37 °C before being diluted with 3× sample buffer and loaded onto a 10% SDS-polyacrylamide gel. Immunoblotting was performed using SALM1 antibodies and processed as described above. Films were scanned for densitometric analysis using a GE Healthcare densitometer.
Immunocytochemistry
HeLa cells or hippocampal neurons were transiently transfected with SALM1 mutant or deletion constructs using calcium phosphate precipitation (Clontech, Mountainview, CA). GFP cDNA was cotransfected with the SALM1 constructs in neurons to visualize their morphology. Hippocampal neurons were cultured as described previously (5, 16), and all experiments involving animals were performed according to National Institutes of Health guidelines. Transfections were performed at 11–12 days in vitro (DIV), and immunocytochemistry was performed at 14–15 DIV.
For surface staining of HeLa cells, cells were washed with cold PBS (supplemented with 1 mm magnesium and 0.1 mm calcium) (PMC) and incubated on ice with primary antibodies for 1 h. Cells were washed, blocked with 10% normal goat serum and 1% bovine serum albumin (BSA) in PMC, and incubated on ice with Alexa Fluor 568 or 647 secondary antibody (Invitrogen/Molecular Probes) for 30 min. Cells were then fixed with 4% paraformaldehyde at room temperature and washed with PBS. For total staining, cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min, blocked with 10% normal goat serum in PBS for 1 h, incubated with primary antibody, and stained with Alexa Fluor 488 secondary antibodies (Invitrogen/Molecular Probes). Total staining was performed at room temperature.
For hippocampal neurons, surface staining was performed for 20 min at room temperature. Neurons were washed with neurobasal media, fixed with 4% paraformaldehyde, blocked for 10 min with 10% normal goat serum in PBS, and stained with Alexa Fluor 555 antibodies. Cells were then processed for total staining by permeabilizing neurons with 0.25% Triton X-100 in PBS for 5 min. Blocking steps and antibody incubations were the same as for HeLa cells. Monoclonal PSD-95 or guinea pig VGLUT1 primary antibodies were labeled with Alexa Fluor 647 secondary antibodies. Coverslips were mounted onto slides using ProLong Antifade mounting reagent (Invitrogen/Molecular Probes).
Fluoresence Microscopy
Images of HeLa cells were acquired using the 60× oil objective of an E-1000 Nikon microscope. Neurons were imaged using an LSM 710 confocal microscope or using a Delta Vision wide field restoration or super resolution microscope. Images from the LSM710 were taken using a 40 × 1.3 numerical aperture or 100 × 1.46 numerical aperture oil objective. Three-dimensional reconstructions used Zeiss software. Peter Franklin from Applied Precision assisted with the microscopy using DeltaVision Widefield Restoration Microscopy, and Leanna Ferrand and Adrian Quintanilla from Applied Precision assisted with microscopy using the DeltaVision OMX three-dimensional structured illumination microscope, which can provide superresolution (resolution beyond the diffraction limit) in all three axes (17). Three-dimensional images of neurons using the latter were examined with Imaris software. Spine density was counted using ImageJ (National Institutes of Health), and dendrite branching was determined using Metamorph Neurite Outgrowth Module version 7.0r3 (Molecular Devices, Sunnyvale, CA) as described previously (9). Surface expression of transfected SALM proteins in neurons was quantified using Metamorph. Images were consistently thresholded, and integrated pixel intensity of surface puncta on axons and dendrites was examined in 20 μm regions.
Electron Microscopy
After surface staining with anti-SALM1 primary antibody, transfected neuronal cultures were fixed in 4% PFA and 0.1% glutaraldehyde and treated with 0.3% H2O2 in PBS and then 0.5 mg/ml fresh sodium borohydride in PBS for 5 min. Immunoperoxidase labeling was performed using the Vectastain ABC kit and 3,3′-diaminobenzidine peroxidase substrate kit (Vector Laboratories). Labeled sections were postfixed in 2% glutaraldehyde, followed by 1% osmium tetroxide and then 1% uranyl acetate during dehydration, and embedded in epon resin. This was followed by removal of glass using hydrofluoric acid and further processing for electron microscopy, as described previously (15, 18).
Laser Scanning Cytometry
Cell surface SALM1 transfected in HeLa cells was measured using an iCys® Research Imaging Cytometer (CompuCyte; Cambridge, MA). Cells were stained for surface SALM1 as described, and Cy5 was used as a secondary antibody. After cells were permeabilized, total staining was performed using anti-Myc and Alexa Fluor 488 antibodies. Slides were imaged with an attached IX71 Olympus microscope and DONPISHA XC-003 3CCD Color Vision Camera module. Standard filter settings were used for detection: green filter (530/30 FITC) and long red filter (650/LP Cy5). Excitation was with a 488-nm argon laser and 633-nm helium-neon laser. Two slides were imaged per sample per experiment, with a minimum of 672 cells analyzed per sample. Samples were analyzed using a ×20 objective and gated via detection of FITC-stained internal SALM. The amount of cell surface SALM1 was then determined on these cells as the mean red integral. Experiments were performed four times.
Statistical Analysis
Data were analyzed using GraphPad Software (San Diego, CA) Prism version 5.0. Significant differences between two groups were determined using two-tailed, unpaired Student's t test. ANOVA, followed by the Newman-Keuls multiple-comparison test, was used to find significant differences among multiple groups. All values are reported as mean ± S.E.
RESULTS
SALM1 Requires PDZ-BD for Surface Expression
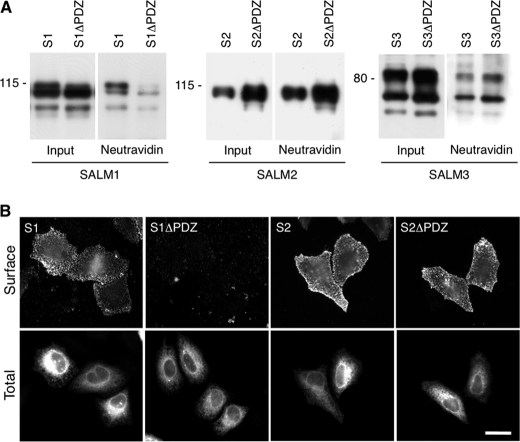
Using a biotinylation assay to determine surface expression of SALM1, -2, and -3 transfected in heterologous cells, we found that both SALM2ΔPDZ and SALM3ΔPDZ are expressed on the cell surface at levels similar to those of the intact molecules, whereas SALM1ΔPDZ is only weakly detected on the cell surface (Fig. 1A and supplemental Fig. 1B). On the other hand, SALM2 and SALM2ΔPDZ had more protein expressed on the cell surface than SALM1 or SALM3 (supplemental Fig. 1B). The levels were further analyzed with immunofluorescence to measure surface labeling of SALM1 and SALM2. Similar to results of the biotinylation experiments, we found that removal of the PDZ-BD of SALM2 had no effect on surface expression in heterologous cells (Fig. 1B), whereas very little SALM1ΔPDZ was detected. These results indicate that SALM1 has a unique trafficking mechanism that regulates its surface expression and therefore surface signaling.
FIGURE 1.
SALM1 surface expression in HeLa cells. A, HeLa cells transfected with Myc-SALM1 (S1), Myc-SALM1ΔPDZ (S1ΔPDZ), Myc-SALM2 (S2), Myc-SALM2ΔPDZ (S2ΔPDZ), SALM3 (S3), and SALM3ΔPDZ (S3ΔPDZ) were labeled with biotin, washed with glycine, and then lysed. Surface-labeled SALM was precipitated with NeutrAvidin beads. SALM1, SALM2, and SALM3 were detected by immunoblotting using antibodies toward their N-terminal region (S1 and S3) or C-terminal region (S2). Very little SALM1ΔPDZ associates with the avidin beads compared with WT; however, similar amounts of SALM2ΔPDZ and SALM3ΔPDZ bind compared with full-length SALM2 and SALM3, respectively (see supplemental Fig. 2). B, Myc-SALM1 (S1), Myc-SALM1ΔPDZ (S1ΔPDZ), Myc-SALM2 (S2), and Myc-SALM2ΔPDZ (S2ΔPDZ) were transfected into HeLa cells, and surface expression was detected using an antibody directed against the SALM1 N-terminal region or the SALM2 Myc tag (top row, Surface). Intracellular SALM1 was detected with anti-Myc antibody, and SALM2 was detected with a SALM2 C-terminal antibody (bottom row, Total). SALM1 is reduced at the surface when the PDZ-binding domain is deleted. Scale bar, 20 μm.
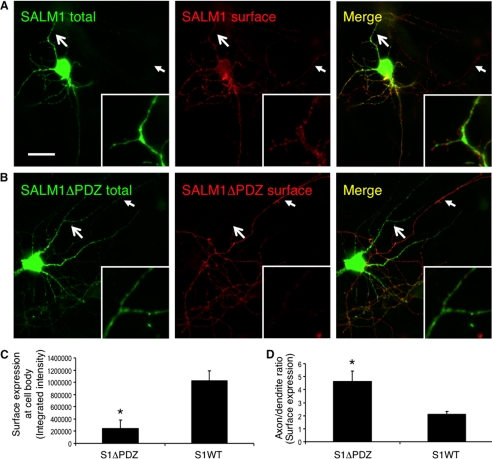
SALM1 Axonal Versus Dendritic Targeting Is Regulated by PDZ-BD
The trafficking of proteins is particularly important in neurons due to their complex architecture. Various proteins depend on PDZ interactions or other intracellular motifs for proper trafficking to specific locations in neurons, such as the pre- or postsynaptic membrane (19). Other studies have shown that removal of the PDZ-BD of CAMs, such as neurexin, reduces the surface expression in both axons and dendrites (20). To examine SALM1 surface targeting and localization, we transfected SALM1 and SALM1ΔPDZ into hippocampal neurons. We found that wild type (WT) SALM1 was detected on the surface of both axons and dendrites (Fig. 2A). On the other hand, SALM1ΔPDZ surface expression was mainly detected in axons (Fig. 2B). Quantification of surface levels of SALM1 detected on the dendrites and cell body was 5 times that of SALM1ΔPDZ (Fig. 2C). Quantification of the axon/dendrite ratio indicated that more SALM1 was present on the surface of axons relative to dendrites (Fig. 2D). However, SALM1ΔPDZ showed a 2-fold increase in axonal surface expression over SALM1 (Fig. 2D). This is in contrast to heterologous cells, where very little SALM1ΔPDZ makes it to the cell surface. When the surface expression of SALM2 and SALM2ΔPDZ was examined in neurons (supplemental Fig. 1B), both were detected on the surface of axons and dendrites in similar amounts (supplemental Fig. 1C). These results indicate that trafficking of SALM1 to the dendrites may be at least partially regulated by the PDZ-BD, whereas axonal targeting may be regulated by a different domain in the SALMs or serve as a default pathway.
FIGURE 2.
SALM1 surface expression is mediated by the PDZ-binding domain. A, primary hippocampal neurons were transfected with Myc-SALM1 or Myc-SALM1ΔPDZ at DIV 12, and immunocytochemistry was performed 48 h later. Transfected SALM1 is expressed throughout the neuron and traffics readily to the surface of axons, dendrites, and the soma as detected by an N-terminal SALM1 (anti-S1NT) antibody (surface). Intracellular SALM1 was detected using an anti-Myc antibody (total). B, transfected SALM1ΔPDZ is expressed throughout the neuron, although the surface expression at somatodendritic sites is significantly reduced. The large arrows indicate dendritic arbors (magnified in insets), whereas the small arrows indicate axonal processes. C, quantification of SALM1 and SALM1ΔPDZ surface expression (integrated intensity) at the cell body. Deletion of the PDZ-BD significantly reduces somatic surface expression of SALM1ΔPDZ (241,944 ± 140,318, n = 6; *, p < 0.05, Student's t test) compared with SALM1 (1,029,640 ± 155,449, n = 7). D, quantification of the surface expression ratio (integrated intensity) of axons and dendrites of SALM1 (2.08 ± 0.27, n = 7) and SALM1ΔPDZ (4.61 ± 0.79, n = 6; *, p < 0.05, Student's t test) transfected neurons indicates that SALM1ΔPDZ has a significantly higher axon/dendrite ratio than SALM1. Values are shown as mean ± S.E. (error bars). Scale bar, 20 μm.
Identification of Retention Signal in C-terminal Domain
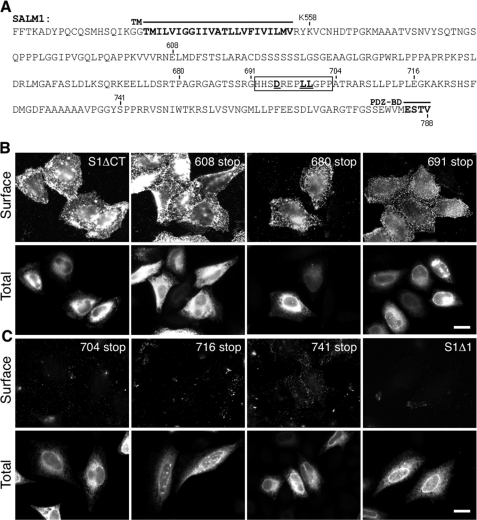
The lack of surface expression of SALM1ΔPDZ in heterologous cells could be due to several factors, including misfolding of the molecule or improper trafficking due to the loss of the PDZ-BD. The latter could arise through activation of a default pathway or could be due to uncovering retention signals that are normally masked in the WT molecule. To address these questions, we asked if removal of the entire C terminus affected surface expression of SALM1. Transient transfection of SALM1 truncated after the transmembrane domain at residue Lys-558 (SALM1ΔCT) (Fig. 3A) showed robust surface expression (Fig. 3B, top left). These results indicate that the PDZ-BD is not the primary determinant for expression on the cell surface and raise the possibility of a signal in the C terminus of SALM1 that is responsible for the intracellular retention of SALM1 in the absence of a PDZ-BD.
FIGURE 3.
SALM1 C-terminal domain regulates surface expression. A, sequence of SALM1 C-terminal region. Myc-SALM1 was truncated by inserting a stop codon at Lys-558 (S1MΔCT), Glu-608 (608 stop), Pro-680 (680 stop), Gly-691 (691 stop), Ala-704 (704 stop), Glu-716 (716 stop), Tyr-741 (741 stop), or Val-788 (S1Δ1), as indicated in the diagram. The motif responsible for intracellular retention is underlined. TM, transmembrane domain. B and C, representative immunofluorescent images of Myc-SALM1 deletion constructs transfected into HeLa cells. Surface staining was detected by anti-S1NT (Surface), and total SALM1 was detected using anti-Myc antibodies (Total). 691 stop is detected on the cell surface (B, last panel), whereas 704 stop is not (C, first panel), suggesting a putative retention motif between residues 691 and 704 (boxed region). Scale bar, 20 μm.
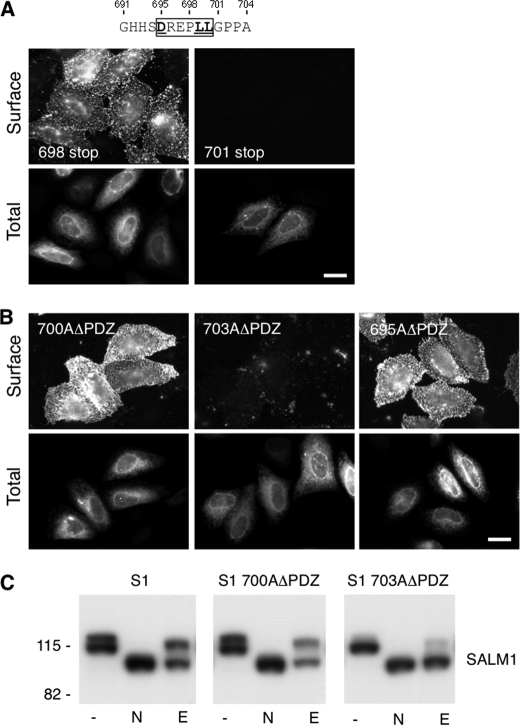
Next, we sought to identify the domain(s) responsible for intracellular retention of SALM1ΔPDZ in heterologous cells. We made a series of truncations (Fig. 3A) in the C terminus and determined if they were expressed on the cell surface of heterologous cells. Surface expression was determined by a region between amino acids 691 and 704; immunostaining indicated that a construct terminated at Gly-691 was robustly expressed on the cell surface (Fig. 3B), whereas one terminated at Pro-704 had little cell surface expression (Fig. 3C). This region contains a dileucine motif corresponding to the consensus sequence (D/E)XXXLL. Dileucine motifs are commonly involved in protein trafficking but usually at the level of the trans-Golgi network, endosomes, plasma membrane, and lysosomes (21). However, dileucine motifs also have been shown to play a role in ER trafficking (22–28).
To determine whether the dileucine motif in this region is responsible for intracellular retention of SALM1ΔPDZ, additional truncation and mutation studies were carried out. The SALM1 C terminus was further truncated at residue Pro-698, immediately before the dileucine motif, and at Gly-701, after the dileucine motif (Fig. 4A, 698 stop and 701 stop, respectively). Whereas 698 stop was present on the cell surface, 701 stop was not detected (Fig. 4A). Expression of 701 stop was confirmed by total staining. Therefore, these results suggest that the dileucine motif at residues 699 and 700 is responsible for the intracellular retention of SALM1ΔPDZ. To confirm the requirement of this domain, an alanine was introduced at residue 700 to replace the leucine in the SALM1ΔPDZ construct. Mutation of residue 700 disrupted the dileucine motif and allowed the detection of the surface expression of the mutant construct (Fig. 4B, top left). However, mutation of residue Pro-703 to alanine, after the dileucine motif, did not result in surface expression of SALM1ΔPDZ (Fig. 4B, top middle). Therefore, the dileucine motif at residues 699 and 700 regulates the intracellular retention of SALM1 in the absence of the PDZ-BD. In some cases, the acidic amino acid preceding the dileucines (Asp or Glu of (D/E)XXXLL) is also required for activity (21). To test this, we mutated Asp-695 to alanine (Fig. 4B, top right). This construct also permitted SALM1ΔPDZ to reach the cell surface, showing that Asp-695 along with the dileucine motif at 699 and 700 are required for the intracellular retention of SALM1ΔPDZ. Therefore, our results show that the motif DREPLL is responsible for the intracellular retention of SALM1ΔPDZ.
FIGURE 4.
Identification of a SALM1 dileucine ER retention motif. A, two additional Myc-SALM1 deletion constructions (698 stop and 701 stop) were made to further isolate the retention motif. Myc-SALM1 truncations (698 stop or 701 stop) were transfected into HeLa cells, and surface staining was detected by anti-S1NT (Surface). Cells were fixed and permeabilized to detect intracellular SALM1 with anti-Myc antibodies (Total). B, mutagenesis was used to mutate Leu-700 and Pro-703 to alanine in Myc-SALM1ΔPDZ (L700AΔPDZ and P703AΔPDZ, respectively), and both surface and total staining of transfected cells was performed. Scale bar, 20 μm. C, lysate from HeLa cells transfected with Myc-SALM1, Myc-SALM1 L700AΔPDZ, or Myc-SALM1 P703AΔPDZ was left untreated (−) or digested with N-glycosidase (N) or Endo H (E). All digests were followed by SDS-PAGE, and SALM1 was detected by immunoblotting with a polyclonal anti-S1NT antibody. Both mutant and WT proteins show more rapid migration following glycosidase cleavage of N-linked oligosaccharides, whereas only SALM1 P703AΔPDZ is fully digested by the high mannose-directed glycosidase Endo H. Molecular mass markers are shown in kDa.
Intracellular ER Retention Site of SALM1
Next, we assessed the glycosylation states of WT SALM1, SALM1 L700AΔPDZ, SALM1 P703AΔPDZ (Fig. 4C), and SALM1ΔPDZ (Fig. 5, A and B). In untreated lysate, WT SALM1 appears as a 116- and 114-kDa doublet on immunoblots (Fig. 4C, lane 1). This doublet was also present in SALM1 L700AΔPDZ lysates. However, the 114 kDa band was the predominant form present in SALM1 P703AΔPDZ (Fig. 4C, third panel) and SALM1ΔPDZ lysates (Fig. 5A, second panel), suggesting that it is a less mature, high mannose glycosylated form of SALM1. The maturation state was further analyzed by using N-glycosidase F, which cleaves both high and complex mannose sugars, resulting in complete deglycosylation of the SALM1 constructs, and by using an Endo H sensitivity assay. WT SALM1 and SALM1 L700AΔPDZ were largely Endo H-resistant, indicating that the protein had passed through the medial Golgi, whereas SALM1 P703AΔPDZ and SALM1ΔPDZ were nearly entirely Endo H-sensitive, indicating an ER localization (Figs. 4C and 5A). As a first step in identifying where the trafficking of SALM1ΔPDZ is disrupted, we determined the intracellular localization of WT SALM1 and SALM1ΔPDZ. In HeLa cells, the intracellular staining of SALM1 produced a pattern similar to the reticular network of the ER. Although WT SALM1 was present in the ER, which is common for many proteins overexpressed in heterologous cells, it was evident that SALM1ΔPDZ was very abundant in the ER (supplemental Fig. 2). Colocalization experiments showed that SALM1 and SALM1ΔPDZ colocalize with the ER markers KDEL (supplemental Fig. 2A) and SERCA (not shown) but not the Golgi marker GM130 (supplemental Fig. 2B).
FIGURE 5.
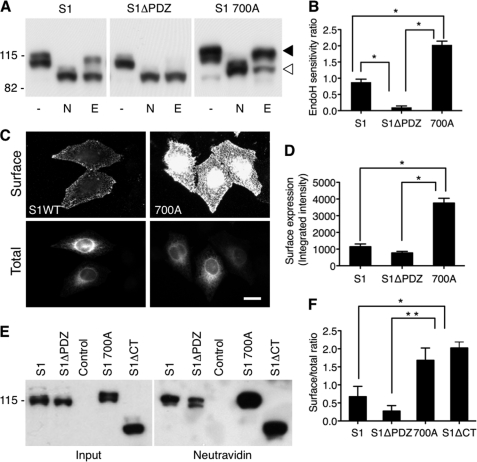
Regulation of SALM1 ER retention and surface expression. A, lysate from HeLa cells transfected with Myc-SALM1, Myc-SALM1ΔPDZ, or Myc-SALM1 L700A was left untreated (−) or digested with N-glycosidase (N) or Endo H (E). Expression of SALM1 constructs was detected by immunoblotting with a polyclonal anti-S1NT antibody. SALM1 L700A is primarily Endo H-insensitive compared with SALM1 (top band, filled arrowhead), whereas most of the SALM1ΔPDZ expressed is Endo H-sensitive (bottom band, open arrowhead), suggesting that it is ER-retained. B, quantification of the ratio of Endo H-insensitive to Endo H-sensitive fraction using a densitometer indicates that SALM1 L700A is significantly less retained in the ER. Histograms show mean ± S.E. (error bars) (S1, 0.88 ± 0.09, n = 6; S1ΔPDZ, 0.10 ± 0.04, n = 6; 700A, 2.03 ± 0.08, n = 5; *, p < 0.05, one-way ANOVA). C, comparison of wild type Myc-SALM1 with Myc-SALM1 L700A indicates that mutation of residue Leu-700 enhances surface expression of SALM1. Surface staining was detected by anti-S1NT (Surface), and total SALM1 was detected using anti-Myc antibody (Total). D, quantification of surface expression of Myc-SALM1, Myc-SALM1ΔPDZ, and Myc-SALM1 L700A using laser scanning cytometry shows a significant increase in surface expression of SALM1 L700A. Histograms show mean ± S.E. (S1, 1145 ± 161, n = 4; S1ΔPDZ, 771 ± 92, n = 4; 700A, 3754 ± 289, n = 4; one-way ANOVA, p < 0.001). E, HeLa cells transfected with Myc-SALM1, Myc-SALM1ΔPDZ, control pCDNA3.1 vector, Myc-SALM1 L700A, or Myc-SALM1ΔCT were labeled with biotin, washed with glycine, and then lysed. Surface-labeled SALM was precipitated with NeutrAvidin beads. SALM1 constructs were detected by immunoblotting with a polyclonal anti-S1NT antibody. F, quantification of the surface (NeutrAvidin pull-down)/total (input) ratio using a densitometer. Histograms show mean ± S.E. (S1, 0.53 ± 0.33, n = 3; S1ΔPDZ, 0.24 ± 0.17, n = 3; 700A, 1.68 ± 0.34, n = 3; S1ΔCT, 2.02 ± 0.17, n = 3; *, p < 0.05; **, p < 0.001, one-way ANOVA).
Regulation of SALM1 Surface Expression
Although our results show that an acidic dileucine motif is responsible for ER retention of SALM1 constructs lacking the PDZ-BD, there is no evidence that this motif plays any role in the trafficking of WT SALM1. To address this, we mutated Leu-700 to alanine in WT SALM1 to determine its effect on surface expression and ER retention, again by Endo H sensitivity. As seen in Fig. 5A (panel 3), the mutation significantly increased the presence of the 116 kDa band in SALM1 L700A lysates and the amount of Endo H-insensitive protein. Similarly, surface expression of SALM1 L700A was increased over that of the WT, as shown by an immunofluorescence (Fig. 5, C and D) or biotinylation assay (Fig. 5, E and F). Quantification of surface expression by laser scanning cytometry showed that SALM1 L700A had significantly higher surface expression than WT SALM1 or SALM1ΔPDZ (Fig. 5D). To test if mutation of the dileucine motif was necessary for this increased surface expression, we used a biotinylation assay to compare SALM1 L700A and SALM1ΔCT surface expression with SALM1 and SALM1ΔPDZ. Expression of both SALM1 L700A and SALM1ΔCT greatly enhanced the amount of biotinylated protein detected on the cell surface (Fig. 5F). These results show that the acidic dileucine motif enhances ER retention of WT SALM1. In the absence of the PDZ-BD, this retention was nearly complete. However, mutation of the dileucine motif allows release of SALM1 from the ER, passage through the trans-Golgi network, and enhanced surface expression of SALM1. Both WT SALM1 and SALM1 L700AΔPDZ have a similar Endo H-insensitive fraction, indicating that the PDZ-BD is necessary and may play a role in SALM1 trafficking from the ER. However, the removal of the entire C-terminal domain of SALM1 enhanced surface expression, suggesting that the PDZ-BD is not required when the ER retention motif is not present.
Enhanced Dendrite Complexity with Increased SALM1 Surface Expression
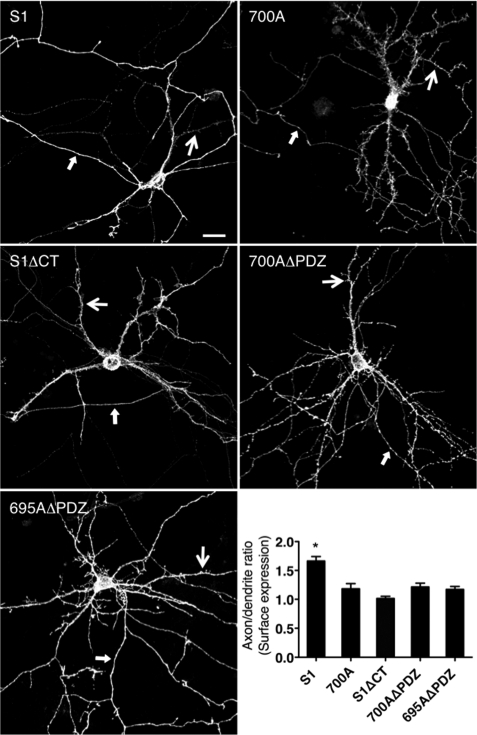
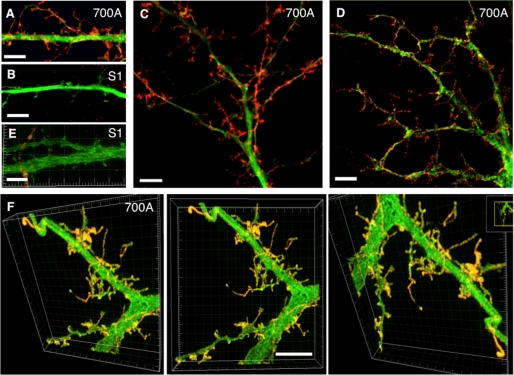
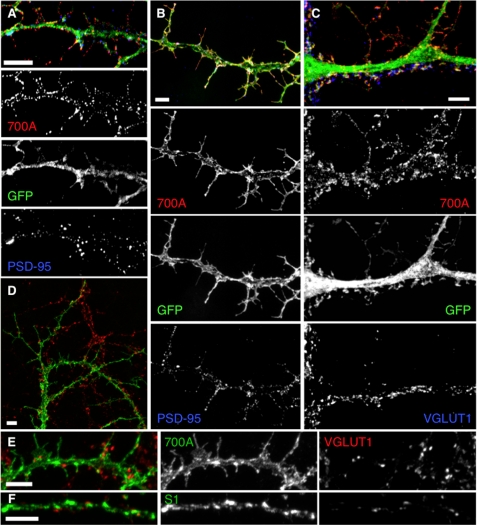
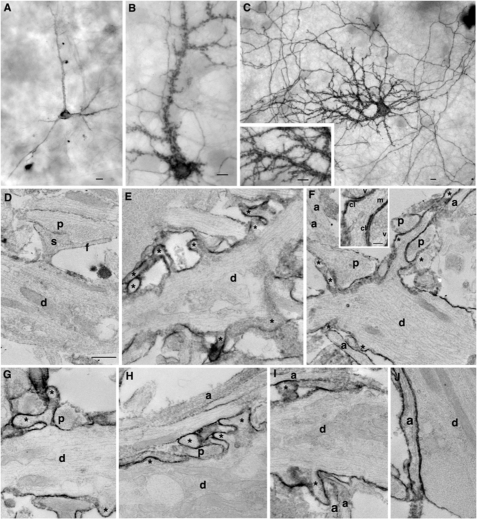
To test the requirement of the dileucine motif and/or PDZ-BD for somatodendritic localization, SALM1, SALM1 L700A, SALM1ΔCT, L700AΔPDZ, and D695AΔPDZ were transfected into neurons. Expression of SALM1 L700A, SALM1ΔCT, L700AΔPDZ, and D695AΔPDZ all had increased somatodendritic surface expression compared with WT SALM1 (Fig. 6), suggesting that loss of the dileucine motif enhances dendritic localization. Furthermore, the SALM1 L700A mutant appeared to have more dendritic filopodia, and the complexity of dendrite branching was also enhanced. We detected a 40% increase in dendrite branching, and spine density was increased from 0.49 ± 0.04 spine/μm (SALM1 WT) to 0.76 ± 0.04 spine/μm (SALM1 L700A, Student's t test, p < 0.001). Therefore, the enhanced dendrite complexity produced by SALM1 L700A expression was examined in more detail using both high resolution light microscopy (Figs. 7 and 8) and electron microscopy (Fig. 9). High resolution light microscopy was performed using confocal microscopy with a 1.46 numerical aperture objective lens (Fig. 7, A–C), with wide field restoration microscopy (Fig. 7D), and with three-dimensional structured illumination superresolution microscopy (Fig. 7, E and F). The latter method can improve the resolution in all three axes and is especially useful for three-dimensional analysis of structures. These methods showed the SALM1 L700A labeling as dense puncta that cover large irregular processes on dendrites. Some processes resembled filopodia, growth cones, or fine dendritic branches, and others may be extensions of abnormally growing spines. Colocalization with the postsynaptic marker PSD-95 or presynaptic marker VGLUT1 showed that only some of these irregular processes are synaptic, and often the processes extend well beyond the synaptic zone (Fig. 8). PSD-95 was present in the filopodial extensions of neurons expressing mutant SALM1 L700A (Fig. 8, A and B). In contrast, VGLUT1 puncta did not colocalize with these extensions but was localized more proximal to the dendrites in these neurons (Fig. 8, C–E). A similar distribution of VGLUT1 was seen in WT SALM1 neurons (Fig. 8F). Actin was present in the dendrites and filopodia, but the endosomal markers EEA1 and transferrin receptor were mainly in the dendrites and excluded from the filopodia (supplemental Fig. 3). We further examined these irregular processes using immunoperoxidase/DAB labeling (Fig. 9). We first examined the hippocampal cultures with light microscopy (Fig. 9, A–C); these showed the same pattern as seen with fluorescence microscopy (Figs. 6–8), with processes on dendrites enlarged and growing irregularly as compared with WT. With electron microscopy (Fig. 9, D–J), most of these processes extended beyond synaptic connections if present; some showed the morphology of immature dendritic branches, whereas others appeared to be irregular extensions of spines or filopodia, as indicated with the light microscope studies.
FIGURE 6.
Loss of dileucine motif enhances SALM1 dendritic localization. Primary hippocampal neurons were transfected with Myc-SALM1, Myc-SALM1 L700A, Myc-SALM1ΔCT, Myc-SALM1 L700AΔPDZ, or Myc-SALM1 D695AΔPDZ at DIV 12 (compare with Myc-SALM1ΔPDZ in Fig. 2). Surface immunocytochemistry was performed 48 h later using an N-terminal SALM1 antibody. Confocal images of neurons transfected with Myc-SALM1 L700A show an increased complexity of the dendritic arbor, with highly prominent irregular spines and filopodia. Quantification of the surface expression (integrated intensity) of the SALM1 constructs indicates that SALM1 has a higher axon/dendrite ratio than SALM1 L700A, SALM1ΔCT, SALM1 L700AΔPDZ, and SALM1 D695AΔPDZ. Results are mean ± S.E. (error bars) (S1, 1.68 ± 0.087, n = 8; 700A, 1.19 ± 0.10, n = 8; S1ΔCT, 1.00 ± 0.04, n = 10; 700AΔPDZ, 1.26 ± 0.07, n = 8; 695AΔPDZ, 1.22 ± 0.05, n = 6; one-way ANOVA; *, p < 0.05). Scale bar, 20 μm; large arrows indicate a dendrite, and small arrows indicate an axon.
FIGURE 7.
Enhanced SALM1 surface expression increases filopodia and dendritic process formation; high resolution light microscopy. Shown are examples of dendrites from hippocampal neurons transfected with Myc-SALM1 or Myc-SALM1 L700A and GFP (green) at DIV 12 and surface-stained with an N-terminal SALM1 antibody (S1NT) 48 h later. Images were taken using laser confocal microscopy with a 1.46-nm objective lens (Zeiss) (A–C), wide field restoration microscopy (DeltaVision) (D), and three-dimensional structured illumination microscope superresolution imaging (DeltaVision) (E and F; profiles are shown from various angles in the same three-dimensional reconstruction) and show SALM1 surface puncta (red, anti-S1NT). High resolution microscopy illustrates the length and shape of the filopodia and processes that form as a result of L700A expression. L700A-transfected neurons (A and C–F) have larger and more irregular processes on the surface of their dendrites compared with SALM1 WT (B and E). Scale bars, 5 μm.
FIGURE 8.
SALM1 L700A localization studies show processes lacking synapses. Representative immunofluorescent images of hippocampal neurons transfected at DIV 12 and immunostained after 48 h. Wide field restoration microscopy (DeltaVision) (A) and confocal microscopy (B) show surface expression of SALM1 L700A (red) and endogenous PSD-95 (blue). The postsynaptic marker PSD-95 (blue) is present in the filopodia that form as a result of L700A expression (red, surface, S1NT antibody; green, GFP). On the other hand, L700A does not necessarily colocalize with the presynaptic marker, VGLUT1 (blue), that is present along the dendrite, and VGLUT1 does not colocalize with the longer filopodia (C). Total staining for L700A (D–E) or WT SALM1 (F) (green, total, S1NT antibody) and endogenous VGLUT1 (red) shows that L700A is present throughout the dendrite and filopodia, whereas VGLUT1 (red) mainly localizes along the dendrite shaft, at the base of the filopodia, and not at their tips. Scale bars, 5 μm.
FIGURE 9.
Enhanced SALM1 surface expression increases dendritic arborization; electron microscopy. Shown is immunoperoxidase/DAB surface labeling of transfected SALM WT (A) or L700A (B–J) with light microscopy (A–C and inset) and EM (D–J) using an N-terminal SALM1 antibody. WT shows relatively smooth dendrites with a few labeled processes and low surface labeling (axons also labeled but not shown). In contrast, light microscopy of L700A shows “rough” dendrites covered in processes and densely labeled. Labeling in axons is also extensive. With EM, compare the relatively smooth, unlabeled surface of dendrites (d; thick, with tapering sides and containing bundles of microtubules, elongate mitochondria, and endosomes and other tubulovesicular structures) from untransfected neurons (D and J) with the irregular and process-laden (asterisks) labeled surface of dendrites of transfected neurons (E–I). Examples of presynaptic terminals (containing a cluster of synaptic vesicles) forming synapses include one on a normal spine (s; plus filopodium, f) in D, whereas another presynaptic terminal makes synapses near the base of the long process in F (synaptic vesicles (v) and synaptic clefts (cl) are labeled in the inset); the latter process appears to have at least one microtubule (m in inset) and is probably a developing dendritic branch. Other presynaptic terminals appear to be making synaptic contact with the dendrite shafts or the base of the adjacent process (a cleft is not distinct in these sections; left p in F and in G and H). Axons (a; thin, with roughly straight, parallel sides and containing microtubules) from untransfected (unlabeled axons in F, H, and I) or transfected (labeled axon in J) neurons often form in bundles on the surface of dendrites (H and I) as seen in normal cultures (14). Unmarked neurites in the micrographs may be dendrites or axons. Scale bars, 10 μm for light microscopy (A–C) and 500 nm for EM (D–J; 100 nm for inset in F) micrographs.
DISCUSSION
In this study, we investigated the intracellular trafficking and surface expression of SALM1. We found that the distal 5 amino acids of its C terminus, which include a type 1 PDZ-BD, are required for exit from the ER in heterologous cells. SALM2 and -3 are highly similar to SALM1 in their N termini and also terminate in PDZ-BDs. However, they do not have enhanced ER retention after distal PDZ-BD deletions, showing that SALM1 has a unique trafficking mechanism. We identified an acidic dileucine motif in the C-terminal domain of SALM1 that, when mutated, reversed the effect of removing the PDZ-BD by allowing surface expression of SALM1ΔPDZ. The dileucine motif functions as a partial ER retention signal in WT SALM1 and nearly completely retains SALM1ΔPDZ. In hippocampal neurons, deletion of the PDZ-BD reduces somatodendritic surface labeling. However, axonal surface expression is still present. Mutation of the dileucine motif in the absence of the PDZ-BD allows dendritic labeling. In addition, the same mutation in wild type SALM1 enhances dendritic targeting and the complexity of dendrites, indicating that the SALM1 PDZ-BD may be at least partially involved in dendritic targeting or surface retention.
Our results show that removing the PDZ-BD of the SALM1 C terminus reduces surface expression of SALM1 in heterologous cells and increases its retention in the ER. SALM1 to -3 bind PSD-95, PSD-93, SAP102, and SAP97; intact PDZ-BDs on these SALMs are necessary for synaptic localization (5, 6) and for enhancing neurite outgrowth (9). SAP97 and SAP102 are present in heterologous cells (29) and could interact with the expressed SALMs. Our results are consistent with the idea that binding of a PDZ protein is required for successful passage through the ER. A number of other studies suggest that PDZ proteins bind to the cytoplasmic domain of membrane proteins while they interact in the ER. The NR1 subunit of the NMDA receptor has eight different splice variants in neurons; these are differentially retained in the ER when expressed without the NR2 subunit. Using chimeras in which the C termini are attached to a single transmembrane protein (Tac or CD8), an RXR retention site was identified as responsible for ER retention of the major splice variant NR1-1 (30–32). The lack of ER retention of the splice variant NR1-3, which also contains the RXR motif, was shown to be due to the presence of a PDZ-BD that masked the RXR retention motif. It was later suggested that an ER export motif, rather than a PDZ-BD, was responsible for overcoming ER retention in the NR1-3 splice variant (33). Although these studies support an early interaction with PDZ proteins, none of them definitively rule out the possibility that binding of a PDZ protein is not the critical component for ER export but rather that a non-PDZ protein, with an entirely different function, interacts with the last four residues of SALM1 in the ER. Therefore, we cannot definitively distinguish between a PDZ-BD and an ER export motif, and we refer to the motif as the PDZ-BD/export motif.
Dileucine motifs are known to play important roles in trafficking of membrane proteins at the level of the trans-Golgi network, endosomes, lysosomes, and the plasma membrane (21). The C-terminal sequences of the SALMs are highly divergent; SALM2 and -3 do not have dileucine motifs that correspond to the acidic dileucine motif in SALM1, shown here to be involved in ER retention, but they do contain other dileucine motifs. Several examples have been demonstrated in which dileucine motifs play a role in ER trafficking, although they are less common. In these cases, the dileucine motifs usually function as export signals (22–26, 28). However, because mutations in the dileucine motif increase Endo H-insensitive WT SALM1 and SALM1ΔPDZ, the dileucine motif appears to be functioning as a retention signal. Similar to SALM1, a dileucine motif has been implicated in the ER retention of GABABR1 (34). GABABR1, which must assemble with GABABR2 to form functional receptors, is retained in the ER when expressed alone. Mutation of an RXR motif in its C terminus only partially eliminates ER retention, whereas mutation of the RXR motif along with a dileucine motif enhances surface expression. Because mutation of the RXR motif alone results in some labeling in the Golgi, the dileucine motif, in this case, may be involved in Golgi trafficking rather than ER trafficking. Because SALM1ΔPDZ was not localized to the Golgi, the dileucine motif of SALM1 does not appear to play a role in Golgi trafficking.
Various proteins depend on PDZ interactions or other intracellular motifs for proper trafficking to specific locations in neurons, such as the pre- or postsynaptic membrane (19). Most similar to our results on SALM1 is a recent study on the trafficking of neurexins (20), which are largely presynaptic and form a trans-synaptic adhesion complex with postsynaptic neuroligins. Although some neurexin is detected in dendrites, it is mainly expressed on the surface of axons (20, 35). Fairless et al. (20) found that mutation of the PDZ-BD on the C terminus of neurexins resulted in their ER retention. Furthermore, they report that an upstream but unidentified motif in the C terminus plays a role in the retention of neurexins lacking the PDZ-BD. Axonal targeting of neurexin requires a PDZ-BD and an upstream retention motif for passage through the ER (20). SALM1, on the other hand, is present in both axons and dendrites (5). However, SALM1ΔPDZ appears to be mainly localized to axons. Therefore, both the PDZ-BD and the dileucine motif of SALM1 may regulate trafficking to the dendrites.
Dendritic targeting in neurons often involves cytoplasmic signals that have dileucine or tyrosine residues (36). For example, Rivera et al. (37) showed that a dileucine motif was essential for the dendritic trafficking of Shal K+ channels in neurons and was sufficient to mediate the dendritic localization of Kv1.3 and Kv1.4 channels, which are normally axonal. The glycine transporter and 5-HT1AR serotonin receptors have dileucine motifs that mediate dendritic localization (26, 38), whereas α7-nicotinic acetylcholine receptors contain both tyrosine and dileucine motifs (39). Other motifs associated with dendritic targeting have been found in neuroligin 1, GluR1, mGluR1a, and mGluR2 (40–43). The LRRTMs (leucine-rich repeat transmembrane neuronal proteins) are another family of CAMs, besides neurexins, that have polarized trafficking in neurons. LRRTM2 localizes to dendrites; however, similar to SALM1, deletion of the C-terminal domain allows expression in both axons and dendrites (44).
Less is known about axonal targeting. However, the axonal localization of NgCAM/L1 is one example of targeting mediated by the extracellular domain of a CAM (45, 46). L1 is important in axon growth and synapse formation, and disruption of its axonal localization in humans causes L1 syndrome (47). Two missense mutants that occur in the extracellular domain of L1 were recently found to reduce L1 surface expression in heterologous cells due to partial ER retention (46). Similar to SALM1, this retention causes a change in the glycosylation pattern of L1. In neurons, these mutants reduce axonal surface localization or mistarget L1 to dendrites (46). Wisco et al. (48) have reported that NgCAM/LI can also be indirectly targeted to axons by transcytosis from dendrites mediated by an endocytosis motif (YRSL). Thus, multiple targeting motifs and trafficking pathways may regulate axonal versus dendritic localization.
Here, we report that the SALM1 L700A mutant enhances the surface expression of SALM1 in dendrites, causing an increase in spine density, branching complexity, and the production of filopodia-like spines. Secreted semaphorin 3F (Sema3F) and its receptor neuropilin-2 (Npn-2) are negative regulators of spine development. Loss of either one results in an increase in spine density and causes the formation of enlarged spine heads, with multiple PSDs (49). These spines make synaptic connections, as shown by EM and by an increase in miniature excitatory postsynaptic current frequency. Like SALM1, the targeting of neuropilin-2 to its appropriate dendritic location is dependent on a PDZ domain-binding motif (49). The overexpression of SALM1 L700A promotes the formation of elongated spines that do not colocalize with the presynaptic maker VGLUT1. EM also shows filopodia that do not meet up with presynaptic terminals. Therefore, the growth of the spines appears to occur very quickly and surpasses presynaptic contacts.
On the other hand, we find PSD-95 and actin in the filopodia. Actin is involved in the formation of spines and filopodial protrusions by generating a propulsive force on the cell membrane (50). As filopodia or protospines form, PSD-95 can move outward from the dendrite into the spine (51). Several actin binding/regulatory proteins are involved in regulation of spine elongation, including α-actinin (52), cortactin (53), actin capping protein (54), and kalirin-7 (55). SALM1 binds PSD-95 and is indirectly linked to the actin cytoskeleton via its interaction with guanylate kinase-associated protein (GKAP) because GKAP binds shank and shank binds cortactin, an F-actin-binding protein (56). Another possible connection to actin is kalirin-7, which also binds PSD-95 and is a GTP/GDP exchanger for Rac1, a Rho GTPase involved in actin polymerization, dendritic growth, and spine morphogenesis (57, 58). The Rho GTPases have been implicated in mental disabilities (59), and expression of activated Rac mutants in organotypic hippocampal slices produces long filopodial-like extensions (58). Loss of the αPIX/ARHGEF6 gene, the guanine nucleotide exchange factor for the Rho GTPases Rac1 and Cdc42, causes X-linked mental retardation in humans. In knock-out mice, there is a loss of functional synapses, identified by EM and a decrease in long-term potentiation (60). Therefore, one possible mechanism for the formation of filopodia by mutant SALM1 is that the increased surface expression of SALM1 L700A may transduce a signal to the actin cytoskeleton that promotes filopodia growth. Another cell adhesion molecule that can induce filopodia formation by a similar mechanism is syndecan-2. The actin-associated protein Ena/VASP is phosphorylated by PKA when syndecan-2 is overexpressed, resulting in actin polymerization (61). Interestingly, unlike SALM1 and neuropilin-2, the type II PDZ-binding motif of syndecan-2 is not required for synaptic targeting but is required for the maturation of spines at later stages of growth (62). Finally, the SALMs may be associated with the actin cytoskeleton via an as yet unknown mechanism through their C-terminal domains (63).
CAMs are important for normal brain development, and disruption of their expression can cause developmental impairment. Several CAMs, such as neurexin, neuroligin, and L1, have been implicated in the acquisition of developmental abnormalities related to autism spectrum disorders (64). Mutations that decrease surface expression of CAMs due to intracellular retention often contribute to more severe phenotypes (65–68). Similarly, our results show that SALM1ΔPDZ has limited dendritic targeting in hippocampal neurons and is retained in the ER of heterologous cells, suggesting that regulated surface expression of SALM1 may affect normal neurodevelopment. Supporting this idea, SALM1/Lrfn2 and SALM5/Lrfn5 have recently been associated with autism spectrum disorders (11–14). Furthermore, many forms of mental retardation involve a change in synaptic densities and spine shape, resulting in the presence of immature spines that might contribute to mental impairment (69, 70). For example, fragile X syndrome causes an increase in elongated spines (71, 72), and autism is associated with modifications of spine morphology and filopodia number (73). Here we find that changes in SALM1 surface expression alter neuronal morphology. Therefore, SALM1 could help regulate normal circuit plasticity during brain development.
In summary, our results show that SALM1, although highly similar to other members of the SALM family, has a unique mechanism of trafficking through the ER. This finding further supports the idea that the SALMs, although very similar in their extracellular N termini, have distinct functions, assembly properties, and distributions that may be regulated, in part, by their highly divergent C termini (5–7, 9, 10). Deletion of the PDZ-BD reduces somatodendritic surface labeling. However, axonal surface expression is still present. Expression of SALM1 dileucine mutants increases dendritic and cell body protrusions, which EM reveals to be irregular, enlarged spines and filopodia. These studies, which identify a novel mechanism involving the interplay between the SALM1 PDZ-BD and a putative ER retention motif, highlight the importance of understanding how trafficking of cell adhesion molecules impacts neuronal cell adhesion and synapse formation.
Supplementary Material
Acknowledgments
We thank Drs. Katherine Roche, Craig Blackstone, Elizabeth Webber, and Catherine Swanwick and members of the Laboratory of Neurochemistry for helpful comments on the manuscript. We also acknowledge the help of Mark Ryherd and Stacie Anderson from the Flow Cytometry Core (National Human Genome Research Institute, National Institutes of Health).
This work was supported, in whole or in part, by the National Institutes of Health, NIDCD, Intramural Research Program.

This article contains supplemental Figs. 1–3.
- CAM
- cell adhesion molecule
- SALM
- synaptic adhesion-like molecule
- PDZ-BD
- PDZ-binding motif
- Endo H
- endoglycosidase H
- DIV
- days in vitro
- ANOVA
- analysis of variance
- ER
- endoplasmic reticulum
- GKAP
- guanylate kinase-associated protein.
REFERENCES
- 1. Yamagata M., Sanes J. R., Weiner J. A. (2003) Synaptic adhesion molecules. Curr. Opin. Cell Biol. 15, 621–632 [DOI] [PubMed] [Google Scholar]
- 2. Scheiffele P. (2003) Cell-cell signaling during synapse formation in the CNS. Annu. Rev. Neurosci. 26, 485–508 [DOI] [PubMed] [Google Scholar]
- 3. Dalva M. B., McClelland A. C., Kayser M. S. (2007) Cell adhesion molecules. Signaling functions at the synapse. Nat. Rev. Neurosci. 8, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson D. L., Huntley G. W. (2010) Building and remodeling synapses. Hippocampus, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang C. Y., Chang K., Petralia R. S., Wang Y. X., Seabold G. K., Wenthold R. J. (2006) A novel family of adhesion-like molecules that interacts with the NMDA receptor. J. Neurosci. 26, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ko J., Kim S., Chung H. S., Kim K., Han K., Kim H., Jun H., Kaang B. K., Kim E. (2006) SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron 50, 233–245 [DOI] [PubMed] [Google Scholar]
- 7. Morimura N., Inoue T., Katayama K., Aruga J. (2006) Comparative analysis of structure, expression, and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene 380, 72–83 [DOI] [PubMed] [Google Scholar]
- 8. Homma S., Shimada T., Hikake T., Yaginuma H. (2009) Expression pattern of LRR and Ig domain-containing protein (LRRIG protein) in the early mouse embryo. Gene Expr. Patterns 9, 1–26 [DOI] [PubMed] [Google Scholar]
- 9. Wang P. Y., Seabold G. K., Wenthold R. J. (2008) Synaptic adhesion-like molecules (SALMs) promote neurite outgrowth. Mol. Cell Neurosci. 39, 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mah W., Ko J., Nam J., Han K., Chung W. S., Kim E. (2010) Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J. Neurosci. 30, 5559–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang K., Zhang H., Ma D., Bucan M., Glessner J. T., Abrahams B. S., Salyakina D., Imielinski M., Bradfield J. P., Sleiman P. M., Kim C. E., Hou C., Frackelton E., Chiavacci R., Takahashi N., Sakurai T., Rappaport E., Lajonchere C. M., Munson J., Estes A., Korvatska O., Piven J., Sonnenblick L. I., Alvarez Retuerto A. I., Herman E. I., Dong H., Hutman T., Sigman M., Ozonoff S., Klin A., Owley T., Sweeney J. A., Brune C. W., Cantor R. M., Bernier R., Gilbert J. R., Cuccaro M. L., McMahon W. M., Miller J., State M. W., Wassink T. H., Coon H., Levy S. E., Schultz R. T., Nurnberger J. I., Haines J. L., Sutcliffe J. S., Cook E. H., Minshew N. J., Buxbaum J. D., Dawson G., Grant S. F., Geschwind D. H., Pericak-Vance M. A., Schellenberg G. D., Hakonarson H. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459, 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Bruijn D. R., van Dijk A. H., Pfundt R., Hoischen A., Merkx G. F., Gradek G. A., Lybæk H., Stray-Pedersen A., Brunner H. G., Houge G. (2010) Severe progressive autism associated with two de novo changes. A 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1;p11.2) translocation with long-range epigenetic silencing of LRFN5 expression. Mol. Syndromol. 1, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voineagu I., Wang X., Johnston P., Lowe J. K., Tian Y., Horvath S., Mill J., Cantor R. M., Blencowe B. J., Geschwind D. H. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mikhail F. M., Lose E. J., Robin N. H., Descartes M. D., Rutledge K. D., Rutledge S. L., Korf B. R., Carroll A. J. (2011) Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am. J. Med. Genet. A 155A, 2386–2396 [DOI] [PubMed] [Google Scholar]
- 15. Seabold G. K., Wang P. Y., Chang K., Wang C. Y., Wang Y. X., Petralia R. S., Wenthold R. J. (2008) The SALM family of adhesion-like molecules forms heteromeric and homomeric complexes. J. Biol. Chem. 283, 8395–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sans N., Wang P. Y., Du Q., Petralia R. S., Wang Y. X., Nakka S., Blumer J. B., Macara I. G., Wenthold R. J. (2005) mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat. Cell Biol. 7, 1179–1190 [DOI] [PubMed] [Google Scholar]
- 17. Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., Agard D. A., Sedat J. W. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petralia R. S., Wang Y. X., Hua F., Yi Z., Zhou A., Ge L., Stephenson F. A., Wenthold R. J. (2010) Organization of NMDA receptors at extrasynaptic locations. Neuroscience 167, 68–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wenthold R. J., Prybylowski K., Standley S., Sans N., Petralia R. S. (2003) Trafficking of NMDA receptors. Annu. Rev. Pharmacol. Toxicol. 43, 335–358 [DOI] [PubMed] [Google Scholar]
- 20. Fairless R., Masius H., Rohlmann A., Heupel K., Ahmad M., Reissner C., Dresbach T., Missler M. (2008) Polarized targeting of neurexins to synapses is regulated by their C-terminal sequences. J. Neurosci. 28, 12969–12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 22. Schülein R., Hermosilla R., Oksche A., Dehe M., Wiesner B., Krause G., Rosenthal W. (1998) A dileucine sequence and an upstream glutamate residue in the intracellular carboxyl terminus of the vasopressin V2 receptor are essential for cell surface transport in COS.M6 cells. Mol. Pharmacol. 54, 525–535 [DOI] [PubMed] [Google Scholar]
- 23. Duvernay M. T., Zhou F., Wu G. (2004) A conserved motif for the transport of G protein-coupled receptors from the endoplasmic reticulum to the cell surface. J. Biol. Chem. 279, 30741–30750 [DOI] [PubMed] [Google Scholar]
- 24. Loo T. W., Bartlett M. C., Clarke D. M. (2005) The dileucine motif at the COOH terminus of human multidrug resistance P-glycoprotein is important for folding but not activity. J. Biol. Chem. 280, 2522–2528 [DOI] [PubMed] [Google Scholar]
- 25. Robert J., Clauser E., Petit P. X., Ventura M. A. (2005) A novel C-terminal motif is necessary for the export of the vasopressin V1b/V3 receptor to the plasma membrane. J. Biol. Chem. 280, 2300–2308 [DOI] [PubMed] [Google Scholar]
- 26. Carrel D., Hamon M., Darmon M. (2006) Role of the C-terminal di-leucine motif of 5-HT1A and 5-HT1B serotonin receptors in plasma membrane targeting. J. Cell Sci. 119, 4276–4284 [DOI] [PubMed] [Google Scholar]
- 27. Dong C., Filipeanu C. M., Duvernay M. T., Wu G. (2007) Regulation of G protein-coupled receptor export trafficking. Biochim. Biophys. Acta 1768, 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ludwig T., Theissen S. M., Morton M. J., Caplan M. J. (2008) The cytoplasmic tail dileucine motif LL572 determines the glycosylation pattern of membrane-type 1 matrix metalloproteinase. J. Biol. Chem. 283, 35410–35418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sans N., Prybylowski K., Petralia R. S., Chang K., Wang Y. X., Racca C., Vicini S., Wenthold R. J. (2003) NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat. Cell Biol. 5, 520–530 [DOI] [PubMed] [Google Scholar]
- 30. Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21, 3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Standley S., Roche K. W., McCallum J., Sans N., Wenthold R. J. (2000) PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28, 887–898 [DOI] [PubMed] [Google Scholar]
- 32. Xia H., Hornby Z. D., Malenka R. C. (2001) An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology 41, 714–723 [DOI] [PubMed] [Google Scholar]
- 33. Mu Y., Otsuka T., Horton A. C., Scott D. B., Ehlers M. D. (2003) Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron 40, 581–594 [DOI] [PubMed] [Google Scholar]
- 34. Restituito S., Couve A., Bawagan H., Jourdain S., Pangalos M. N., Calver A. R., Freeman K. B., Moss S. J. (2005) Multiple motifs regulate the trafficking of GABA(B) receptors at distinct checkpoints within the secretory pathway. Mol. Cell Neurosci. 28, 747–756 [DOI] [PubMed] [Google Scholar]
- 35. Taniguchi H., Gollan L., Scholl F. G., Mahadomrongkul V., Dobler E., Limthong N., Peck M., Aoki C., Scheiffele P. (2007) Silencing of neuroligin function by postsynaptic neurexins. J. Neurosci. 27, 2815–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lasiecka Z. M., Yap C. C., Vakulenko M., Winckler B. (2009) Compartmentalizing the neuronal plasma membrane from axon initial segments to synapses. Int. Rev. Cell Mol. Biol. 272, 303–389 [DOI] [PubMed] [Google Scholar]
- 37. Rivera J. F., Ahmad S., Quick M. W., Liman E. R., Arnold D. B. (2003) An evolutionarily conserved dileucine motif in Shal K+ channels mediates dendritic targeting. Nat. Neurosci. 6, 243–250 [DOI] [PubMed] [Google Scholar]
- 38. Poyatos I., Ruberti F., Martínez-Maza R., Giménez C., Dotti C. G., Zafra F. (2000) Polarized distribution of glycine transporter isoforms in epithelial and neuronal cells. Mol. Cell Neurosci. 15, 99–111 [DOI] [PubMed] [Google Scholar]
- 39. Xu J., Zhu Y., Heinemann S. F. (2006) Identification of sequence motifs that target neuronal nicotinic receptors to dendrites and axons. J. Neurosci. 26, 9780–9793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stowell J. N., Craig A. M. (1999) Axon/dendrite targeting of metabotropic glutamate receptors by their cytoplasmic carboxyl-terminal domains. Neuron 22, 525–536 [DOI] [PubMed] [Google Scholar]
- 41. Ruberti F., Dotti C. G. (2000) Involvement of the proximal C terminus of the AMPA receptor subunit GluR1 in dendritic sorting. J. Neurosci. 20, RC78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Francesconi A., Duvoisin R. M. (2002) Alternative splicing unmasks dendritic and axonal targeting signals in metabotropic glutamate receptor 1. J. Neurosci. 22, 2196–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosales C. R., Osborne K. D., Zuccarino G. V., Scheiffele P., Silverman M. A. (2005) A cytoplasmic motif targets neuroligin-1 exclusively to dendrites of cultured hippocampal neurons. Eur. J. Neurosci. 22, 2381–2386 [DOI] [PubMed] [Google Scholar]
- 44. Linhoff M. W., Laurén J., Cassidy R. M., Dobie F. A., Takahashi H., Nygaard H. B., Airaksinen M. S., Strittmatter S. M., Craig A. M. (2009) An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron 61, 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sampo B., Kaech S., Kunz S., Banker G. (2003) Two distinct mechanisms target membrane proteins to the axonal surface. Neuron 37, 611–624 [DOI] [PubMed] [Google Scholar]
- 46. Schäfer M. K., Nam Y. C., Moumen A., Keglowich L., Bouché E., Küffner M., Bock H. H., Rathjen F. G., Raoul C., Frotscher M. (2010) L1 syndrome mutations impair neuronal L1 function at different levels by divergent mechanisms. Neurobiol. Dis. 40, 222–237 [DOI] [PubMed] [Google Scholar]
- 47. Maness P. F., Schachner M. (2007) Neural recognition molecules of the immunoglobulin superfamily. Signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 10, 19–26 [DOI] [PubMed] [Google Scholar]
- 48. Wisco D., Anderson E. D., Chang M. C., Norden C., Boiko T., Fölsch H., Winckler B. (2003) Uncovering multiple axonal targeting pathways in hippocampal neurons. J. Cell Biol. 162, 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tran T. S., Rubio M. E., Clem R. L., Johnson D., Case L., Tessier-Lavigne M., Huganir R. L., Ginty D. D., Kolodkin A. L. (2009) Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature 462, 1065–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mogilner A., Oster G. (1996) Cell motility driven by actin polymerization. Biophys. J. 71, 3030–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marrs G. S., Green S. H., Dailey M. E. (2001) Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat. Neurosci. 4, 1006–1013 [DOI] [PubMed] [Google Scholar]
- 52. Nakagawa T., Engler J. A., Sheng M. (2004) The dynamic turnover and functional roles of α-actinin in dendritic spines. Neuropharmacology 47, 734–745 [DOI] [PubMed] [Google Scholar]
- 53. Hering H., Sheng M. (2003) Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J. Neurosci. 23, 11759–11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fan Y., Tang X., Vitriol E., Chen G., Zheng J. Q. (2011) Actin capping protein is required for dendritic spine development and synapse formation. J. Neurosci. 31, 10228–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Penzes P., Johnson R. C., Sattler R., Zhang X., Huganir R. L., Kambampati V., Mains R. E., Eipper B. A. (2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229–242 [DOI] [PubMed] [Google Scholar]
- 56. Naisbitt S., Kim E., Tu J. C., Xiao B., Sala C., Valtschanoff J., Weinberg R. J., Worley P. F., Sheng M. (1999) Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23, 569–582 [DOI] [PubMed] [Google Scholar]
- 57. Threadgill R., Bobb K., Ghosh A. (1997) Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron 19, 625–634 [DOI] [PubMed] [Google Scholar]
- 58. Nakayama A. Y., Harms M. B., Luo L. (2000) Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 20, 5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Newey S. E., Velamoor V., Govek E. E., Van Aelst L. (2005) Rho GTPases, dendritic structure, and mental retardation. J. Neurobiol. 64, 58–74 [DOI] [PubMed] [Google Scholar]
- 60. Ramakers G. J., Wolfer D., Rosenberger G., Kuchenbecker K., Kreienkamp H. J., Prange-Kiel J., Rune G., Richter K., Langnaese K., Masneuf S., Bosl M. R., Fischer K. D., Krugers H. J., Lipp H. P., van Galen E., Kutsche K. (2011) Dysregulation of Rho GTPases in the αPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Hum. Mol. Genet. 21, 268–286 [DOI] [PubMed] [Google Scholar]
- 61. Lin Y. L., Lei Y. T., Hong C. J., Hsueh Y. P. (2007) Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-Ena/VASP pathway. J. Cell Biol. 177, 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ethell I. M., Yamaguchi Y. (1999) Cell surface heparan sulfate proteoglycan syndecan-2 induces the maturation of dendritic spines in rat hippocampal neurons. J. Cell Biol. 144, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Konakahara S., Saitou M., Hori S., Nakane T., Murai K., Itoh R., Shinsaka A., Kohroki J., Kawakami T., Kajikawa M., Masuho Y. (2011) A neuronal transmembrane protein LRFN4 induces monocyte/macrophage migration via actin cytoskeleton reorganization. FEBS Lett. 585, 2377–2384 [DOI] [PubMed] [Google Scholar]
- 64. Betancur C., Sakurai T., Buxbaum J. D. (2009) The emerging role of synaptic cell adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 32, 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. De Angelis E., Watkins A., Schäfer M., Brümmendorf T., Kenwrick S. (2002) Disease-associated mutations in L1 CAM interfere with ligand interactions and cell surface expression. Hum. Mol. Genet. 11, 1–12 [DOI] [PubMed] [Google Scholar]
- 66. Comoletti D., De Jaco A., Jennings L. L., Flynn R. E., Gaietta G., Tsigelny I., Ellisman M. H., Taylor P. (2004) The Arg451Cys-neuroligin-3 mutation associated with autism reveals a defect in protein processing. J. Neurosci. 24, 4889–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chih B., Afridi S. K., Clark L., Scheiffele P. (2004) Disorder-associated mutations lead to functional inactivation of neuroligins. Hum. Mol. Genet. 13, 1471–1477 [DOI] [PubMed] [Google Scholar]
- 68. Gauthier J., Siddiqui T. J., Huashan P., Yokomaku D., Hamdan F. F., Champagne N., Lapointe M., Spiegelman D., Noreau A., Lafreniere R. G., Fathalli F., Joober R., Krebs M. O., DeLisi L. E., Mottron L., Fombonne E., Michaud J. L., Drapeau P., Carbonetto S., Craig A. M., Rouleau G. A. (2011) Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 130, 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Purpura D. P. (1974) Dendritic spine “dysgenesis” and mental retardation. Science 186, 1126–1128 [DOI] [PubMed] [Google Scholar]
- 70. Dierssen M., Ramakers G. J. (2006) Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 5 Suppl 2, 48–60 [DOI] [PubMed] [Google Scholar]
- 71. Hinton V. J., Brown W. T., Wisniewski K., Rudelli R. D. (1991) Analysis of neocortex in three males with the fragile X syndrome. Am. J. Med. Genet. 41, 289–294 [DOI] [PubMed] [Google Scholar]
- 72. Portera-Cailliau C. (2011) Which comes first in fragile X syndrome, dendritic spine dysgenesis or defects in circuit plasticity? Neuroscientist, in press [DOI] [PubMed] [Google Scholar]
- 73. Durand C. M., Perroy J., Loll F., Perrais D., Fagni L., Bourgeron T., Montcouquiol M., Sans N. (2012) SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol. Psychiatry 17, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.