Background: The budding yeast transcription factor Aft1 has been implicated in chromosome stability.
Results: Aft1 interacts with the kinetochore protein Iml3 and is required for promoting pericentromeric cohesin.
Conclusion: We have identified a new factor impacting the pericentric chromatin spring.
Significance: This discovery explains why Aft1 mutants display chromosome segregation defects and sheds light on how the Ctf19 complex promotes pericentric cohesin.
Keywords: Centromeres, Cohesin, Kinetochore, Mitotic Spindle, Yeast, Aft1, Iml3, Pericentromeric Chromatin
Abstract
The Saccharomyces cerevisiae iron-responsive transcription factor, Aft1, has a well established role in regulating iron homeostasis through the transcriptional induction of iron-regulon genes. However, recent studies have implicated Aft1 in other cellular processes independent of iron regulation such as chromosome stability. In addition, chromosome spreads and two-hybrid data suggest that Aft1 interacts with and co-localizes with kinetochore proteins; however, the cellular implications of this have not been established. Here, we demonstrate that Aft1 associates with the kinetochore complex through Iml3. Furthermore, like Iml3, Aft1 is required for the increased association of cohesin with pericentric chromatin, which is required to resist microtubule tension, and aft1Δ cells display chromosome segregation defects in meiosis. Our work defines a new role for Aft1 in chromosome stability and transmission.
Introduction
In the budding yeast, Saccharomyces cerevisiae, iron levels are regulated by the transcription factor Aft1 (reviewed in Ref. 1). Iron depletion activates Aft1 to induce the expression of ∼25 genes referred to as the “iron-regulon” by binding the consensus sequence 5′-YRCACCCR-3′ present in iron-responsive elements of these target genes (2, 3). This subset of genes is involved in the uptake, compartmentalization, and utilization of iron with the end result of increasing iron levels and remodeling cellular metabolism to survive low iron conditions.
Although Aft1 has an established role as an iron-responsive transcription factor, many studies have implicated Aft1 in other cellular functions, including chromosome stability, cell cycle regulation, DNA damage, cell wall stability, and mitochondrial function (4–10). Evidence implicating Aft1 in chromosome stability first originated from synthetic genetic array studies that determined aft1Δ mutants could not tolerate either overexpression or loss-of-function of kinetochore genes (6). Moreover, chromosome transmission fidelity assays determined that aft1Δ mutant cells have defects in maintaining artificial chromosome fragments (4, 6, 7), and aft1Δ cells were shown to be hypersensitive to benomyl treatment (4). Benomyl is a microtubule-destabilizing drug, and many mutants with defects in chromosome stability share the same sensitivity (11). In contrast to most chemical sensitivities and phenotypes of aft1Δ cells, neither the benomyl sensitivity nor chromosome transmission fidelity defects of aft1Δ mutant cells can be rescued by increased extracellular iron in media (4). Furthermore, the paralog of AFT1, AFT2, and other iron-regulon genes were not identified in the genome-wide studies that implicated Aft1 in chromosome stability (4, 6, 7) nor are aft2Δ mutant cells sensitive to benomyl treatment (4). Overall, this suggests that the role of Aft1 in chromosome stability is not related to regulation of the iron-regulon or iron homeostasis.
Many lines of evidence suggest that Aft1 has a role at the kinetochore. The kinetochore complex consists of many multiprotein subcomplexes that assemble in a hierarchical fashion onto centromeric (CEN) DNA physically linking the chromosomes to microtubule spindles (reviewed in Ref. 12). Aft1 was found to co-localize with kinetochore proteins using chromosome spreads (6), and Aft1 has been shown to interact with the kinetochore protein Iml3 by yeast-two hybrid assays (13, 14). Iml3 is a peripheral member of the Ctf19 complex that is composed of the COMA subcomplex (Ctf19, Okp1, Mcm21, and Ame1) along with Chl4 and Iml3 (15, 16). Studies investigating the hierarchical dependences of the kinetochore proteins have established that Iml3 localization to the kinetochore is dependent on its interacting partner, Chl4, which in turn is dependent on Ctf19 (Fig. 1A) (15). Although very little is known about Iml3 or associated Ctf19 complex function, it has been shown that these proteins are required for the increased association of cohesin (reviewed in Refs. 17, 18) at the pericentromere and centromere regions in both mitosis and meiosis (5, 19–21). Contrary to its role in embracing sister chromatids at the arms, cohesin, along with condensin, functions at pericentric DNA to form an intramolecular loop with the centromere as its apex (22). This establishes a pericentromeric chromatin spring-like structure that counterbalances microtubule forces during metaphase (23) and ensuring sister chromatid bi-orientation (reviewed in 24). Despite the importance of pericentric cohesin, the molecular mechanism by which the Ctf19 complex recruits cohesin has yet to be ascertained.
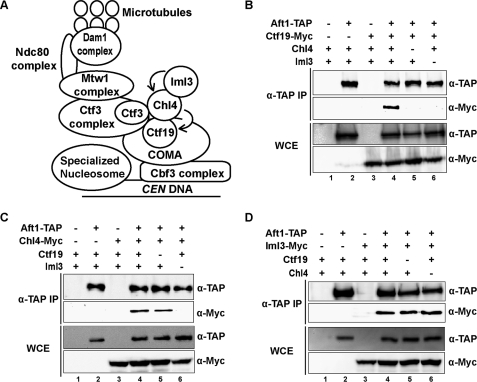
FIGURE 1.
Aft1 interaction with Ctf19 kinetochore complex is dependent on Iml3. A, schematic diagram of the budding yeast kinetochore highlighting the hierarchical dependences of the Ctf19 subcomplex. B, Aft1 co-purification with the kinetochore protein Ctf19 is dependent on Chl4 and Iml3. Anti-TAP immunoprecipitations were performed with strains containing different combinations of TAP-tagged Aft1 and Myc-tagged Ctf19 in the presence or absence of Chl4 and Iml3. C, Aft1 co-purification with the kinetochore protein Chl4 is dependent on Iml3 but not Ctf19. Anti-TAP immunoprecipitations were performed with strains containing different combinations of TAP-tagged Aft1 and Myc-tagged Chl4 in the presence or absence of Ctf19 and Iml3. D, Aft1 co-purification with the kinetochore protein Iml3 is independent of Ctf19 and Chl4. Anti-TAP immunoprecipitations were performed with strains containing different combinations of TAP-tagged Aft1 and Myc-tagged Iml3 in the presence or absence of Ctf19 and Chl4. For all immunoprecipitations, 10% of the eluate was loaded in the TAP blot, and 90% of the eluate was loaded in the Myc blot. IP, immunoprecipitations; WCE, whole-cell extract.
In this study we further investigate the role of Aft1 in chromosome stability and its interaction with the kinetochore. Surprisingly, we discover that Aft1 interacts with Iml3 and has a role in promoting pericentric cohesin.
EXPERIMENTAL PROCEDURES
Yeast Strains
Yeast strains used in this study are listed in Table 1. Deletion strains and epitope tag integrations made for this study were designed using a standard PCR-mediated gene insertion technique (25) and confirmed by PCR analysis.
TABLE 1.
Strains used in this study
| Strain | Auxotrophies | Ref. or Source |
|---|---|---|
| YKB779 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 51 |
| YKB780 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 51 |
| YKB2020 | MATα his3 leu2 ura3 AFT1-TAP::HIS | This study |
| YKB2019 | MATa ade2-101 his3-Δ200 lys2-801 leu2-Δ1 ura3-52 trp1-Δ63 IML3-Myc::kanMX | This study |
| YKB2023 | MATa his3 leu2 ura3 AFT1-TAP::HIS IML3-Myc::kanMX | This study |
| YKB2285 | MATa leu2 ura3 AFT1-TAP::HIS IML3-Myc::kanMX ctf19Δ::HIS3 | This study |
| YKB2639 | MATa his3 leu2 ura3 AFT1-TAP::HIS IML3-Myc::kanMX chl4Δ::TRP1 | This study |
| YKB2281 | MATa his3 leu2 ura3 AFT1-TAP::HIS CHL4-Myc::TRP1 | This study |
| YKB2284 | MATa his3 leu2 ura3 CHL4-Myc::TRP1 | This study |
| YKB2287 | MATa his3 leu2 ura3 AFT1-TAP::HIS CHL4-Myc::TRP1 iml3Δ::kanMX | This study |
| YKB2640 | MATa his3 leu2 ura3 AFT1-TAP::HIS CHL4-Myc::TRP1 ctf19Δ::kanMX | This study |
| YKB2283 | MATa his3 leu2 ura3 CTF19-Myc::TRP1 | This study |
| YKB2282 | MATα his3 leu2 ura3 AFT1-TAP::HIS CTF19-Myc::TRP1 | This study |
| YKB2289 | MATa his3 leu2 ura3 AFT1-TAP::HIS CTF19-Myc::TRP1 iml3Δ::kanMX | This study |
| YKB2641 | MATα his3 leu2 ura3 AFT1-TAP::HIS CTF19-Myc::TRP1 chl4Δ::kanMX | This study |
| YKB2766 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 IML3-TAP::HIS | TAP collection |
| YKB2767 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 IML3-TAP::HIS aft1Δ::kanMX | This study |
| YKB2768 | MATα ade2-101 his3-Δ200 lys2-801 leu2-Δ1 ura3-52 trp1-Δ63 CTF19-Myc::kanMX | This study |
| YKB2769 | MATα ade2-101 his3-Δ200 lys2-801 leu2-Δ1 ura3-52 trp1-Δ63 CHL4-Myc::kanMX | This study |
| YKB2770 | MATα his3 leu2 ura3 IML3-TAP::HIS CTF19-Myc::kanMX | This study |
| YKB2772 | MATa his3 leu2 ura3 IML3-TAP::HIS CTF19-Myc::kanMX aft1Δ::kanMX | This study |
| YKB2777 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 trp1-Δ63 IML3-3HA::kanMX | 15 |
| YKB2776 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 trp1- Δ63 IML3-3HA::kanMX CHL4-Myc::TRP1 | 15 |
| YKB2902 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 trp1-Δ63 IML3-3HA::kanMX CHL4-Myc::TRP1 aft1Δ::kanMX | This study |
| YKB236 | MATa pep4Δ::LEU2 ura trp ade his SCC1–6HA::HIS3 | 52 |
| YKB2764 | MATa ura trp ade his SCC1–6HA::HIS3 aft1Δ::kanMX | This study |
| YKB2765 | MATa ura trp his SCC1–6HA::HIS3 iml3Δ::kanMX | This study |
| Strains used in growth assays | ||
| YKB1095 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 aft1Δ::kanMX | This study |
| YKB157 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 trp1-Δ63 scc1-73 | 32 |
| YKB90 | MATa trp1-1 ade2-101 can1-100 leu2-3,112 his3-11,15 ura3 scc2-4 | 32 |
| YKB227 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 smc3-42 | 32 |
| YKB2138 | MATa pep4Δ::G418 ura3-52 leu2-3,112 his3-11,15 bar1 GAL+ eco1-203 | 33 |
| YKB2274 | MATα his3 leu2 ura3 iml3Δ::kanMX | This study |
| YKB2749 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 trp1-Δ63 scc1-73 aft1Δ::kanMX | This study |
| YKB2752 | MATα his3 ura3 scc1-73 iml3Δ::kanMX | This study |
| YKB2753 | MATα trp ade2-101 leu2 his3 ura3 scc2-4 aft1Δ::kanMX | This study |
| YKB2755 | MATα his3 leu2 ura3 scc2-4 iml3Δ::kanMX | This study |
| YKB2757 | MATα ade2 trp1 leu2 his3 ura3 smc3-42 aft1Δ::kanMX | This study |
| YKB2759 | MATα his3 leu2 ura3 smc3-42 iml3Δ::kanMX | This study |
| YKB2761 | MATα ura3-52 leu2 his3 eco1-203 aft1Δ::kanMX | This study |
| YKB2762 | MATα his3 leu2 ura3 eco1-203 iml3Δ::kanMX | This study |
| Strains used in mitotic cohesion assay | ||
| YKB2635 | MATa ura3 trp1 leu2 his3-11 MET-CDC20::URA3 promURA3::tetR::GFP::LEU2, cenIV::tetOx448::URA3 | 19 |
| YKB2636 | MATa ura3 trp1 leu2 his3-11 MET-CDC20::URA3 promURA3::tetR::GFP::LEU2, cenIV::tetOx448::URA3 iml3Δ::kanMX | 19 |
| YKB2637 | MATa ura3 trp1 leu2 his3-11 MET-CDC20::URA3 promURA3::tetR::GFP::LEU2, cenIV::tetOx448::URA3 aft1Δ::kanMX | This study |
| YKB2638 | MATa ura3 trp1 leu2 his3-11 MET-CDC20::URA3 promURA3::tetR::GFP::LEU2, cenIV::tetOx448::URA3 iml3Δ::kanMX6 aft1Δ::TRP1 | This study |
| YKB2903 | MATa ura3 trp1 leu2 his3-11 MET-CDC20::URA3 promURA3::tetR::GFP::LEU2, cenIV::tetOx448::URA3 aft1-TADΔ-Myc::TRP1 | This study |
| Strains used in meiotic cohesion assay | ||
| YKB2589 | MATa/α ho::LYS2 (lys2?) ura3 leu2 his4X trp1::hisG promURA3-TetR-GFP::LEU2 TETOx224-URA3 | 19 |
| YKB2590 | MATa/α ho::LYS2 (lys2?) ura3 leu2 his4X trp1::hisG promURA3-TetR-GFP::LEU2 TETOx224-URA3 aft1Δ::kanMX | This study |
Whole-cell Extract Co-immunoprecipitation
Cells were grown in 200 ml of YPD at 30 °C to mid-log phase (A600 ∼0.6–0.8) and harvested. Cell pellets were resuspended in Tackett extraction buffer (20 mm HEPES, pH 7.4, 0.1% Tween 20, 2 mm MgCl2, 200 mm NaCl, and protease inhibitors (Sigma, P-8215)) and an equal volume of glass beads (BioSpec Products, 11079105). Cells were lysed through vortexing (five 1-min blasts with incubation on ice between vortexing) followed by centrifugation at 3000 rpm for 15 min to isolate whole-cell extract. Samples were normalized by protein concentration and incubated end-over-end for 2 h at 4 °C with magnetic Dynabeads (Dynal, Invitrogen, 143-01) crossed-linked to rabbit immunoglobulin G (IgG) (Chemicon, PP64) or with magnetic protein G Dynabeads (Invitrogen, 100.03D) cross-linked to mouse HA.11 monoclonal antibody (Covance, MMS-101P). Dynabeads were collected with a magnet, washed three times with 1 ml of cold Tackett extraction buffer, and resuspended in 25 μl of modified 1× loading buffer (50 mm Tris, pH 6.8, 2% SDS, 0.1% bromphenol blue, 10% glycerol). Purified and co-purified proteins were eluted from the beads at 65 °C for 10 min and transferred to a new tube. β-Mercaptoethanol was added to a final concentration of 200 mm, and samples were placed at 95 °C for 10 min. Immunoprecipitates were subjected to SDS-PAGE and Western blotting. Primary antibodies used are as follows: anti-TAP (Thermo Scientific, CAB1001, 1:5000), anti-Myc (Roche Applied Science, 11667149001, 1:800), and anti-HA (Covance, MMS-101P, 1:1000). Secondary antibodies were HRP-linked, goat anti-rabbit IgG (Chemicon, AP307P, 1:5000), and goat anti-mouse IgG (Bio-Rad, 170–6516, 1:5000).
Dot Assays and Growth Conditions
Yeast strains were grown in YPD at 25 °C to mid-log phase and then spotted in 10-fold serial dilutions (A600 = 0.1, 0.01, 0.001, and 0.0001) onto YPD plates and incubated at 25, 30, and 33 °C for 2 days before epi-white imaging using the Molecular Imager ChemiDoc XRS System (Bio-Rad). Dot assay experiments were repeated in triplicate using different isolates of each strain.
Chromatin Immunoprecipitation (ChIP)-Multiplex PCR and qPCR3
ChIP was carried out as described previously (15). Immunoprecipitated DNA was amplified using multiplex-PCR with the following primer pairs: CEN3 forward (5′-GCGATCAGCGCCAAACAATATGG) and CEN3 reverse (5′-GAGCAAAACTTCCACCAGTAAACG) were obtained from Ref. 26; FET3 forward (5′-GGTCCCTACAGTACGCTGAG) and FET3 reverse (5′-GGATCGACTGTTTGAGTGCATCC). PCR products were resolved on a 3% agarose gel and visualized with ethidium bromide. For qPCR-ChIP, cells were grown at 25 °C to mid-log phase and arrested as described previously (19). qPCR was performed in a 10-μl EvaGreen (Bio-Rad, 172-5201) reaction using the Bio-Rad MiniOpticon real time PCR system. ChIP enrichment/input values were calculated as described previously (19). Primer sequences were obtained from Ref. 19, and the pericentromere, centromere, and arm primers correspond to primer pairs P2, C1, and A3, respectively. The primer sequences used are as follows: pericentromere forward (5′-ATTGTTTAGAAACGGGAACA) and reverse (5′GTTCAACTCTCTGCATCTCC); centromere forward (5′-ACACGAGCCAGAAATAGTAAC) and reverse (5′-TGATTATAAGCATGTGACCTTT); and arm forward (5′-GAAAGCGACCAGCTAGATTA) and reverse (5′-CAAACGCTTTAACACACAAG).
Cohesion Assays and Microscopy
The mitotic cohesion assay was performed as described previously (19). For the meiotic cohesion assay, sporulation was performed at 30 °C as described previously (27). Cells for both assays were fixed as described previously (28). Microscopy was performed on a Leica DMI6000B fluorescent microscope (Leica Microsystems GmbH, Wetzler Germany), equipped with a Sutter DG4 light source (Sutter Instruments), Ludl emission filter wheel with Chroma band pass emission filters (Ludl Electronic Products Ltd.), and Hamamatsu Orca AG camera (Hamamatsu Photonics, Herrsching am Ammersee, Germany). Images were collected and analyzed using Velocity 4.3.2 Build 23 (PerkinElmer Life Sciences). Analysis was performed on images collapsed into two dimensions using the “extended focus” in Velocity.
RESULTS
Aft1 Interacts with the Kinetochore Protein Iml3
To elucidate the function of Aft1 in chromosome stability, we first sought to identify the individual kinetochore protein or protein subcomplex that interacts with Aft1. As Iml3 was found to interact with Aft1 by yeast two-hybrid (13, 14), we decided to confirm this physical interaction by a secondary method. Co-immunoprecipitation experiments with strains containing TAP-tagged Aft1 and Myc-tagged Iml3, Chl4, or Ctf19 confirmed that Aft1-TAP can co-purify proteins of the Ctf19 subcomplex (Fig. 1, B–D, lanes 4).
To further dissect the Aft1-kinetochore interaction, we systematically created deletion mutant strains based on the established hierarchical dependences of the Ctf19 complex (Fig. 1A) (15). We first investigated whether the Aft1-Ctf19 interaction (Fig. 1B, lane 4) is dependent on Chl4 or Iml3. In a chl4Δ background that abolishes the Iml3-Ctf19 interaction (15), the Aft1-Ctf19 interaction was disrupted (Fig. 1B, lane 5) indicating that this interaction was either through Chl4 or Iml3. In an iml3Δ background that maintains the Chl4-Ctf19 interaction, albeit in a reduced fashion (15, 19), the Aft1-Ctf19 interaction was also disrupted (Fig. 1B, lane 6). These results suggest that Aft1 is co-purifying Chl4 and Ctf19 through Iml3. To confirm this, we next examined the effect of ctf19Δ and iml3Δ mutations on the Aft1-Chl4 interaction (Fig. 1C, lane 4). In a ctf19Δ background that maintains the Iml3-Chl4 complex (15), the Aft1-Chl4 interaction was preserved (Fig. 1C, lane 5) indicating that Aft1 interacts with either Chl4 or Iml3 but not Ctf19. In an iml3Δ background that maintains the Chl4-Ctf19 interaction, Aft1 could not co-immunoprecipitate Chl4 anymore (Fig. 1C, lane 6), indicating that this interaction was through Iml3. Our last experiment looked directly at the consequence of ctf19Δ and chl4Δ mutations on the Aft1-Iml3 interaction (Fig. 1D, lane 4). These results confirmed the findings of the previous two experiments, as we show that in both ctf19Δ and chl4Δ mutant backgrounds Aft1 can still co-immunoprecipitate Iml3 (Fig. 1D, lanes 5 and 6). Therefore, our results confirmed the Aft1-Iml3 two-hybrid interaction and indicated that Aft1 is interacting with kinetochore COMA complex subunits through Iml3.
Iml3 Kinetochore Localization Is Not Dependent on Aft1
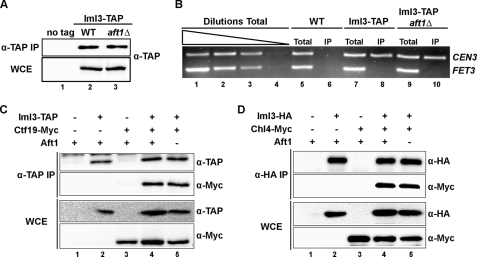
To determine whether Aft1 impacts the localization of Iml3 to the CEN, we performed a chromatin immunoprecipitation (ChIP) assay using an untagged control, Iml3-TAP, and Iml3-TAP aft1Δ strains (Fig. 2A). TAP-tagged Iml3 was immunoprecipitated from formaldehyde cross-linked whole-cell extracts, and co-precipitated DNA was subjected to multiplex PCR with primers specific to the centromere of chromosome 3 (CEN3) and the promoter of FET3 (negative control). Our results indicate that Aft1 does not impact the centromeric localization of Iml3 (Fig. 2B). To further confirm our results, we carried out protein co-immunoprecipitation experiments to investigate if Iml3-Ctf19 and Iml3-Chl4 interactions are dependent on Aft1. Our results demonstrate that the Iml3-Ctf19 and Iml3-Chl4 interactions are not disrupted in an aft1Δ background (Fig. 2, C and D). Taken together, our results indicate that Aft1 is not regulating Iml3 localization to CENs or interaction with COMA proteins.
FIGURE 2.
Aft1 does not impact the kinetochore localization and protein interactions of Iml3. A, presence or absence of Aft1 does not impact the purification of Iml3-TAP. Western blot analysis of Iml3-TAP purified by ChIP (IP) and whole-cell extract (WCE). B, Iml3 localization to the centromere is not affected by Aft1. Traditional ChIP performed using untagged, Iml3-TAP, and Iml3-TAP aft1Δ strains. Total or immunoprecipitated DNA was subjected to multiplex PCR amplification using primers specific to the centromere of chromosome 3 (CEN3) and the negative control FET3. C, Iml3-Ctf19 co-purification is not affected by Aft1. Anti-TAP immunoprecipitations were performed with strains containing different combinations of TAP-tagged Iml3 and Myc-tagged Ctf19 in the presence or absence of Aft1. D, Iml3-Chl4 interaction is not affected by Aft1. Anti-HA immunoprecipitations were performed with strains containing different combinations of HA-tagged Iml3 and Myc-tagged Chl4 in the presence or absence of Aft1. For all immunoprecipitations, 50% of the eluate was loaded in the TAP or HA blot, whereas 50% of the eluate was loaded in the Myc-blot.
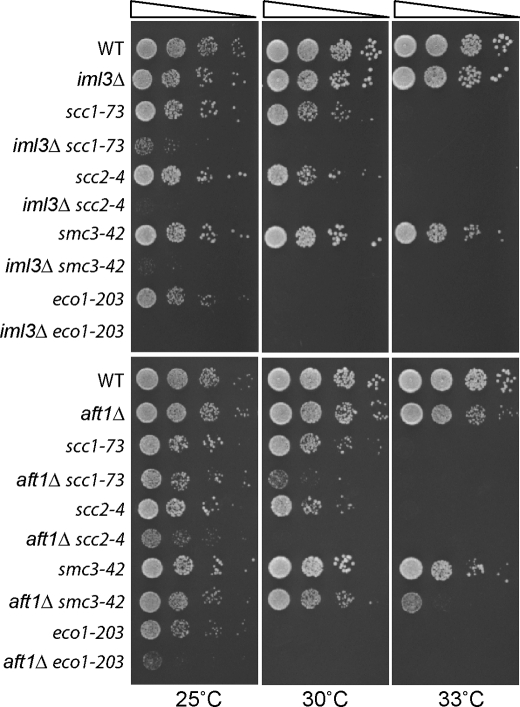
aft1Δ and iml3Δ Display Similar Synthetic Genetic Interactions with Cohesion Mutants
Given that Iml3 has been implicated to function in the cohesion pathway (5, 19, 29, 30) and that genetic interactions predict functional relationships (31), we wanted to determine whether aft1Δ and iml3Δ mutants display synthetic genetic interactions with temperature-sensitive mutants of the sister chromatid cohesion pathway: scc1-73, scc2-4, smc3-42 (32), and eco1-203 (33). Scc1 and Smc3 are part of the tripartite cohesin ring that physically links sister chromatids until the onset of anaphase. Scc2 is part of the Scc2-Scc4 cohesin loader complex, whereas Eco1 functions through acetylation of Smc3 to establish sister chromatid cohesion (reviewed in Ref. 18). As expected, we found that iml3Δ mutants display negative genetic interactions with all the temperature-sensitive cohesion mutants at 25 °C. Similarly, we found that aft1Δ in combination with the cohesion mutants also resulted in synthetic genetic interactions; however, the strength of the interactions varied by temperature and was not as pronounced as with iml3Δ (Fig. 3). This suggests that like Iml3, Aft1 may have a role in sister chromatid cohesion.
FIGURE 3.
aft1Δ and iml3Δ mutants display negative genetic interactions with cohesion mutants. Cells were plated in 10-fold serial dilutions (A600 = 0.1, 0.01, 0.001, and 0.0001) onto YPD plates and incubated for 2 days at the indicated temperatures.
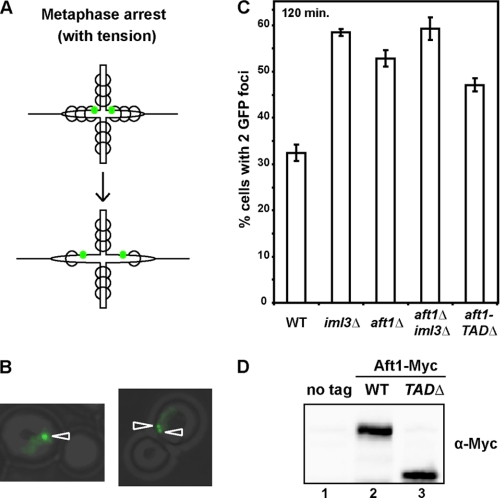
Defective Mitotic Pericentromeric Cohesion in aft1Δ Mutants
It has been shown previously that Iml3 and other Ctf19 complex proteins exhibit defects in resisting microtubule pulling forces (19) and increased spindle length in metaphase (23). To determine whether Aft1 displays similar defects, we used strains that express TetR-GFP and have tetO arrays integrated 2.4 kb from the CEN of chromosome 4 (+2.4CEN4-GFP), and we asked if sister chromatids are more frequently separated in a metaphase arrest (Fig. 4A). Wild-type, iml3Δ, aft1Δ, and aft1Δiml3Δ cells under the control of a methionine-repressible promoter (MET-CDC20) were released from a G1 α-factor block into methionine-rich media. This induces a metaphase arrest through depletion of Cdc20 (required for the metaphase to anaphase transition) (34). Sister chromatids in this metaphase arrest remain under tension from the microtubule pulling forces (Fig. 4A), and the separation of sister pericentric regions were scored by counting the number of cells that have 1 or 2 GFP foci (Fig. 4B). Consistent with previous studies (19), our results show that ∼30% of wild-type cells exhibit GFP dot separation, and this increases to 60% in iml3Δ cells that exhibit defects in counter-balancing spindle forces (Fig. 4C). Approximately 55% of aft1Δ cells have separated GFP foci indicating that Aft1, like Iml3, is required for establishing pericentric cohesion in mitosis. As aft1Δiml3Δ cells did not display additive defects, it suggests that Aft1 and Iml3 function together to establish pericentric chromatin that can balance mitotic forces.
FIGURE 4.
aft1Δ mutants exhibit pericentromeric cohesion defects in mitosis. A, schematic showing the separation of GFP-marked chromosomes 2.4 kb to the right of CEN4 in a metaphase arrest while maintaining microtubule tension in a strain with adequate pericentric cohesin (top panel) and decreased pericentric cohesin (bottom panel). Spindle axis is along the horizontal plane. GFP is represented by green dots. Cohesin is represented by circles. B, representative images of cells at 120 min after release from G1 showing one or two GFP foci (left and right panel, respectively). C, sister centromeres are more frequently separated in aft1Δ, iml3Δ, and aft1-TADΔ mutants. Wild-type (WT), aft1Δ, iml3Δ, and aft1-TADΔ cells that have GFP-labeled chromosome 2.4 kb to the right of CEN4 and MET-CDC20 were arrested in G1 with α-factor and released into media containing methionine to induce a metaphase arrest while maintaining tension between sister chromatids. Metaphase arrest was confirmed by bud count, and the frequency of GFP separation was assayed by microscopy. Results are the mean of three experiments in which 200 cells were scored. D, removal of the transactivation domain does not impact protein levels. Western analysis of whole-cell extract from wild-type (no tag), Aft1-Myc, and aft1-TADΔ-Myc cells.
We were then interested to determine which domain of Aft1 is responsible for this phenotype. Previous studies have mapped the transactivation domain of Aft1 to the C-terminal region and within amino acids 413–690 (35), and the N-terminal region contains the DNA binding domain (36). As such, we deleted the transactivation domain of Aft1, or amino acids 413–690, to create the truncated aft1-TADΔ (Fig. 4D) and asked whether we observed the same phenotype as aft1Δ cells. Our results show that the aft1-TADΔ strain displays defects in resisting microtubule tension similar to aft1Δ cells (Fig. 4C) indicating that the transactivation domain of Aft1 is contributing to regulate spindle length dynamics.
Aft1 Is Required for Faithful Chromosome Segregation during Meiosis
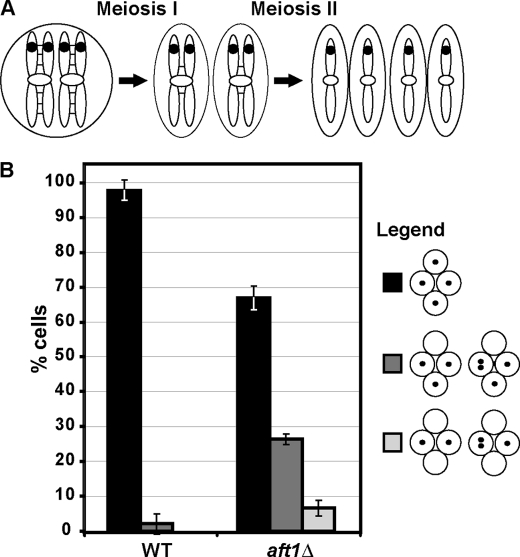
After the first meiotic division in which homologous chromosomes are segregated, sister chromatids lack all arm cohesion and depend only on pericentromeric cohesion (reviewed in Ref. 37). As such, defects in pericentromeric cohesin results in chromosome nondisjunction following the second meiotic division. Previous reports have shown that iml3Δ mutants display defects in chromosome segregation in meiosis (5, 19, 29, 30), and genome-wide screens identified aft1Δ as having mild chromosome segregation defects during meiosis (5). To confirm the role of Aft1 in meiotic chromosome segregation, we monitored the fate of GFP-labeled chromosomes in wild-type and aft1Δ diploids after sporulation (Fig. 5A). The strains used have GFP labels located at URA3 (38.4 kb from CEN5) on both copies of chromosome 5 (tetR-GFP::LEU2 URA3::tetOx224). Using the same strains, a previous report showed the following GFP segregation pattern for iml3Δ mutants as follows: ∼60% had GFP dots in all four spores, ∼32% in three of four spores, and ∼8% in two of four spores (19). Our results indicate that aft1Δ mutants display GFP dot segregation patterns similar to iml3Δ mutants after meiosis (Fig. 5B). We demonstrate that aft1Δ mutants have the following GFP segregation pattern: ∼65% had GFP dots in all four spores, ∼27% in three of four spores, and ∼8% in two of four spores. Therefore, Aft1, like Iml3, is required for proper chromosome segregation during meiosis, suggesting a role for Aft1 in promoting pericentric cohesin like Iml3.
FIGURE 5.
Aft1 is required for proper chromosome segregation during meiosis. A, schematic showing the segregation of chromosome 5 labeled with GFP. Sister chromatid cohesion is represented by black lines between sister chromatids. GFP is represented by black dot. B, aft1Δ cells display meiotic chromosome segregation defects. Wild-type (WT) and aft1Δ diploids that have GFP-labeled chromosome 5 (tetR-GFP::LEU2 URA3::tetOx224) at URA3 (38.4 kb from the centromere) were induced to sporulate at 30 °C. GFP dot segregation patterns in tetra-nucleate cells were scored and represented as percentages. Results are the mean of three experiments in which 100 tetra-nucleate cells were scored for GFP segregation patterns.
Aft1 Is Required for Increased Cohesin at the Pericentromere and Centromere
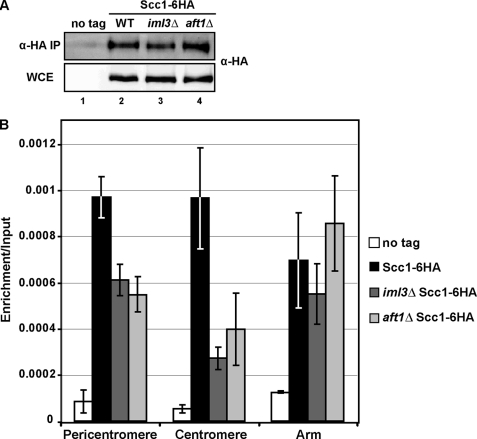
Increased separation of sister chromatids at the pericentromere are indicative of cohesion defects that are required to resist the microtubule pulling forces (19). To assess whether Aft1 is required for increased cohesin binding at the pericentromere, we employed ChIP followed by real time qPCR to examine the enrichment level of the cohesin subunit Scc1-HA at three different sites on chromosome 4 that had previously been shown to be enriched for cohesin as follows: along the chromosomal arm, pericentromere, and centromere regions (19). Wild-type, iml3Δ, and aft1Δ cells that express HA-tagged Scc1 along with an untagged control strain were arrested in metaphase following treatment with the microtubule-depolymerizing drugs nocodazole and benomyl. This generated a metaphase arrest whereby sister chromatids are not under tension from the microtubule-pulling forces. HA-tagged Scc1 was then immunoprecipitated from formaldehyde cross-linked extracts (Fig. 6A) upon which the co-precipitated DNA was subjected to qPCR analysis, and enrichment/input values were calculated as described previously (19). Our results indicate that there is reduced Scc1 enrichment at the pericentromere and centromere regions in iml3Δ and aft1Δ mutants as compared with wild-type cells (Fig. 6B). In contrast, Scc1 levels were comparable between wild-type, iml3Δ, and aft1Δ cells at the chromosomal arm location. Therefore, Aft1, like Iml3, plays a role in promoting cohesin association at the centromere and pericentromere.
FIGURE 6.
Levels of Scc1–6HA are reduced at the pericentromere and centromere in aft1Δ mutants. A, presence or absence of Aft1 does not impact the amount of Scc1–6HA purified by ChIP. Western blot analysis of Scc1–6HA purified by ChIP (IP) and whole-cell extract (WCE) in wild-type, aft1Δ, and iml3Δ mutants. B, analysis of Scc1–6HA association in cells arrested in metaphase of mitosis in the absence of microtubules. Wild-type (no tag), SCC1–6HA, iml3Δ SCC1–6HA, and aft1Δ SCC1–6HA cells were arrested in metaphase in the presence of microtubule-depolymerizing drugs nocodazole and benomyl. Metaphase arrest was confirmed by bud count. qPCR analysis of Scc1–6HA levels were performed at three different regions of chromosome 4, and results are the mean of three independent experiments; error bar indicates 1 S.D.
DISCUSSION
Several genome-wide studies identified Aft1 as a potential regulator in chromosome stability (4, 5, 7, 13, 14). In this study, we demonstrate that Aft1 interacts with kinetochore proteins Ctf19 and Chl4 through Iml3, and like these members of the Ctf19 complex, we found that Aft1 contributes to the enrichment of cohesin at the pericentromere. The mechanism by which Iml3 and other Ctf19 complex members promote cohesin enhancement at the centromere and pericentromere remains unknown. It is speculated that the Ctf19 complex enriches the Scc2-Scc4 cohesin loader complex at the centromere, which in turn loads the cohesin complex that subsequently spreads bidirectionally throughout the pericentromere (19–21). As Iml3 is the most peripheral Ctf19 complex subunit (15), and pericentromeric cohesion defects of Ctf19 complex mutants are not additive (19), Iml3 is likely the key protein controlling this process. The pericentromeric separation of mitotic sister chromatids displayed by aft1Δiml3Δ mutant cells is comparable with the single mutants (Fig. 4C). As Aft1 does not impact the interaction of Iml3 with the kinetochore (Fig. 2), it suggests that one role Iml3 may be playing is to recruit Aft1 to the kinetochore, which in turn aids in the recruitment of pericentric cohesin. However, as the strength of the genetic interactions of aft1Δ mutants with the cohesion pathway mutants is not as pronounced as iml3Δ mutants (Fig. 3), it suggests that Iml3 plays additional roles in the cohesion pathway not shared by Aft1.
Although Aft1 has not been linked to the transcription of genes implicated in chromosome stability or benomyl resistance (4), we cannot eliminate the possibility that part of the role Aft1 plays in chromosome stability is mediated through transcription. With that said, the physical interaction between Aft1 and Iml3 (Fig. 1) (13, 14) and the co-localization of Aft1 with kinetochore proteins (6) strongly argues for a role for Aft1 directly at the kinetochore. Aft1 has not been identified in the numerous proteomic studies of the kinetochore (12, 16) but only via two-hybrid screens (13, 14) suggesting that Aft1 interaction with the kinetochore may be transient (38). Indeed, we may have been able to detect the Aft1-Iml3 interaction because our purifications utilized low speed centrifugation, which has been shown to increase the solubility of chromatin-associated proteins (39).
What are the possible roles for Aft1 at the kinetochore? One possibility is that Aft1 is physically interacting with and recruiting members of the cohesin complex or the Scc2-Scc4 loader complex to the centromere; however, like Iml3, no physical interaction between Aft1 and these subunits has been reported. An alternative possibility is that the kinetochore is harnessing the ability of transcription factors to recruit chromatin remodelers or modifiers to the centromere where they specifically position or add epigenetic marks to pericentromeric histones which in turn recruit Scc2-Scc4. There is precedent for this idea as during DNA damage repair cohesin is enriched at sites flanking the DNA break in a manner dependent on histone H2A phosphorylation (40, 41). Indeed, histone modifications dependent on a functional kinetochore have been reported at the centromere, and these modifications contribute to chromosome segregation (42). Also, establishment of cohesion has been linked to numerous chromatin regulators and histone modifications (43–47), and chromatin-remodeling complexes were also found to interact with kinetochore proteins and localize to the centromere (13, 48, 49). Could Aft1 be recruiting such factors to the centromere? The transactivation domain of Aft1, which is believed to recruit chromatin remodelers and modifiers to iron-regulon promoters (35), is required for resisting microtubule pulling forces (Fig. 4C). Furthermore, affinity-capture mass spectrometry studies have demonstrated Aft1 co-purifying with protein subunits of the SWI/SNF chromatin remodeler (39) and the lysine acetyltransferases SAGA (50) and NuA4 (39); all three have been implicated in chromosome stability and/or cohesion defects (5, 26, 49). Exactly how Aft1 contributes to the structure and function of the pericentric chromatin spring will require additional study. However, as numerous other transcription factors have been shown to interact with kinetochore proteins (13), the role of transcription factors in pericentromeric chromatin may not be limited to Aft1 but rather to a general mechanism that aids in the establishment of the unique chromatin environment of the pericentromere.
Acknowledgments
We thank Adele Marston for kindly providing strains, Alexander Blais for advice on ChIP-qPCR, and members of Baetz laboratory for critical reading of the manuscript.
This work was supported in part by a Natural Sciences and Engineering Research Council of Canada Discovery grant (to K. B.).
- qPCR
- quantitative PCR.
REFERENCES
- 1. Philpott C. C., Protchenko O. (2008) Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rutherford J. C., Jaron S., Winge D. R. (2003) Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278, 27636–27643 [DOI] [PubMed] [Google Scholar]
- 3. Yamaguchi-Iwai Y., Stearman R., Dancis A., Klausner R. D. (1996) Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15, 3377–3384 [PMC free article] [PubMed] [Google Scholar]
- 4. Berthelet S., Usher J., Shulist K., Hamza A., Maltez N., Johnston A., Fong Y., Harris L. J., Baetz K. (2010) Functional genomics analysis of the Saccharomyces cerevisiae iron-responsive transcription factor Aft1 reveals iron-independent functions. Genetics 185, 1111–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marston A. L., Tham W. H., Shah H., Amon A. (2004) A genome-wide screen identifies genes required for centromeric cohesion. Science 303, 1367–1370 [DOI] [PubMed] [Google Scholar]
- 6. Measday V., Baetz K., Guzzo J., Yuen K., Kwok T., Sheikh B., Ding H., Ueta R., Hoac T., Cheng B., Pot I., Tong A., Yamaguchi-Iwai Y., Boone C., Hieter P., Andrews B. (2005) Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 102, 13956–13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuen K. W., Warren C. D., Chen O., Kwok T., Hieter P., Spencer F. A. (2007) Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 3925–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee W., St Onge R. P., Proctor M., Flaherty P., Jordan M. I., Arkin A. P., Davis R. W., Nislow C., Giaever G. (2005) Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 1, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubacq C., Chevalier A., Courbeyrette R., Petat C., Gidrol X., Mann C. (2006) Role of the iron mobilization and oxidative stress regulons in the genomic response of yeast to hydroxyurea. Mol. Genet. Genomics 275, 114–124 [DOI] [PubMed] [Google Scholar]
- 10. Philpott C. C., Rashford J., Yamaguchi-Iwai Y., Rouault T. A., Dancis A., Klausner R. D. (1998) Cell-cycle arrest and inhibition of G1 cyclin translation by iron in AFT1–1(up) yeast. EMBO J. 17, 5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sora S., Lucchini G., Magni G. E. (1982) Meiotic Diploid Progeny and Meiotic Nondisjunction in Saccharomyces cerevisiae. Genetics 101, 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Westermann S., Drubin D. G., Barnes G. (2007) Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76, 563–591 [DOI] [PubMed] [Google Scholar]
- 13. Wong J., Nakajima Y., Westermann S., Shang C., Kang J. S., Goodner C., Houshmand P., Fields S., Chan C. S., Drubin D., Barnes G., Hazbun T. (2007) A protein interaction map of the mitotic spindle. Mol. Biol. Cell 18, 3800–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) High quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pot I., Measday V., Snydsman B., Cagney G., Fields S., Davis T. N., Muller E. G., Hieter P. (2003) Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 14, 460–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Wulf P., McAinsh A. D., Sorger P. K. (2003) Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17, 2902–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ocampo-Hafalla M. T., Uhlmann F. (2011) Cohesin loading and sliding. J. Cell Sci. 124, 685–691 [DOI] [PubMed] [Google Scholar]
- 18. Nasmyth K., Haering C. H. (2009) Cohesin. Its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 [DOI] [PubMed] [Google Scholar]
- 19. Fernius J., Marston A. L. (2009) Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 5, e1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ng T. M., Waples W. G., Lavoie B. D., Biggins S. (2009) Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell 20, 3818–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eckert C. A., Gravdahl D. J., Megee P. C. (2007) The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 21, 278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. (2008) Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stephens A. D., Haase J., Vicci L., Taylor R. M., 2nd, Bloom K. (2011) Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J. Cell Biol. 193, 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verdaasdonk J. S., Bloom K. (2011) Centromeres. Unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 12, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 26. Krogan N. J., Baetz K., Keogh M. C., Datta N., Sawa C., Kwok T. C., Thompson N. J., Davey M. G., Pootoolal J., Hughes T. R., Emili A., Buratowski S., Hieter P., Greenblatt J. F. (2004) Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. U.S.A. 101, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marston A. L., Lee B. H., Amon A. (2003) The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell 4, 711–726 [DOI] [PubMed] [Google Scholar]
- 28. Klein F., Mahr P., Galova M., Buonomo S. B., Michaelis C., Nairz K., Nasmyth K. (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 [DOI] [PubMed] [Google Scholar]
- 29. Ghosh S. K., Sau S., Lahiri S., Lohia A., Sinha P. (2004) The Iml3 protein of the budding yeast is required for the prevention of precocious sister chromatid separation in meiosis I and for sister chromatid disjunction in meiosis II. Curr. Genet. 46, 82–91 [DOI] [PubMed] [Google Scholar]
- 30. Kiburz B. M., Reynolds D. B., Megee P. C., Marston A. L., Lee B. H., Lee T. I., Levine S. S., Young R. A., Amon A. (2005) The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 19, 3017–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 32. Michaelis C., Ciosk R., Nasmyth K. (1997) Cohesins. Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45 [DOI] [PubMed] [Google Scholar]
- 33. Skibbens R. V., Corson L. B., Koshland D., Hieter P. (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uhlmann F., Wernic D., Poupart M. A., Koonin E. V., Nasmyth K. (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375–386 [DOI] [PubMed] [Google Scholar]
- 35. Yamaguchi-Iwai Y., Ueta R., Fukunaka A., Sasaki R. (2002) Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277, 18914–18918 [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi-Iwai Y., Dancis A., Klausner R. D. (1995) AFT1. A mediator of iron-regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14, 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marston A. L., Amon A. (2004) Meiosis. Cell cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5, 983–997 [DOI] [PubMed] [Google Scholar]
- 38. Brückner A., Polge C., Lentze N., Auerbach D., Schlattner U. (2009) Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lambert J. P., Fillingham J., Siahbazi M., Greenblatt J., Baetz K., Figeys D. (2010) Defining the budding yeast chromatin-associated interactome. Mol. Syst. Biol. 6, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J. E., Koshland D. (2004) DNA damage response pathway uses histone modification to assemble a double strand break-specific cohesin domain. Mol. Cell 16, 991–1002 [DOI] [PubMed] [Google Scholar]
- 41. Ström L., Lindroos H. B., Shirahige K., Sjögren C. (2004) Postreplicative recruitment of cohesin to double strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 42. Choy J. S., Acuña R., Au W. C., Basrai M. A. (2011) A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics 189, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baetz K. K., Krogan N. J., Emili A., Greenblatt J., Hieter P. (2004) The ctf13–30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24, 1232–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hakimi M. A., Bochar D. A., Schmiesing J. A., Dong Y., Barak O. G., Speicher D. W., Yokomori K., Shiekhattar R. (2002) A chromatin remodeling complex that loads cohesin onto human chromosomes. Nature 418, 994–998 [DOI] [PubMed] [Google Scholar]
- 45. Huang J., Hsu J. M., Laurent B. C. (2004) The RSC nucleosome-remodeling complex is required for Cohesin's association with chromosome arms. Mol. Cell 13, 739–750 [DOI] [PubMed] [Google Scholar]
- 46. Ogiwara H., Enomoto T., Seki M. (2007) The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle 6, 1090–1095 [DOI] [PubMed] [Google Scholar]
- 47. Oum J. H., Seong C., Kwon Y., Ji J. H., Sid A., Ramakrishnan S., Ira G., Malkova A., Sung P., Lee S. E., Shim E. Y. (2011) RSC facilitates Rad59-dependent homologous recombination between sister chromatids by promoting cohesin loading at DNA double strand breaks. Mol. Cell. Biol. 31, 3924–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu J. M., Huang J., Meluh P. B., Laurent B. C. (2003) The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 23, 3202–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gkikopoulos T., Singh V., Tsui K., Awad S., Renshaw M. J., Scholfield P., Barton G. J., Nislow C., Tanaka T. U., Owen-Hughes T. (2011) The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 30, 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee K. K., Sardiu M. E., Swanson S. K., Gilmore J. M., Torok M., Grant P. A., Florens L., Workman J. L., Washburn M. P. (2011) Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]