Background: IcmF is a fusion between a coenzyme B12-dependent isobutyryl-CoA/n-butyryl-CoA isomerase and a G-protein chaperone.
Results: IcmF also isomerizes isovaleryl-CoA to pivalyl-CoA and is partially protected from inactivation in the presence of GTP.
Conclusion: The isovaleryl-CoA mutase activity of IcmF might be important in leucine catabolism where isovaleric acid is an intermediate.
Significance: IcmF might be critical for microbial bioremediation of the anthropogenic compound pivalic acid.
Keywords: Adenosylcobalamin, Bacterial Metabolism, Enzyme Inactivation, Fusion Protein, GTPase
Abstract
5′-Deoxyadenosylcobalamin (AdoCbl)-dependent isomerases catalyze carbon skeleton rearrangements using radical chemistry. We have recently characterized a fusion protein that comprises the two subunits of the AdoCbl-dependent isobutyryl-CoA mutase flanking a G-protein chaperone and named it isobutyryl-CoA mutase fused (IcmF). IcmF catalyzes the interconversion of isobutyryl-CoA and n-butyryl-CoA, whereas GTPase activity is associated with its G-protein domain. In this study, we report a novel activity associated with IcmF, i.e. the interconversion of isovaleryl-CoA and pivalyl-CoA. Kinetic characterization of IcmF yielded the following values: a Km for isovaleryl-CoA of 62 ± 8 μm and Vmax of 0.021 ± 0.004 μmol min−1 mg−1 at 37 °C. Biochemical experiments show that an IcmF in which the base specificity loop motif NKXD is modified to NKXE catalyzes the hydrolysis of both GTP and ATP. IcmF is susceptible to rapid inactivation during turnover, and GTP conferred modest protection during utilization of isovaleryl-CoA as substrate. Interestingly, there was no protection from inactivation when either isobutyryl-CoA or n-butyryl-CoA was used as substrate. Detailed kinetic analysis indicated that inactivation is associated with loss of the 5′-deoxyadenosine moiety from the active site, precluding reformation of AdoCbl at the end of the turnover cycle. Under aerobic conditions, oxidation of the cob(II)alamin radical in the inactive enzyme results in accumulation of aquacobalamin. Because pivalic acid found in sludge can be used as a carbon source by some bacteria and isovaleryl-CoA is an intermediate in leucine catabolism, our discovery of a new isomerase activity associated with IcmF expands its metabolic potential.
Introduction
Methylmalonyl-CoA mutase (MCM)2 and isobutyryl-CoA mutase (ICM) are two closely related 5′-deoxyadenosylcobalamin (AdoCbl)-dependent isomerases that catalyze 1,2 rearrangements of methylmalonyl-CoA to succinyl-CoA and isobutyryl-CoA to n-butyryl-CoA, respectively (Fig. 1) (1, 2). MCMs are widely distributed in nature, ranging from bacteria and Archaea to animals, including humans. ICMs were initially believed to be restricted to the genus Streptomyces, which belongs to the Actinobacteria phylum (1, 2). With our discovery of IcmF, the fusion protein between ICM and its G-protein chaperone, the known distribution of ICM has expanded to include four more phyla: Proteobacteria, Bacteroidetes, Firmicutes, and Spirochaetes (3). The only known function of ICM in the genus Streptomyces is its participation in polyketide biosynthesis (4). In contrast, the relatively wide distribution of IcmF in diverse organisms points to a broader range of roles in bacterial metabolism that remain to be elucidated.
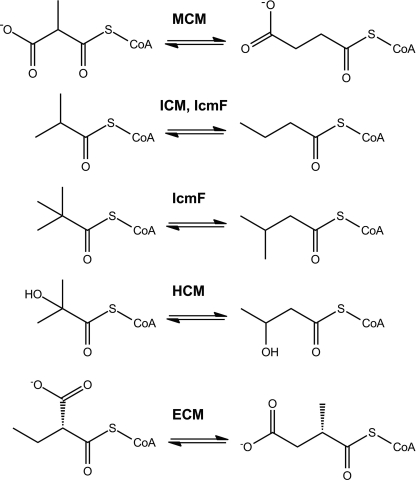
FIGURE 1.
Reactions catalyzed by MCM, ICM, IcmF, HCM, and ECM.
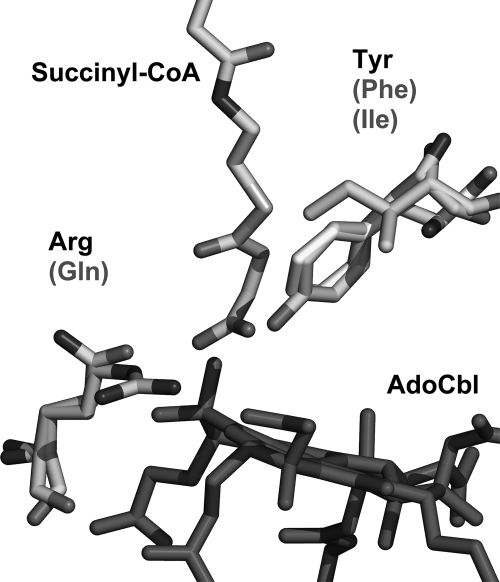
A family of enzymes that are similar in their primary sequence to MCM catalyzes AdoCbl-dependent carbon skeleton rearrangements and include, in addition to ICM, 2-hydroxyisobutyryl-CoA mutase (HCM) (5) and ethylmalonyl-CoA mutase (ECM) (6) (Fig. 1). The three-dimensional structure of these proteins is expected to resemble the overall structure of MCM (7, 8). The B12-binding domain of MCM exhibits a typical Rossman-like fold, and the AdoCbl cofactor is bound in a “base-off/His-on” conformation (9). The substrate-binding domain is a triose-phosphate isomerase barrel comprising a core of eight (αβ)-repeats (8). A limited number of key amino acid substitutions in the active site distinguish these closely related enzymes and accommodate chemical differences in the substrate (6, 10). Previous studies from our laboratory had pointed out that the two key substitutions that accommodate the switch from a carboxylate to a methyl group in the substrates of MCM versus ICM are Tyr → Phe and Arg → Gln, respectively (Figs. 2 and 3) (3). The Phe and Arg substitutions are conserved in all ICM/IcmFs (Figs. 2 and 3). In contrast, in HCM, the corresponding amino acids that interact with substrate are Ile and Gln, respectively. Thus, although the Gln residue is conserved in both ICM/IcmF and HCM, the Phe is substituted by Ile to accommodate the bulkier 2-hydroxyisobutyryl-CoA substrate (Figs. 2 and 3).
FIGURE 2.
Comparison of active site residues in related AdoCbl-dependent mutases. The MCM structure from Propionibacterium shermanii (Protein Data Bank code 4REQ) was used as a template to show that Tyr and Arg in MCM/ECM correspond to Phe and Gln in ICM/IcmF. In HCM, this pair of residues is Ile and Gln.
FIGURE 3.
Multiple sequence alignment of substrate-binding domains of different AdoCbl-dependent mutases. IcmF from G. kaustophilus (YP_149244), ICM from S. cinnamonensis (AAC08713), MCM from Methylobacterium extorquens (YP_001642233), MeaA from M. extorquens (YP_002961419), ECM from Rhodobacter sphaeroides (YP_354045), and HCM from M. petroleiphilum (YP_001023546) are shown. Four residues recognized to be important for substrate binding are highlighted in gray and indicated with asterisks. Two residues, Phe and Gln, found in ICM and IcmF are substituted by Tyr and Arg in MCM and ECM or Ile and Gln in HCM. ECM is different from other acyl-CoA mutases by the substitution of His and Asp to Gly and Pro, respectively. ECM was previously known as MeaA in M. extorquens. All accession numbers are from NCBI Proteins database.
In contrast to ICM, ECM, which catalyzes the interconversion of ethylmalonyl-CoA and methylsuccinyl-CoA, is very similar to MCM (6). It has been proposed that two key substitutions in the active site of ECM dictate specificity for the bulkier ethylmalonyl-CoA substrate: a His and an Asn residue in MCM are replaced by Gly and Pro, respectively (6) (Fig. 3). Notably, the Tyr and Arg residues in the active site of MCM that interact with the carboxylate moiety of the substrate are also conserved in ECM (Fig. 3).
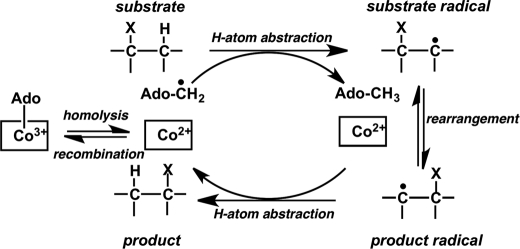
In B12-dependent isomerases, AdoCbl serves as a radical reservoir, and a common first step that initiates the isomerization reactions is homolytic cleavage of the cobalt-carbon bond (Fig. 4), leading to formation of the 5′-deoxyadenosyl radical and a paramagnetic cob(II)alamin species (1, 11). Inadvertent side reactions of the reactive radical intermediates render AdoCbl-dependent enzymes susceptible to inactivation (12). Alternatively, inactivation can result from escape of the 5′-deoxyadenosine intermediate during the catalytic cycle (13). In both cases, inactivation results from the inability to reform AdoCbl from the 5′-deoxyadenosyl and cob(II)alamin radicals at the end of the turnover cycle. MeaB, the G-protein chaperone of MCM, protects against inactivation in the presence of GTP (13, 14). A similar role for the homologous G-protein chaperone of ICM, MeaI, has not been studied. In IcmF, the MeaI domain is sandwiched between the AdoCbl- and substrate-binding domains (3). In this study, we report a novel AdoCbl-dependent 1,2 rearrangement reaction catalyzed by IcmF and demonstrate that in the presence of GTP the isovaleryl-CoA mutase activity of IcmF is partially protected from the inactivation.
FIGURE 4.
Generalized scheme for 1,2 rearrangement reaction catalyzed by AdoCbl-dependent mutases. The binding of substrate to the enzyme induces homolytic cleavage of the cobalt-carbon bond of AdoCbl, resulting in formation of a pair of radicals, the 5′-deoxyadenosyl radical and cob(II)alamin. The 5′-deoxyadenosyl radical abstracts a hydrogen atom from the substrate to form 5′-deoxyadenosine and a substrate radical, which subsequently undergoes isomerization to the product radical. The latter reabstracts a hydrogen atom from the methyl group of 5′-deoxyadenosine to form the product and regenerates the 5′-deoxyadenosyl radical. Finally, cob(II)alamin and the 5′-deoxyadenosyl radicals recombine to give AdoCbl.
EXPERIMENTAL PROCEDURES
Materials
AdoCbl, ADP, AMPPNP, ATP, GDP, GTP, isobutyryl-CoA, n-butyryl-CoA, isovaleryl-CoA, dl-3-hydroxybutyryl-CoA, and pivalic acid were purchased from Sigma. Tris(2-carboxyethyl)phosphine hydrochloride was from GoldBio (St. Louis, MO). Valeryl-CoA was purchased from Crystal Chem (Downers Grove, IL). A pET28a vector expressing IcmF from Cupriavidus metallidurans was a generous gift from the laboratory of Dr. Catherine Drennan (Massachusetts Institute of Technology).
DNA Manipulations
Cloning of IcmF
In a previous study, we used IcmF from Geobacillus kaustophilus cloned into pET30 Ek/LIC expression vector (3). The S-tag, which is located just downstream of the N-terminal His tag in pET30 Ek/LIC, was removed by subcloning the full-length IcmF DNA into the ligation-independent cloning vector pMCSG7 (15). The insert was amplified with the following primers: forward, 5′-TACTTCCAATCCAATGCCATGGCGCACATTTACCGTCC-3′; and reverse, 5′-TTATCCACTTCCAATGCTATTACATATTCGCCGGTATTGTCC-3′. In pMCSG7, which is a derivative of the pET21 vector, the N-terminal His tag can be cleaved using tobacco etch virus protease. The His tag was not removed in this study because its presence does not affect IcmF activity.
Construction of K213A Mutant of IcmF from G. kaustophilus (Gk)
The K213A mutation was created using the QuikChange XL site-directed mutagenesis kit (Agilent) and the following sense primer: 5′-CCGGCACAGGCGGAGCTGGGGCAAGCTCGCTCACCGATG-3′. The sequence of the reverse primer was complementary to the sequence of the forward primer. The Gk IcmF cloned in pMCSG7 was used as a template. All constructs were confirmed by nucleotide sequence determination at The University of Michigan DNA Sequencing Core.
Protein Expression and Purification
Purification of Gk Wild Type and K213A IcmF
Escherichia coli BL21(DE3) cells containing plasmids expressing the IcmF constructs were grown at 37 °C in Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin to an absorbance at 600 nm of 0.5–0.6. Cells were grown for 12–14 h at 15 °C after induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Proteins were purified as described previously for wild-type IcmF (3) with the following difference. Gel filtration chromatography was performed with 50 mm HEPES, pH 7.5 containing 100 mm NaCl (Buffer A).
Purification of C. metallidurans (Cm) IcmF
E. coli BL21(DE3) cells containing plasmid expressing IcmF were grown in LB medium supplemented with 50 μg/ml kanamycin and induced at 15 °C with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested 12–14 h after induction. Cm IcmF was purified as described previously for Gk IcmF (3). Gel filtration chromatography was also performed on this enzyme using Buffer A.
ATPase/GTPase Assays
NTPase activity of IcmF was measured by HPLC as described previously (3) or by a modification of the malachite green assay involving inclusion of citrate in the reaction mixture (16). Briefly, the color reagent was prepared by mixing 4.2% ammonium molybdate in 4 n HCl with 0.045% malachite green hydrochloride (1:3, v/v). The resulting solution was shaken for 30 min at 250 rpm in a Falcon tube and filtered through a 0.2-μm Millipore filter. After that, 10% Tween 20 solution (v/v) was added (200 μl for every 100 ml of color reagent). This solution was stable for a week at 4 °C with only a minor increase in the background absorbance at 650 nm. The reaction was performed in Buffer A in a total volume of 0.6–1 ml containing 0.5–2.5 μm IcmF or 10–20 μm K213A IcmF, 10 mm MgCl2, and nucleotides (0.02–1.2 mm GTP or 0.02–6 mm ATP). At the desired time points, 200-μl aliquots were removed, and the reaction was quenched with 20 μl of 2 m trichloroacetic acid (TCA). After centrifugation, 150 μl of supernatant was added to 750 μl of color reagent. After 3 min, 100 μl of 34% sodium citrate was added to the sample. The color was allowed to develop for 30 min at room temperature, and the absorbance was measured at 650 nm. The calibration curve for phosphate was prepared using a 5–200 μm solution of monobasic potassium phosphate, which was dried in an oven (to remove traces of moisture) for 4 h at 120 °C.
IcmF Assay
A GC-based assay was used to measure both ICM activity and the new, isovaleryl-CoA/pivalyl-CoA mutase activity (3). In all assays, apoenzyme was preincubated with AdoCbl ± nucleotides, and the reaction was started by addition of substrate.
Pivalyl-CoA Mutase Activity
The reaction was performed in Buffer A in a total volume of 0.8–1.4 ml containing 600–2500 μg of Gk IcmF or Cm IcmF, 100 μm AdoCbl, 20–2000 μm isovaleryl-CoA, and 15 mm MgCl2 ± 5 mm GTP. For every concentration of substrate, two to six aliquots (200 μl each) were removed and treated as described below.
Isobutyryl-CoA Mutase Activity
The reaction was performed in Buffer A in a total volume of 0.8–1.4 ml containing 10–60 μg of Gk IcmF or Cm IcmF, 100 μm AdoCbl, a saturating concentration of substrates (600–2000 μm isobutyryl-CoA or n-butyryl-CoA), and 10 mm MgCl2 ± 3–6 mm GTP or ATP. At various time points (0.5–30 min), 200-μl aliquots were removed and quenched with 100 μl of 2 n KOH containing 0.18 mm valeric acid used as an internal standard. Following addition of 100 μl of H2SO4 (15%, v/v), the reaction mixture was saturated with NaCl and extracted with ethyl acetate (250 μl). The extract was analyzed directly by GC using a DB-FFAP (30-m × 0.25-mm-inner diameter, 0.25-μm) capillary column (Agilent). A 5-μl sample was injected in the pulsed splitless mode. The oven temperature was initially at 80 °C. Following sample injection, the temperature was raised to 150 °C at a rate of 10 °C/min and maintained at 150 °C for 2 min. Retention times for the compounds of interest was as follows: isobutyric acid, 5.85 min; pivalic acid, 5.99 min; n-butyric acid, 6.5 min; isovaleric acid, 6.96 min; and valeric acid, 7.78 min.
IcmF Assays with Alternative Substrates
We used an HPLC-based assay to evaluate whether 3-hydroxybutyryl-CoA can be converted by IcmF to 2-hydroxyisobutyryl-CoA. The assay mixture contained in a total volume of 0.5 ml Buffer A, 10 mm MgCl2, 100 μm AdoCbl, 2 mm GTP, 0.4–1 mm dl-β-hydroxybutyryl-CoA, and 0.2–0.4 mg Gk IcmF. The reaction was initiated by addition of enzyme, and the reaction mixture was incubated at 37 °C. At different time points (0.5–10 min), 60-μl aliquots were removed, quenched with 2 m TCA (10%, v/v), centrifuged, and subjected to HPLC analysis.
The acyl-CoA esters were separated using an HPLC system equipped with an Alltima HP 5-μm C18 (250 × 4.6-mm) column (Grace). The detector was set at 254 nm. Solvent A was 50 mm monobasic potassium phosphate, pH 5.4. Solvent B was prepared as follows. 500 ml of MeOH was added to 500 ml of 100 mm monobasic potassium phosphate, pH 5.4 (to give a final concentration of 50 mm potassium phosphate and 50% MeOH). Initial conditions used for separation were 10% solvent B and a flow rate of 1.0 ml/min. Between 5 and 30 min, solvent B was increased to 100% and then held at 100% solvent B for 5 min. At 36 min, solvent B was decreased to 10% and held for 10 min at that composition to equilibrate the column between injections. Under these conditions, the retention time for 3-hydroxybutyryl-CoA was 20.1 min. An authentic standard of 2-hydroxyisobutyryl-CoA was not available, but based on the known performance of the C18 column, branched acyl-CoAs elute earlier than linear isomers.
Enzyme-monitored Turnover of IcmF
Changes in the spectra of Gk IcmF-bound AdoCbl were monitored by UV-visible spectroscopy at 24 °C in Buffer A containing 5–15 mm MgCl2. Substrates (final concentration of 0.5–4 mm) were added to 20–65 μm apoIcmF loaded with 1 or 2 eq of AdoCbl. To check the influence of the MeaI domain on catalytic turnover, the reaction was also supplemented with 1–5 mm GTP or ATP. The amount of cob(II)alamin formed under steady-state turnover conditions was calculated from the decrease in absorbance at 525 nm upon substrate addition using a value of Δϵ525 nm of −4.8 mm−1 cm−1 (14). During the course of the reaction, AdoCbl was gradually converted to enzyme-bound aquacobalamin (OH2Cbl) as indicated by the appearance of a 350 nm absorption peak. The increase at 350 nm was fitted to a single exponential function, A = A0 + A1(1 − e−bt) where A is the absorbance at 350 nm, b is the observed rate constant for inactivation, A0 is the initial absorbance at 350 nm, t is time in minutes, and A1 is the amplitude.
Enzyme-monitored Turnover of Gk IcmF under Anaerobic Conditions
To assess the effect of oxygen on enzyme inactivation, enzyme-monitored turnover experiments were performed under anaerobic conditions. For this, Buffer A containing 10 mm MgCl2 was bubbled with N2 for 3 h before use and introduced into an anaerobic chamber (containing <0.3 ppm O2). Stock solutions of AdoCbl, n-butyryl-CoA, isobutyryl-CoA, and isovaleryl-CoA were prepared in the chamber using anaerobic buffer. The enzyme solution was deoxygenated by blowing a stream of N2 over its surface at 4 °C for 40 min. UV-visible spectra were collected using a Mikropack DH-2000 UV-visible light source connected with fiber optics to a cuvette holder inside the glove box.
HPLC Characterization of Inactivation Products
A solution containing 64 μm Gk holo-IcmF (containing 2 eq of AdoCbl) in Buffer A and 10 mm MgCl2 ± 5 mm GTP was incubated with 1 mm isobutyryl-CoA at 37 °C in the dark. At various times (0–60 min), 15-μl aliquots were removed and immediately quenched with 60 μl of 0.5% trifluoroacetic acid (TFA). The aerobic inactivation products of AdoCbl, 5′-deoxyadenosine and OH2Cbl, were monitored by HPLC using an Alltima HP 5-μm C18 (250 × 4.6-mm) column (Grace). All steps of sample preparation and HPLC analysis were performed in the dark. Initial buffer conditions were 92% Solvent C (0.1% TFA in water) and 8% Solvent D (0.1% TFA in acetonitrile) at a flow rate of 1.0 ml/min. Between 10 and 35 min, Solvent D was increased to 32%. Between 35 and 36 min, Buffer D was decreased to 8% and held for 5 min at that composition to equilibrate the column between injections. 50 μl of the sample was injected, and elution was monitored at 254 and 350 nm. Under these conditions, the following retention times were obtained: 6.92 min for 5′-deoxyadenosine, 22.77 min for OH2Cbl, and 29.27 min for AdoCbl. The control reaction was performed in the absence of isobutyryl-CoA. Calibration curves were generated for all three compounds prepared and treated similarly to the assay samples. An extinction coefficient of ϵ260 nm = 15 mm−1 cm−1 was used to estimate the concentration of 5′-deoxyadenosine (17). The data were well fitted by a single exponential function for the disappearance of AdoCbl and appearance of 5′-deoxyadenosine and OH2Cbl. To improve recovery of OH2Cbl, proteinase K (Roche Applied Science) was used to digest the protein sample for 1–2 h at 37 °C before precipitation with acid.
Bioinformatics Analysis
Operon and regulon browsers on the Microbes Online web site were used for the elucidation of functional predictions for the genes of interest (18).
RESULTS
Gene Neighborhood Analysis for icmF
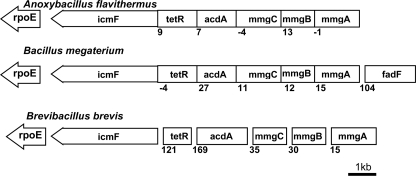
In several bacteria, the icmF gene is located in the same operon with or in close proximity to genes encoding enzymes involved in fatty acid metabolism (3). For example, enzymes found in the operon with icmF in several bacteria (Bacillus selenitireducens, Lysinibacillus sphaericus, Anoxybacillus flavithermus, Bacillus megaterium, Bacillus halodurans, Bacillus pseudofirmus, Bacillus coagulans, Bacillus sp. NRRL B-14911, Geobacillus sp. WCH70, and Brevibacillus brevis) are annotated as enzymes in the mother cell metabolic gene (mmg) operon, which has been described in Bacillus subtilis (19) (Fig. 5). Three genes in this operon are annotated as mmgA (acetyl-CoA transferase), mmgB (3-OH-butyryl-CoA dehydrogenase), and mmgC (acetyl-CoA dehydrogenase) (19). A similar set of enzymes in Ralstonia eutropha encoded by the H16_A0459–H16_A0464 operon allows growth on long-chain fatty acids. The absence of this operon together with the H16_A1526–H16_A1532 operon renders R. eutropha unable to grow on plant oils or long-chain fatty acids as a carbon source (20). The gene acdH from Streptomyces coelicolor and Streptomyces avermitilis that is homologous to mmgC is an acyl-CoA dehydrogenase and plays a role in the catabolism of branched-chain amino acids (21). Finally, the presence of the rpoE gene (which encodes the σ subunit of RNA polymerase) just downstream of icmF strongly suggests that IcmF is linked to fatty acid metabolism (22) (Fig. 5).
FIGURE 5.
Organization of genes in mmgABC operon harboring icmF gene. Top, A. flavithermus; middle, B. megaterium; bottom, B. brevis. The following genes are found in the same operon with icmF: tetR (TetR/Acr-like family transcriptional regulator), acdA (acyl-CoA dehydrogenase), mmgA (acetyl-CoA transferase), mmgB (3-OH-butyryl-CoA dehydrogenase), mmgC (acetyl-CoA dehydrogenase), and fadF (medium-/long-chain fatty acyl-CoA dehydrogenase). rpoE gene, which encodes the σ subunit of RNA polymerase, is found just downstream of icmF. The numbers below the genes indicate the distance in nucleotides between two adjacent genes. Negative numbers indicate overlapping genes. For a complete list of bacteria showing similar operonic organization, see the text.
Alternative Substrates for IcmF
Based on the above gene neighborhood analysis, we sought to assess whether IcmF plays a role in the metabolism of branched fatty acids and can catalyze the isomerization of substrates other than isobutyryl-CoA and n-butyryl-CoA. We decided to use isovaleryl-CoA, a building block for iso odd branched fatty acids, and 2-methylbutyryl-CoA, a building block for anteiso branched fatty acids, as potential substrates for IcmF. Isovaleryl-CoA and 2-methylbutyryl-CoA are synthesized from the branched-chain amino acids leucine and isoleucine, respectively, in reactions catalyzed by the branched-chain ketoacid dehydrogenase complex (21).
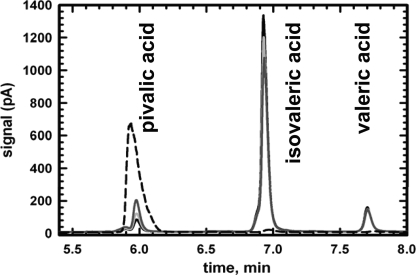
Gk IcmF was mixed with isovaleryl-CoA in the presence of AdoCbl and GTP/MgCl2, and the products were hydrolyzed and analyzed by GC. A time-dependent decrease in the isovaleric acid peak at 6.96 min was accompanied by the appearance of a peak at 5.99 min, suggesting conversion of isovaleryl-CoA to a new compound (Fig. 6). The expected product of AdoCbl-dependent 1,2 rearrangement of isovaleryl-CoA (or 3-methyl-butyryl-CoA) is pivalyl-CoA (or 2,2-dimethylpropionyl-CoA) (Fig. 1) (1). The retention time of a standard pivalic acid sample (5.99 min) exactly coincided with that of the product formed from isovaleryl-CoA (Fig. 6). The isomerization of isovaleryl-CoA to pivalyl-CoA by both Gk IcmF and Cm IcmF is slow in comparison with the isomerization of n-butyryl-CoA to isobutyryl-CoA (Table 1). The Km value for isovaleryl-CoA for Gk IcmF is 62 ± 8 μm, and the specific activity of the Gk IcmF with isovaleryl-CoA is 0.021 ± 0.004 μmol min−1 mg−1, which is ∼150-fold lower than the activity with n-butyryl-CoA (3.25 ± 0.35 μmol min−1 mg−1). With the Cm IcmF, an ∼2200-fold difference in the specific activities was obtained with the two substrates (Table 1).
FIGURE 6.
Pivalyl-CoA mutase activity of IcmF. A representative GC chromatogram showing separation of isovaleric and pivalic acids following enzymatic conversion of isovaleryl-CoA to pivalyl-CoA is shown. The reaction mixture contained in Buffer A 15 mm MgCl2, 2000 μg of Gk IcmF, 100 μm AdoCbl, 5 mm GTP, and 1.56 mm isovaleryl-CoA. At different time points, aliquots were removed, the esters were hydrolyzed, and the corresponding acids were separated by GC as described under “Experimental Procedures.” The traces represent 1 min (black line), 5 min (light gray line), 25 min (dark gray line), and pivalic acid standard (dashed line). Valeric acid was used as an internal standard.
TABLE 1.
Isobutyryl-CoA mutase and pivalyl-CoA mutase activities of recombinant IcmFs
Values represent the average of at least three independent experiments. In all experiments, a saturating concentration (2 mm) of substrate was used.
| Substrate | Specific activity |
|
|---|---|---|
| Gk IcmF | Cm IcmF | |
| μmol min−1mg−1 | ||
| Isovaleryl-CoA | 0.021 ± 0.004 | 0.015 ± 0.004 |
| Isobutyryl-CoA | 1.2 ± 0.1 | 13.8 ± 1.1 |
| n-Butyryl-CoA | 3.3 ± 0.4 | 33.0 ± 1.4 |
Isomerization of valeryl-CoA is expected to produce 2-methylbutyryl-CoA. However, consumption of valeryl-CoA was not observed in the presence of IcmF, AdoCbl, and GTP/MgCl2 in the GC-based assay (data not shown). Because HCM (5, 23) from Methylibium petroleiphilum is very similar to the standalone ICM from Streptomyces cinnamonensis and catalyzes the interconversion of 3-hydroxybutyryl-CoA and 2-hydroxyisobutyryl-CoA (5, 24), we decided to examine their substrate specificity overlap. However, conversion of dl-3-hydroxybutyryl-CoA to 2-hydroxyisobutyryl-CoA was not observed as judged by an HPLC-based assay (data not shown).
Absorption Spectrum of Gk IcmF during Steady-state Turnover
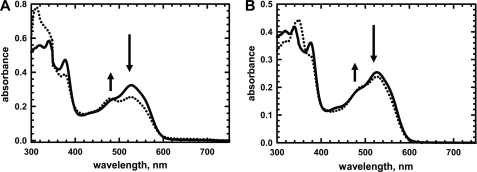
The binding sites for AdoCbl in the Gk IcmF dimer are nonidentical and exhibit an ∼25-fold difference in affinities (KD1 = 0.08 ± 0.01 μm and KD2 = 1.98 ± 0.4 μm) (3). For enzyme-monitored turnover experiments, Gk IcmF was reconstituted with 1 eq of AdoCbl to avoid the presence of free cofactor. The absence of free AdoCbl was confirmed by analyzing the spectrum of the flow-through obtained upon concentrating the reaction mixture using a Microcon 30 kDa concentrator. Addition of substrate (isobutyryl-CoA or n-butyryl-CoA) to holo-IcmF resulted in cob(II)alamin formation as evidenced by the decrease in absorbance at 525 nm and increase at 480 nm (Fig. 7A). Using a Δϵ525 nm of −4.8 mm−1 cm−1 (14), the ratio of AdoCbl:cob(II)alamin under steady-state turnover conditions was estimated to be ∼2:1 when isobutyryl-CoA was used as a substrate. With isovaleryl-CoA, the ratio of AdoCbl:cob(II)alamin under steady-state turnover conditions was 9:1 (Fig. 7B).
FIGURE 7.
Spectral changes in Gk holo-IcmF in presence of isobutyryl-CoA and isovaleryl-CoA. A, spectra of holo-IcmF (40 μm) in buffer A at 24 °C (solid line) and after addition of 4.1 mm isobutyryl-CoA, which resulted in formation of cob(II)alamin (dotted line). B, spectra of holo-IcmF (31 μm) in Buffer A at 24 °C (solid line) and after addition of 1.5 mm isovaleryl-CoA (dotted line).
Inactivation of IcmF and Effect of Nucleotides
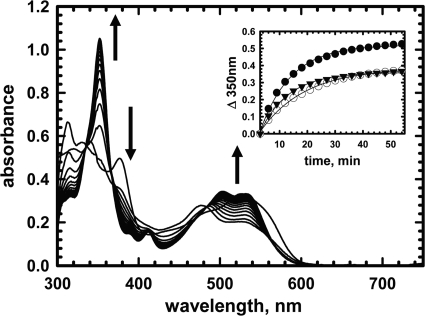
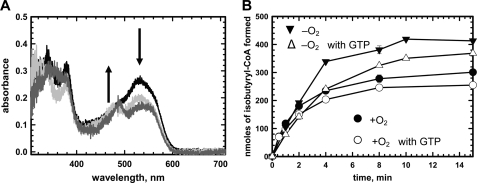
Cobalamin spectra under turnover conditions were used for monitoring the effect of nucleotides on the IcmF-catalyzed reaction (Fig. 8). Addition of isobutyryl-CoA results in formation of cob(II)alamin, which is converted over time to OH2Cbl (Fig. 8). The increase in absorption at 350 nm (kobs = 0.11 ± 0.01 min−1) (Fig. 8, inset) and the distinctive double peaks at 505 and 540 nm indicated formation of OH2Cbl, a hallmark of inactivation for AdoCbl-dependent enzymes (12, 25, 26). The amplitude but not the rate of OH2Cbl formation was diminished in the presence of GTP (kobs = 0.063 ± 0.002 min−1) and ATP (kobs = 0.10 ± 0.01 min−1) (Fig. 8, inset).
FIGURE 8.
Inactivation of Gk IcmF during turnover with isobutyryl-CoA. Rapid oxidation of cob(II)alamin to OH2Cbl was observed during reaction of holo-IcmF (with 1 eq; 41 μm AdoCbl bound) with 1.4 mm isobutyryl-CoA in Buffer A with 5 mm MgCl2 at 24 °C in the dark. The spectra were recorded between 0 and 60 min. Inset, the time-dependent increase at 350 nm was fitted to a single exponential function in the absence of nucleotides (filled circles) (kobs = 0.11 ± 0.01 min−1) or in the presence of 5 mm GTP (open circles) (kobs = 0.063 ± 0.002 min−1) or 5 mm ATP (triangles) (kobs = 0.10 ± 0.01 min−1).
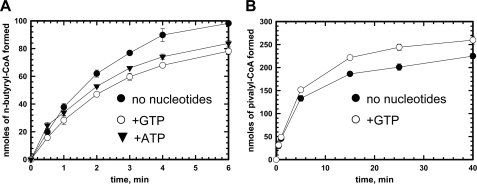
Under standard in vitro assay conditions, Gk IcmF exhibits a linear reaction time course for only 40–60 s before the activity plateaus (Fig. 9A). The specific activity with isobutyryl-CoA (1.1 ± 0.1 μmol min−1mg−1) was calculated using the linear portion of the curve and was comparable with values obtained previously in the coupled enzyme assay (3). When n-butyryl-CoA was used as a substrate, a specific activity of 3.25 ± 0.35 μmol min−1 mg−1 was obtained. With both isobutyryl-CoA and n-butyryl-CoA, inactivation of Gk IcmF resulted in termination of the reaction in ∼6–10 min during which time only ∼7% of substrate was consumed (Fig. 9A). Very similar behavior was also observed with Cm IcmF (data not shown). Protein stability, the presence of metal ions (K+ and Ca2+), reductants (DTT and tris(2-carboxyethyl)phosphine hydrochloride), and different buffers (50 mm sodium phosphate, pH 7.5, 100 mm NaCl; 50 mm potassium phosphate, pH 7.5, 100 mm KCl; and 50 mm HEPES, pH 7.5, 100 mm NaCl) were assessed and ruled out as contributors to the observed inactivation (data not shown). Nucleotides (ATP and GTP) decreased the activity of Gk IcmF (Fig. 9A) as reported previously with GDP and GTP (3).
FIGURE 9.
Effect of nucleotides on time course of reactions catalyzed by IcmF. A, time course of the isobutyryl-CoA mutase reaction catalyzed by Gk IcmF at 37 °C. The assay mixture in Buffer A with 10 mm MgCl2 contained 40 μg of holo-IcmF, 100 μm AdoCbl, 1.5 mm isobutyryl-CoA, and either no nucleotides (black circles), 6 mm ATP (triangles), or 3 mm GTP (open circles). B, time course of the isovaleryl-CoA mutase reaction catalyzed by Gk IcmF. The reaction mixture in Buffer A with 15 mm MgCl2 contained 2500 μg of IcmF, 100 μm AdoCbl, and 1.5 mm isovaleryl-CoA with (white circles) or without 5 mm GTP (black circles) at 37 °C. Aliquots of the reactions were removed at different time points and analyzed by GC as described under “Experimental Procedures.” Data represent the average of three independent experiments. The data represent the mean ± S.D. of three independent experiments.
With isovaleryl-CoA, the Gk IcmF reaction was almost completely inhibited after 25–30 min (Fig. 9B). However, in the presence of GTP, the enzyme was inactivated to a lesser extent in comparison with the reaction without GTP (Fig. 9B). Similar behavior was seen for Cm IcmF (data not shown). Interestingly, for the first 2 min, no difference was seen in the isovaleryl-CoA/pivalyl-CoA mutase activity ±GTP (Fig. 9B).
Loss of 5′-Deoxyadenosine Leads to Inactivation of IcmF
The steady formation of OH2Cbl during IcmF turnover under aerobic conditions suggests that inactivation might result from oxidative interception of cob(II)alamin (27). An alternative possibility is that inactivation is associated with loss of 5′-deoxyadenosine from the active site, and the uncoupled cob(II)alamin is subsequently oxidized to OH2Cbl. To distinguish between these possibilities, enzyme-monitored turnover experiments were conducted under anaerobic conditions (Fig. 10A). Addition of n-butyryl-CoA to holo-IcmF resulted in formation of cob(II)alamin, which was not further converted to OH2Cbl even after 60 min (Fig. 10A).
FIGURE 10.
Inactivation of Gk IcmF under anaerobic conditions. A, spectral changes upon incubation of 33 μm holo-IcmF (containing 1 eq of bound AdoCbl) with 4.8 mm n-butyryl-CoA in Buffer A with 5 mm MgCl2 under anaerobic conditions at 24 °C in the dark. Spectra were recorded at time 0 (black), immediately after addition of substrate (light gray), and 60 min after addition of substrate (dark gray). Formation of cob(II)alamin (480 nm peak) without further conversion to OH2Cbl was observed. B, comparison of time courses for the reaction catalyzed by Gk IcmF at 37 °C under aerobic (circles) and anaerobic (triangles) conditions in the presence of GTP. The aerobic and anaerobic assay mixtures in Buffer A with 15 mm MgCl2 contained 40 μg of IcmF, 2 mm n-butyryl-CoA, and either no nucleotides (solid symbols) or 4.3 mm GTP (open symbols). The data represent the mean ± S.D. of three independent experiments.
Next, we compared the time courses of the reaction catalyzed by Gk IcmF with n-butyryl-CoA under aerobic and anaerobic conditions (Fig. 10B). Under anaerobic conditions, the reaction exhibited a linear dependence for a longer time period (2 versus 1 min) but eventually ceased after 10 min. Because spectroscopic analysis of the reaction time course shows that OH2Cbl is not formed under these conditions, it demonstrates that inactivation is not directly correlated with OH2Cbl formation. As seen under aerobic conditions, GTP also led to decreased OH2Cbl formation under anaerobic conditions (Fig. 10B).
Characterization of Inactivation Products by HPLC
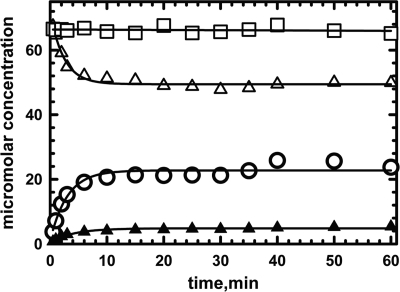
The reaction of 64 μm Gk holo-IcmF (containing 2 eq of AdoCbl) with 4 mm isobutyryl-CoA was monitored as described under “Experimental Procedures” (Fig. 11). Under these conditions, AdoCbl disappeared with a kobs of 0.40 ± 0.04 min−1 (Table 2). The rate of appearance of 5′-deoxyadenosine (kobs = 0.32 ± 0.01 min−1) corresponded to the rate of AdoCbl disappearance. The concentration of 5′-deoxyadenosine recovered (20.6 ± 0.8 μm) was equal to the concentration of AdoCbl consumed (23 ± 1 μm). In contrast, the rate of OH2Cbl formation (kobs = 0.23 ± 0.02 min−1) was slower, and only 4.34 ± 0.01 μm OH2Cbl was recovered (Table 2). Treatment of inactivated enzyme with proteinase K increased OH2Cbl recovery only marginally (6.3 ± 0.2 μm). The reason for the low yield of OH2Cbl is presently not clear. To ensure that AdoCbl is not cleaved non-enzymatically to 5′-deoxyadenosine during sample preparation and/or chromatographic separation, a control reaction was performed in which substrate was omitted from the reaction mixture (Fig. 11).
FIGURE 11.
Formation of OH2Cbl and 5′-deoxyadenosine during enzyme-monitored turnover. Holo-IcmF (64 μm IcmF active site concentration containing 2 eq of AdoCbl) was mixed with 1.5 mm isobutyryl-CoA in Buffer A at 37 °C. All manipulations with the samples and HPLC were performed in the dark. The decay of AdoCbl (open triangles) and the appearance of OH2Cbl (solid triangles) and of 5′-deoxyadenosine (open circles) were monitored over 50 min. As a control for AdoCbl stability during sample handling, the analysis was repeated without addition of isobutyryl-CoA (open squares). The data were fitted by a single exponential function for the disappearance of AdoCbl and appearance of 5′-deoxyadenosine and OH2Cbl.
TABLE 2.
Kinetics of OH2Cbl and 5′-deoxyadenosine formation
Values represent the average of at least two independent experiments.
| Nucleotide | AdoCbl |
OH2Cbl |
5′-Deoxyadenosine |
|||
|---|---|---|---|---|---|---|
| kobs | Concentration | kobs | Concentration | kobs | Concentration | |
| min−1 | μm | min−1 | μm | min−1 | μm | |
| None | 0.4 ± 0.04 | 23 ± 1 | 0.23 ± 0.02 | 4.34 ± 0.01 | 0.32 ± 0.01 | 20.6 ± 0.8 |
| GTP | 0.37 ± 0.04 | 20.65 ± 1.02 | 0.25 ± 0.04 | 5.9 ± 1.4 | 0.35 ± 0.05 | 19.9 ± 1.3 |
IcmF appears to show half-of-sites activity because reconstitution with 1 versus 2 eq of AdoCbl resulted in the same steady-state concentration of cob(II)alamin (data not shown). Surprisingly, in the inactivation experiments, not all the AdoCbl bound to Gk IcmF was converted to OH2Cbl and 5′-deoxyadenosine. Instead, ∼66% of the AdoCbl at one site (i.e. 32 μm) was converted to 5′-deoxyadenosine; this corresponded to the concentration of cob(II)alamin (∼21 μm) under steady-state turnover conditions. This provides further evidence for half-of-sites activity in Gk IcmF. It is unclear why complete inactivation at one of the two active sites was not observed.
ATPase Activity of IcmF
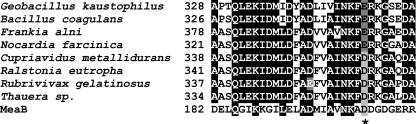
In the MeaI domains of many IcmFs, the base specificity loop motif NKXD is modified to NKXE (3). In the Gk IcmF, the NKXE sequence is present, whereas in the Cm IcmF, the sequence is NKXD (Fig. 12). The Lys residue in the NKXD motif interacts via hydrophobic interactions with the plane of the guanine ring, whereas the Asp coordinates two nitrogen atoms in the purine ring. Gk IcmF catalyzes the hydrolysis of both GTP and ATP (Table 3). The kinetic parameters for IcmF measured in the presence of various nucleotides are comparable: kcat with ATP is 19 ± 1 min−1, and kcat with GTP is 10 ± 1 min−1. Although the Km for ATP is high (1290 ± 300 μm) relative to that for GTP (51 ± 3 μm), the concentration of ATP (3–5 mm) is higher than of GTP (1 mm) in bacterial cells (28).
FIGURE 12.
Multiple sequence alignment of IcmFs and MeaB from M. extorquens showing base specificity loop NKX(D/E). Accession numbers are as follows: G. kaustophilus, YP_149244; C. metallidurans CH34, YP_582365; R. eutropha H16, YP_724799; Frankia alni, YP_716016; Nocardia farcinica, YP_117245; B. coagulans, ZP_01696637; Thauera sp., ZP_02841697; Rubrivivax gelatinosus, ZP_00242991; and MeaB, AAL86727. Asp (D) substitution by Glu (E) is indicated by an asterisk. All accession numbers are from the NCBI protein database.
TABLE 3.
Comparison of GTPase and ATPase Activities of Gk IcmF
| Enzyme | GTPa |
ATPa |
||||
|---|---|---|---|---|---|---|
| kcat | Km(GTP) | kcat/KGTP | kcat | Km(ATP) | kcat/KATP | |
| min−1 | μm | m−1min−1 | min−1 | μm | m−1min−1 | |
| Wild type | 10 ± 2 | 51 ± 3 | (1.96 ± 0.37) × 105 | 19 ± 1 | 1290 ± 300 | (1.47 ± 0.03) × 104 |
| K213Ab | ND | ND | ||||
a All experiments were performed in Buffer A with 20 mm MgCl2 at 37 °C as described under “Experimental Procedures.” Values represent the average of at least five independent experiments. ND, not detected.
b When a 40-fold higher concentration of the K213A mutant was used in the assay, GTPase/ATPase activity above the detection limit of 5 μm inorganic phosphate released in the malachite green assay was not observed.
To rule out the possibility that the ATPase activity of Gk IcmF is due to a contaminant that co-purifies with the mutase, first we tested the effect of introducing the K213A mutation in the GtgGaGKSS sequence of the P-loop motif, which is important for phosphate binding (29). The K213A Gk IcmF mutant was devoid of both GTPase and ATPase activities. Notably, the mutase activity of the K213A mutant was similar to that of wild-type protein (0.6 μmol min−1 mg−1 with isobutyryl-CoA). Furthermore, in the presence of 3 mm AMPPNP, the kcat for the GTPase activity was inhibited ∼3.6-fold to 2.7 ± 0.14 min−1. Taken together, the above results are consistent with the ATPase and GTPase activities being intrinsic to Gk IcmF.
Although it has been suggested that replacement of Asp by Glu in the NKXD motif has no effect on nucleotide specificity (30), our results suggest the contrary. Thus, in Cm IcmF, the base specificity loop sequence is NKFD, and although this protein does not exhibit ATPase activity, it is an active GTPase (kcat = 18 ± 1.3 min−1 and KGTP = 40 ± 8 μm). These results provide an interesting illustration of a G-protein losing its specificity due to a single substitution in the NKXD motif.
DISCUSSION
Previously, it was believed that ICM-like activity was restricted to the Streptomyces genus where it is involved in monensin A and B production (4). Our discovery of ICM activity in the IcmF fusion protein, which is widely distributed in bacteria, suggests its involvement in metabolic processes beyond polyketide synthesis (3). In our efforts to elucidate the possible roles of IcmF in bacterial metabolism, we noticed that icmF genes co-localize with genes encoding enzymes involved in β-oxidation of fatty acids. Subsequently, we probed branched organic acids, which are synthesized from the branched amino acids valine, leucine, and isoleucine, as substrates for IcmF. This in turn led to the discovery of a new IcmF activity, i.e. isomerization of isovaleryl-CoA/pivalyl-CoA (Figs. 1 and 6).
Although AdoCbl-dependent pivalyl-CoA mutase activity has been predicted to exist (10, 31), an enzyme with this catalytic activity has not been identified previously. In this study, we report that IcmFs from G. kaustophilus and C. metallidurans can convert isovaleryl-CoA to pivalyl-CoA (Fig. 1). Depending on the organism, the isovaleryl-CoA/pivalyl-CoA mutase activity of IcmF was ∼150–2,200-fold lower than the conversion of n-butyryl-CoA to isobutyryl-CoA (Table 1). Recently, it was shown that pivalic acid can be incorporated as a starter unit in fatty acids in several bacteria (32). Thus, the production of pivalyl-CoA catalyzed by IcmF might be important in bacteria that use this starter unit for the biosynthesis of branched fatty acids containing a quaternary carbon. Our discovery of the isovaleryl-CoA/pivalyl-CoA mutase activity adds to the growing list of carbon skeleton rearrangements catalyzed by AdoCbl-dependent isomerases (Fig. 1). Thus, it appears that the “mutase core” is quite versatile for two reasons. First, substitutions of a limited number of key active site residues alter substrate specificity, and second, relaxed substrate specificity allows alternative reactions to be catalyzed by the same active site as exemplified by IcmF.
We also report relaxed substrate specificity in the MeaI domain of Gk IcmF. It has been reported that some G-proteins have either lost or switched nucleotide specificity (33). For example, in centaurin γ-1 GTPase where the sequence of the base specificity loop NKXD is modified to TQDR (34), nucleotide specificity is lacking, and it is described as a general NTPase (34). Proteins belonging to the YchF subfamily of the Obg family of G-proteins harbor the NXXE motif instead of the NKXD (35). For example, human OLA1, which belongs to the Obg family, hydrolyzes ATP more efficiently than GTP.
In our study, we show that in a subset of the MeaI chaperone domains of IcmF the NKXD motif is modified to NKXE. This constitutes an interesting example where only some members of a protein family have lost specificity for GTP. We note that although the MeaI domain of Gk IcmF exhibits ATPase activity it is functionally distinct from the ATP-dependent chaperones for AdoCbl-dependent eliminases, e.g. diol dehydratases (36, 37).
A remarkable feature of AdoCbl-dependent enzymes is that they catalyze reactions involving radical intermediates under aerobic conditions. However, this comes with a price, i.e. their susceptibility to inactivation (12). Under standard in vitro assay conditions, IcmF is inactivated quite rapidly with either isobutyryl-CoA or isovaleryl-CoA as substrate (Fig. 9). In a subclass of AdoCbl-dependent enzymes, reactivating factors mediate an ATP-dependent exchange of enzyme-bound OH2Cbl with free AdoCbl (36–39). These reactivases have sequence similarity to DnaK and to other members of the Hsp70 family of molecular chaperones and lower sequence similarity to the large subunits of corresponding mutases (37). In contrast, the G-protein chaperones associated with AdoCbl-dependent mutases belong to the SIMIBI (signal recognition particle, MinD, and BioD) subclass of the G3E family of P-loop metallochaperones (3, 13, 29). In MCM where the role of the G-protein chaperone MeaB is best characterized (14), the chaperone uses GTP hydrolysis to power the expulsion of cob(II)alamin when 5′-deoxyadenosine is lost from the active site (13). Unlike the ATP-dependent reactivases, MeaB is unable to release OH2Cbl bound to MCM (13, 14). Instead, MeaB exerts a protective effect on the MCM reaction by reducing the inactivation rate in the presence of nucleotides. In numerous bacterial genomes, MeaB-like proteins are found in the same operon as the mutases (40). Mutations in the MeaB ortholog in humans results in methylmalonic aciduria, an inborn error of metabolism (41, 42), pointing to the important role of this auxiliary protein in maintaining MCM function.
The presence of nucleotides has virtually no effect on the inactivation kinetics of IcmF with either isobutyryl-CoA or n-butyryl-CoA as substrate (Fig. 9A). In contrast, when isovaleryl-CoA is used as substrate, protection, albeit modest, was seen in the presence of GTP (Fig. 9B). The activity of Gk IcmF with isobutyryl-CoA is reduced in the presence of nucleotides (3). This is unexpected because MeaB increases the kcat of MCM 1.8-fold in addition to protecting it from inactivation (14). The rate of IcmF inactivation (0.11 ± 0.01 min−1) in the presence of isobutyryl-CoA is ∼14-fold higher than inactivation of MCM in the presence of methylmalonyl-CoA (0.0072 min−1) (14).
Under anaerobic conditions, the IcmF reaction with n-butyryl-CoA was linear for a longer duration than in the presence of oxygen (Fig. 10B). Because OH2Cbl formation is observed only under aerobic conditions, it argues against an internal electron transfer from cob(II)alamin to the substrate being responsible for inactivation as described for lysine 5,6-aminomutase (26). Because IcmF is also inactivated under anaerobic conditions where OH2Cbl is not formed, we conclude that inactivation is primarily signaled by the loss of 5′-deoxyadenosine from the active site.
In conclusion, we report that IcmF catalyzes the formation pivalyl-CoA from isovaleryl-CoA. There is virtually no information on bacterial metabolism of pivalic acid, which has a mostly anthropogenic origin (10). We suggest that pivalyl-CoA mutase activity of IcmF might be important for biodegradation of branched compounds where pivalic acid is a central intermediate (10, 43). The activity of IcmF would reduce branching of compounds with a quaternary carbon. It will be important to follow the fate of pivalyl-CoA in IcmF-containing bacteria and test whether this compound is incorporated in fatty acids and/or other compounds.
Acknowledgment
We gratefully acknowledge Dr. Thore Rohwerder (Helmholtz Center for Environmental Research) for helpful discussions on the distribution of acyl-CoA mutases in nature and their potential roles in microbial metabolism.
This work was supported, in whole or in part, by National Institutes of Health Grant DK45776.
- MCM
- methylmalonyl-CoA mutase
- AdoCbl
- 5′-deoxyadenosylcobalamin
- IcmF
- isobutyryl-CoA mutase fused
- ICM
- isobutyryl-CoA mutase
- AMPPNP
- adenosine 5′-(β,γ-imido)triphosphate
- LIC
- ligase-independent cloning
- OH2Cbl
- aquacobalamin
- HCM
- 2-hydroxyisobutyryl-CoA mutase
- ECM
- ethylmalonyl-CoA mutase
- Gk
- G. kaustophilus
- Cm
- C. metallidurans.
REFERENCES
- 1. Banerjee R. (2003) Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 103, 2083–2094 [DOI] [PubMed] [Google Scholar]
- 2. Ratnatilleke A., Vrijbloed J. W., Robinson J. A. (1999) Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J. Biol. Chem. 274, 31679–31685 [DOI] [PubMed] [Google Scholar]
- 3. Cracan V., Padovani D., Banerjee R. (2010) IcmF is a fusion between the radical B12 enzyme isobutyryl-CoA mutase and its G-protein chaperone. J. Biol. Chem. 285, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vrijbloed J. W., Zerbe-Burkhardt K., Ratnatilleke A., Grubelnik-Leiser A., Robinson J. A. (1999) Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: influence on polyketide antibiotic biosynthesis. J. Bacteriol. 181, 5600–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rohwerder T., Breuer U., Benndorf D., Lechner U., Müller R. H. (2006) The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl. Environ. Microbiol. 72, 4128–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erb T. J., Rétey J., Fuchs G., Alber B. E. (2008) Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J. Biol. Chem. 283, 32283–32293 [DOI] [PubMed] [Google Scholar]
- 7. Mancia F., Evans P. (1998) Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism. Structure 6, 711–720 [DOI] [PubMed] [Google Scholar]
- 8. Mancia F., Keep N. H., Nakagawa A., Leadlay P. F., McSweeney S., Rasmussen B., Bösecke P., Diat O., Evans P. R. (1996) How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 4, 339–350 [DOI] [PubMed] [Google Scholar]
- 9. Drennan C. L., Huang S., Drummond J. T., Matthews R. G., Lidwig M. L. (1994) How a protein binds B12: a 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science 266, 1669–1674 [DOI] [PubMed] [Google Scholar]
- 10. Rohwerder T., Müller H. (2008) in Vitamin B: New Research (Elliot C. M., ed.) pp. 81–98, Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 11. Gruber K., Puffer B., Kräutler B. (2011) Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem. Soc. Rev. 40, 4346–4363 [DOI] [PubMed] [Google Scholar]
- 12. Toraya T. (2000) Radical catalysis of B12 enzymes: structure, mechanism, inactivation, and reactivation of diol and glycerol dehydratases. Cell. Mol. Life Sci. 57, 106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Padovani D., Banerjee R. (2009) A G-protein editor gates coenzyme B12 loading and is corrupted in methylmalonic aciduria. Proc. Natl. Acad. Sci. U.S.A. 106, 21567–21572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Padovani D., Banerjee R. (2006) Assembly and protection of the radical enzyme, methylmalonyl-CoA mutase, by its chaperone. Biochemistry 45, 9300–9306 [DOI] [PubMed] [Google Scholar]
- 15. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 16. Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]
- 17. Babior B. M., Carty T. J., Abeles R. H. (1974) The mechanism of action of ethanolamine ammonia-lyase, a B12-dependent enzyme. The reversible formation of 5′-deoxyadenosine from adenosylcobalamin during the catalytic process. J. Biol. Chem. 249, 1689–1695 [PubMed] [Google Scholar]
- 18. Alm E. J., Huang K. H., Price M. N., Koche R. P., Keller K., Dubchak I. L., Arkin A. P. (2005) The MicrobesOnline web site for comparative genomics. Genome Res. 15, 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reddick J. J., Williams J. K. (2008) The mmgA gene from Bacillus subtilis encodes a degradative acetoacetyl-CoA thiolase. Biotechnol. Lett. 30, 1045–1050 [DOI] [PubMed] [Google Scholar]
- 20. Brigham C. J., Budde C. F., Holder J. W., Zeng Q., Mahan A. E., Rha C., Sinskey A. J. (2010) Elucidation of β-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol. 192, 5454–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y. X., Denoya C. D., Skinner D. D., Fedechko R. W., McArthur H. A., Morgenstern M. R., Davies R. A., Lobo S., Reynolds K. A., Hutchinson C. R. (1999) Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 145, 2323–2334 [DOI] [PubMed] [Google Scholar]
- 22. Matsuoka H., Hirooka K., Fujita Y. (2007) Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 282, 5180–5194 [DOI] [PubMed] [Google Scholar]
- 23. Rohwerder T., Müller R. H. (2010) Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microb. Cell Fact. 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller R. H., Rohwerder T., Harms H. (2007) Carbon conversion efficiency and limits of productive bacterial degradation of methyl tert-butyl ether and related compounds. Appl. Environ. Microbiol. 73, 1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toraya T., Tamura N., Watanabe T., Yamanishi M., Hieda N., Mori K. (2008) Mechanism-based inactivation of coenzyme B12-dependent diol dehydratase by 3-unsaturated 1,2-diols and thioglycerol. J. Biochem. 144, 437–446 [DOI] [PubMed] [Google Scholar]
- 26. Tang K. H., Chang C. H., Frey P. A. (2001) Electron transfer in the substrate-dependent suicide inactivation of lysine 5,6-aminomutase. Biochemistry 40, 5190–5199 [DOI] [PubMed] [Google Scholar]
- 27. Maiti N., Widjaja L., Banerjee R. (1999) Proton transfer from histidine 244 may facilitate the 1,2 rearrangement reaction in coenzyme B12-dependent methylmalonyl-CoA mutase. J. Biol. Chem. 274, 32733–32737 [DOI] [PubMed] [Google Scholar]
- 28. Gaal T., Bartlett M. S., Ross W., Turnbough C. L., Jr., Gourse R. L. (1997) Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278, 2092–2097 [DOI] [PubMed] [Google Scholar]
- 29. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 30. Pandit S. B., Srinivasan N. (2003) Survey for G-proteins in the prokaryotic genomes: prediction of functional roles based on classification. Proteins 52, 585–597 [DOI] [PubMed] [Google Scholar]
- 31. Probian C., Wülfing A., Harder J. (2003) Anaerobic mineralization of quaternary carbon atoms: isolation of denitrifying bacteria on pivalic acid (2,2-dimethylpropionic acid). Appl. Environ. Microbiol. 69, 1866–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rezanka T., Siristova L., Schreiberová O., Rezanka M., Masák J., Melzoch K., Sigler K. (2011) Pivalic acid acts as a starter unit in a fatty acid and antibiotic biosynthetic pathway in Alicyclobacillus, Rhodococcus and Streptomyces. Environ. Microbiol. 13, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 33. Wittinghofer A., Vetter I. R. (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 [DOI] [PubMed] [Google Scholar]
- 34. Soundararajan M., Yang X., Elkins J. M., Sobott F., Doyle D. A. (2007) The centaurin γ-1 GTPase-like domain functions as an NTPase. Biochem. J. 401, 679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koller-Eichhorn R., Marquardt T., Gail R., Wittinghofer A., Kostrewa D., Kutay U., Kambach C. (2007) Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 282, 19928–19937 [DOI] [PubMed] [Google Scholar]
- 36. Kajiura H., Mori K., Shibata N., Toraya T. (2007) Molecular basis for specificities of reactivating factors for adenosylcobalamin-dependent diol and glycerol dehydratases. FEBS J. 274, 5556–5566 [DOI] [PubMed] [Google Scholar]
- 37. Mori K., Bando R., Hieda N., Toraya T. (2004) Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J. Bacteriol. 186, 6845–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zelder O., Beatrix B., Leutbecher U., Buckel W. (1994) Characterization of the coenzyme-B12-dependent glutamate mutase from Clostridium cochlearium produced in Escherichia coli. Eur. J. Biochem. 226, 577–585 [DOI] [PubMed] [Google Scholar]
- 39. Chang C. H., Frey P. A. (2000) Cloning, sequencing, heterologous expression, purification, and characterization of adenosylcobalamin-dependent d-lysine 5,6-aminomutase from Clostridium sticklandii. J. Biol. Chem. 275, 106–114 [DOI] [PubMed] [Google Scholar]
- 40. Dobson C. M., Wai T., Leclerc D., Wilson A., Wu X., Doré C., Hudson T., Rosenblatt D. S., Gravel R. A. (2002) Identification of the gene responsible for the cblA complementation group of vitamin B12-responsive methylmalonic acidemia based on analysis of prokaryotic gene arrangements. Proc. Natl. Acad. Sci. U.S.A. 99, 15554–15559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hubbard P. A., Padovani D., Labunska T., Mahlstedt S. A., Banerjee R., Drennan C. L. (2007) Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria. J. Biol. Chem. 282, 31308–31316 [DOI] [PubMed] [Google Scholar]
- 42. Lerner-Ellis J. P., Dobson C. M., Wai T., Watkins D., Tirone J. C., Leclerc D., Doré C., Lepage P., Gravel R. A., Rosenblatt D. S. (2004) Mutations in the MMAA gene in patients with the cblA disorder of vitamin B12 metabolism. Hum. Mutat. 24, 509–516 [DOI] [PubMed] [Google Scholar]
- 43. Solano-Serena F., Marchal R., Heiss S., Vandecasteele J. P. (2004) Degradation of isooctane by Mycobacterium austroafricanum IFP 2173: growth and catabolic pathway. J. Appl. Microbiol. 97, 629–639 [DOI] [PubMed] [Google Scholar]