Background: Plant resistance proteins are molecular switches that respond to plant pathogens.
Results: A subset of resistance proteins has a nucleotide phosphatase activity.
Conclusion: The modes of signaling behavior in resistance proteins are more diverse than previously suspected.
Significance: Novel biochemical activities might have evolved in land plants to resist pathogen attack.
Keywords: Nucleoside, Nucleotide, Phosphatase, Plant Biochemistry, Recombinant Protein Expression, Resistance Protein
Abstract
Plant resistance proteins (R-proteins) are key components of the plant immune system activated in response to a plethora of different pathogens. R-proteins are P-loop NTPase superfamily members, and current models describe their main function as ATPases in defense signaling pathways. Here we show that a subset of R-proteins have evolved a new function to combat pathogen infection. This subset of R-proteins possesses a nucleotide phosphatase activity in the nucleotide-binding domain. Related R-proteins that fall in the same phylogenetic clade all show the same nucleotide phosphatase activity indicating a conserved function within at least a subset of R-proteins. R-protein nucleotide phosphatases catalyze the production of nucleoside from nucleotide with the nucleotide monophosphate as the preferred substrate. Mutation of conserved catalytic residues substantially reduced activity consistent with the biochemistry of P-loop NTPases. Kinetic analysis, analytical gel filtration, and chemical cross-linking demonstrated that the nucleotide-binding domain was active as a multimer. Nuclear magnetic resonance and nucleotide analogues identified the terminal phosphate bond as the target of a reaction that utilized a metal-mediated nucleophilic attack by water on the phosphoester. In conclusion, we have identified a group of R-proteins with a unique function. This biochemical activity appears to have co-evolved with plants in signaling pathways designed to resist pathogen attack.
Introduction
Plants possess an innate immune system in which individual cells autonomously mount a defensive response to pathogen attack (1). There are two parallel mechanisms that mediate this defensive response; (i) Plasma membrane receptors (MAMP receptors)2 that recognize broadly conserved microbial molecules and induce a response capable of protecting against colonization by most microbial species, (ii) Resistance proteins (R-proteins), that have evolved under a selection pressure to defend against specific pathogen proteins (designated Avr proteins) that have arisen to elude MAMP receptor mediated immunity. Knowledge of the activity of components of these signaling pathways is crucial to understand how the plant immune system functions and how it can be modified for crop protection.
R-proteins are the most numerous members of the NB-ARC family of proteins that is a subset of the larger group of highly diverse P-loop NTPases. The NB-ARC family also includes the mammalian pro-apoptotic regulator Apaf-1 and its Caenorhabditis elegans homologue CED-4 (2, 3). R-proteins are components of the innate immune system and activate a defense response on detection, whether direct or indirect, of pathogen produced Avr proteins (4). The P-loop containing NBD and associated tandem ARC motifs of R-proteins are proposed to function as an ATP-hydrolyzing switch regulating downstream signaling events (5). R-proteins are under a constant evolutionary pressure to adapt to co-evolving pathogens leading to considerable amino acid polymorphism and new detection capabilities (6). Here we describe the first report of the production of key R-protein switch domains as soluble and active homogenous recombinant proteins. We use these proteins as tools to demonstrate a new, and unexpected, biochemical activity in the NBD of plant R-proteins, a nucleotide phosphatase activity that might have co-evolved with land plants to integrate into signaling pathways that protect plants from pathogen invasion.
Current research supports the role of the NBD of NB-ARC proteins as an NTPase activated through a structural switch. For example, in it quiescent state, the pro-apoptotic mammalian Apaf-1 protein binds (d)ATP but on activation by cytochrome c hydrolyzes the nucleotide to (d)ADP. Exchange of (d)ADP for (d)ATP permits formation of the apoptosome and initiation of apoptosis through recruitment of caspase-9 (7). A refolded preparation of the CC-NB-ARC domain of the I-2 R-protein of tomato was shown to bind ATP and had an ATPase activity (8). The importance of the ATPase activity was confirmed in further work in which mutations of I-2 that activate a pathogen-independent hypersensitive response in planta are compromised in their ATPase activity in vitro (9).
The identity of the bound nucleotide is not the sole determinant maintaining R-proteins in an inactive state. Parts of the LRR and the ARC2 subdomains are also essential for autoinhibition of the NBD. Hence, R-protein activation was proposed to involve a controlled change in R-protein interdomain interactions (10, 11). Together, this has led to the formulation of a central hypothesis for R-protein activation where ADP bound to the NBD represents the “off” state. A conformational change and subsequent nucleotide exchange for ATP, bought about by effecter recognition, switches the R-protein into the “on” state. ATP hydrolysis returns the R-protein to the “off” state (5). A key role for ADP binding and nucleotide exchange in the NBD in the maintenance of an autoinhibited state is supported by work on the comparable mechanism in Apaf-1 (12, 13). Work on multiple related enzymes from different kingdoms therefore seems to support a general principle of R-proteins being strict ATPases.
R-proteins are under an evolutionary pressure caused by Avr proteins constantly evolving to enable pathogens to circumvent activation of immune responses. R-protein domains have been demonstrated to be under considerable diversifying selection to maintain a high amino acid variability to allow the evolution of new specificities (1). Here we demonstrate a new signaling specificity in the NBD of R-proteins. We present the completely unsuspected finding that a subset of NBDs from rice, maize, and Arabidopsis R-proteins are nucleotide phosphatases. This finding demonstrates that the potential signaling mechanisms available to R-proteins could be more diverse than previously suspected.
EXPERIMENTAL PROCEDURES
Plasmids
DNA corresponding to amino acids 197–334 of Os02g_25900 of Oryza sativa ssp. japonica, amino acids 177–519 of Rpm1 of Arabidopsis thaliana, and amino acids 178–505 of PSiP of Zea mays were isolated by PCR with specific primers. Mutant constructs were generated by site-directed mutagenesis. PCR product for Os02g_25900 was cloned into the PacI and SbfI sites of pETStrp3 and fitted with an N-terminal MASWSHPQFEKGLINH tag for affinity purification of recombinant protein (14). PCR product for amino acids 177–519 of Rpm1 and 178–505 of PSiP were cloned into the PacI and XhoI sites of pETStrp3 and fitted with N-terminal MASWSHPQFEKGLINH tags for affinity purification of recombinant protein. PSiP was also fitted with a C-terminal LEHHHHHH tag. Primer sequences are provided in supplemental Table S1.
Protein Expression and Purification
pETStrp3-Os02g_25900197–334 (R1-NB), pETStrp3-Rpm1177–519 (Rpm1-NBARC), and pETStrp3-PsiP178–505 (PSiP-NBARC) were expressed in Escherichia coli Tuner(DE3)pRosetta at 22 °C for 16 h with 100 μm isopropyl-β-d-thiogalactoside. Pelleted cells were washed with 50 mm Tris/HCl (pH 7.5)/1 mm EDTA, resuspended in 50 mm Tris/HCl (pH 7.5)/1.5 M NaCl/2 mm MgCl2, lysed by sonication (150 s), and protein was purified from the supernatant with Strep-Tactin Superflow (IBA). Eluted protein was dialyzed into 20 mm Tris/HCl (pH 7.5)/50 mm NaCl/2 mm MgCl2, loaded onto a MonoQ anion exchange column (Amersham Biosciences), and eluted using a 50 mm to 1 m NaCl gradient. Protein was sensitive to freeze-thaw and was therefore aliquoted into single use fractions and stored in 20% (v/v) glycerol at −150 °C. Protein molecular weight was determined by electrospray ionization in positive ion mode using linear quadruple ion trap-Fourier transform mass spectrometry. The sample was pre-cleaned using a 5–95% acetonitrile gradient.
Nucleotide Phosphatase Assay
Nucleotide phosphatase assays were performed in a final volume of 5 μl and typically contained 50 mm 1,3-bis(tris(hydroxymethyl)methylamino)propane pH 7.5, 30 mm MgCl2, nucleotide substrate, and protein unless indicated otherwise. Reactions were spiked with 2,8-3H-labeled adenine nucleotides for quantitative biochemistry. Assays were adjusted to maintain substrate utilization at less than 15%. Kinetic constants were determined over a concentration range of substrate (Mg2+-ADP) of 0.01–5 mm. All error bars correspond to the standard error of the mean. If absent, error bars are smaller than the symbol used to depict the data point.
Chemical Cross-linking
A 100:1 molar ratio mix of 3,3′-Dithiobis(sulfosuccinimidylpropionate) to protein was incubated for 30 min at room temperature before analysis by SDS-PAGE. Reactions were reduced by boiling in 25 mm dithiothreitol for 15 min.
Thin Layer Chromatography
A portion of a nucleotide phosphatase reaction was spotted onto a silica TLC plate with adenosine spotted on top after drying to act as marker and carrier. The plates were run in 9:1 isopropanol: 33% (v/v) aqueous ammonia. After drying, spots were visualized at 256 nm and excised for scintillation counting.
High Pressure Liquid Chromatography
Nucleotides were separated by reverse phase HPLC in a gradient of 80% buffer A (50 mm KH2PO4, 5 mm tetrabutylammonium bisulfate, pH 6.0)/20% buffer B (30% methanol) run over 9 min to 80% buffer B/20% buffer A. Eluate was analyzed by a photodiode array detector (190–500 nm) followed by mass spectrometry on a Waters Q-TOF Premier mass spectrometer operating in positive electrospray mode, externally calibrated with sodium formate and adjusted for drift using a lock spray of leucine enkephalin. Ions of interest were fragmented using a collision cell with argon using an energy gradient of 10 eV to 35 eV.
Nuclear Magnetic Resonance
31P NMR spectra were recorded on a Varian VNMRS 700 system with an observe frequency of 283.257 MHz. Spectra were recorded with a 45° pulse angle, proton decoupling during acquisition, and 3 s relaxation delay. 1024 scans were recorded, and spectra were zero-filled and Fourier transformed with 0.5 Hz line broadening. Spectra were deconvoluted with Varian VNMR software.
Comparative Modeling
The homology model of R1-NB was calculated using MODELLER (15). Amino acids 197–334 of R1-NB was used in the most exhaustive search for similar sequences and resulted in a top hit with 21% sequence identity with part of the crystal structure of Apaf-1 (PDB code 1Z6T) (13). Models were analyzed and least-square superpositions were performed with COOT (16).
RESULTS
Two distinct biochemical activities have previously been reported for NB-ARC family proteins in plants. The I-2 and Mi-1 R-proteins of tomato are ATPases whose signaling capability is proposed to be determined by a structural state corresponding to the bound nucleotide (8). The Pollen Signaling Protein (PSiP) of maize has a domain structure similar to R-proteins and has been proposed as an AC (17). These findings suggest that the modes of biochemical activity among this class of proteins might be more diverse than previously suspected and we chose to investigate this intriguing possibility through an in depth analysis of NBD biochemistry.
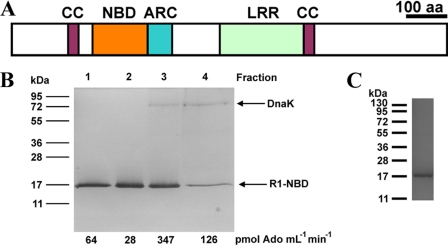
A purified recombinant protein was required to investigate the biochemistry of plant R-proteins but a natively soluble active plant recombinant NBD has never been reported. We therefore tested a range of R-protein NBDs for expression of soluble recombinant protein in E. coli. R-protein NBDs tested included those from ORFs At3g14460, At3g14470, and At3g50950 of A. thaliana, AAX89832 of Glycine max, CAD45035 of Hordeum vulgare, AAX61322 of Phaseolus vulgaris, and Os09g_13820 of Oryza sativa. Among the proteins tested was Os02g_25900, an 809 amino acid orphan R-protein of rice carrying an ∼200 amino acid N-terminal extension. This N-terminal extension contains a CC domain, a domain commonly associated with the N terminus of R-proteins (Fig. 1A) (5). Amino acids 197–334 of Os02g_25900, corresponding to the NBD (hereafter called R1-NB), was expressed at relatively low levels (∼10 μg of purified protein/liter bacterial culture) and co-purified with a single contaminating protein after affinity purification. MALDI-TOF analysis of tryptic digests identified this protein as the Hsp70-family member DnaK, a chaperone involved in protein folding (Fig. 1B). DnaK could be removed through a combination of high salt treatment and anion exchange chromatography to give the homogenous R1-NB protein (Fig. 1C).
FIGURE 1.
Expression of a R-protein NBD. A, protein domains of Os02g_25900. CC, coiled coil domain; NBD, nucleotide binding domain; ARC, Apaf-1, R-protein, CED-4 domain, LRR, leucine-rich repeat. B, analysis of protein co-purifying with R1-NB (SDS/PAGE analysis and Coomassie Blue staining). 10 μl aliquots of protein from successive 1 ml of anion exchange column fractions were assayed for nucleotidase activity using ADP as substrate and applied to an SDS-PAGE gel. Molecular mass standards (in kDa) are indicated. Eluted proteins were identified by trypsin digest and MALDI-TOF. C, purification of recombinant R1-NB (SDS/PAGE analysis and Coomassie Blue staining). A 1.5 μg portion of protein was applied and molecular mass standards (in kDa) are indicated.
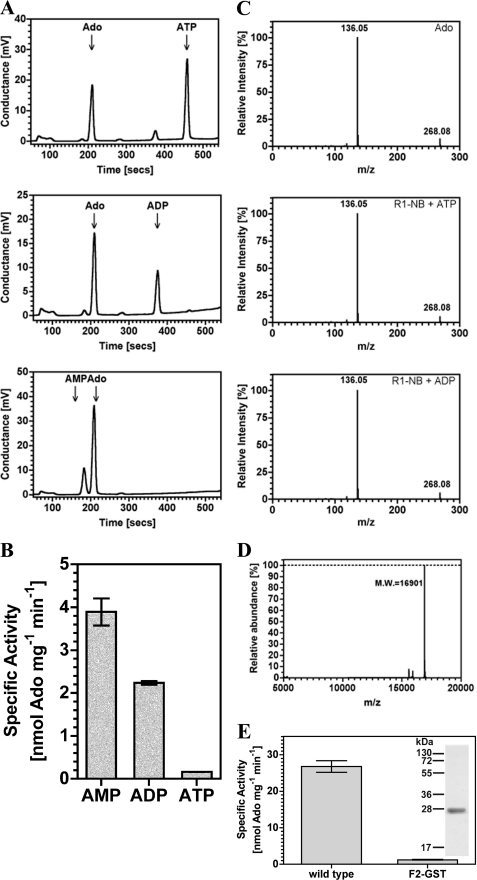
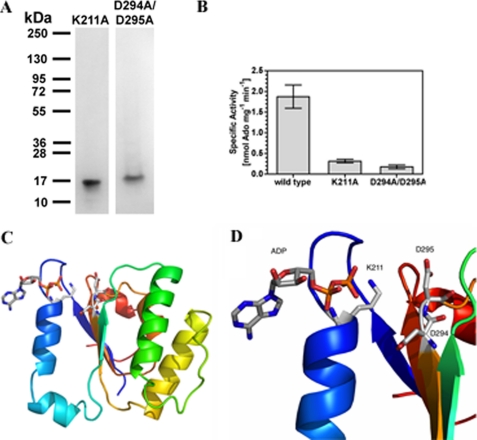
We examined the range of nucleotides utilized by R1-NB. R1-NB catalyzed the conversion of all adenine nucleotides to adenosine (Ado) with a substrate preference order of AMP>ADP>ATP (Fig. 2A; the ratios of nucleoside product to unused substrate as measured by peak areas are 2.9, 1.6, and 0.6 for AMP, ADP, and ATP, respectively). Radiolabeled substrates and analysis by TLC were used to validate this finding (Fig. 2B). The identity of the product as Ado was confirmed by ion fragmentation in an electrospray ionization mass spectrometer. Product generated from R1-NB was indistinguishable from an Ado standard (Fig. 2C). No cAMP was produced indicating that Os02g_25900 is not an AC as proposed for PSiP (14). The Ado-generating activity was not due to another protein as no contaminants were visible by SDS-PAGE and electrospray ionization mass spectrometry in positive ion mode (Fig. 2D). To prove that the observed nucleotidase activity was not an experimental artifact we used an unrelated plant protein (Arabidopsis glutathione S-transferase; (14)) expressed from the same vector and purified and assayed in exactly the same manner as R1-NB as a control. This preparation gave no significant nucleoside generating activity (Fig. 2E). As it may be formally possible that the activity observed is due to a minor contaminating protein that does not ionize in electrospray mass spectrometry we mutated a conserved amino acid (K211A) in the Walker-A/P-loop motif homologous to residues presumed to bind the β and γ phosphate moieties in P-loop ATPases (2) and expressed as a recombinant protein (Fig. 3A). We noted a significantly reduced specific activity in the K211A mutant compared with wild type protein (Fig. 3B). To be absolutely certain that the catalytic activity observed for R1-NB is not due to a contaminant we chose to mutate additional catalytic residues. Modeling the structure of R1-NB using Apaf-1 as template identified Asp-294 and Asp-295 as likely residues to co-ordinate a catalytic metal ion (Fig. 3, C and D). R1-NB D294A/D295A was expressed as a recombinant protein (Fig. 3A) and assay revealed, again, a significantly lowered specific activity compared with wild type (Fig. 3B).
FIGURE 2.
R1-NB generates Ado from adenine nucleotides. A, 5.2 μm R1-NB was assayed in 100 μl reactions in the presence of 100 μm ATP (upper panel), ADP (middle panel), or AMP (lower panel). Reactions were analyzed by HPLC. B, 9.75 μm R1-NB was assayed in the presence of 100 μm substrate. Reactions were analyzed by TLC. C, an Ado standard and product from part A were analyzed by ion fragmentation in an electrospray mass spectrometer. 2. D, electrospray ionization analysis of recombinant R1-NB. The mass/charge ratio (m/z) of the observed protein is consistent with cleavage of the N-terminal methionine as previously observed with proteins expressed from pETStrp3 (14). E, nucleotidase activity in a control protein. 10 μm R1-NB and Arabidopsis F2 glutathione S-transferase (F2-GST) (14) were assayed with 100 μm ADP as substrate (n = 4). Inset shows the purification of recombinant F2-GST (SDS/PAGE analysis and Coomassie Blue staining). A 2.0 μg portion of protein was applied, and molecular mass standards (in kDa) are indicated.
FIGURE 3.
R1-NB P-loop mutants show defective nucleotidase activity. A, purification of recombinant R1-NB K211A and R1-NB D294A/D295A (SDS/PAGE analysis and Coomassie Blue staining). 1.5 μg portions of protein were applied, and molecular mass standards (in kDa) are indicated. B, 1.2 μm R1-NB wild type, K211A mutant, and D294A/D295A mutant were assayed in the presence of 50 μm ADP (n = 3), and the reactions analyzed by TLC. C, ribbon diagram of the homology model of R1-NB with a rainbow color scheme from the N terminus (blue) to C terminus (red). The ADP was positioned by least-squares superposition of the P-loop from the crystal structure of the Apaf-1 protein (13). D, close-up of homology model showing the ADP binding site with Lys-211 responsible for phosphate binding and Asp-294 and Asp-295 responsible for Mg2+ binding.
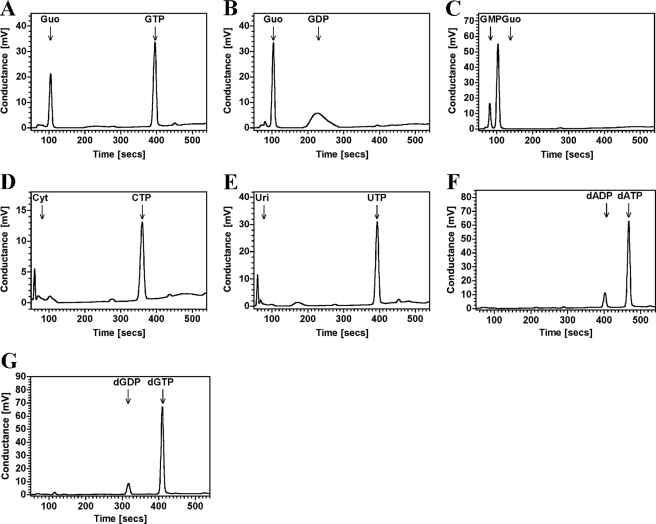
R1-NB is therefore not an AC nor is it an ATPase similar to other characterized NB-ARC domain proteins. Unexpectedly, R1-NB was found to have a nucleoside generating catalytic activity. R1-NB was also able to utilize guanine nucleotides to generate guanosine with a similar substrate preference order as observed for adenine nucleotides (Fig. 4, A–C; the ratios of nucleoside product to unused substrate as measured by peak areas are 3.7, 0.9, and 0.5 for GMP, GDP, and GTP, respectively). R1-NB was able to utilize UTP and CTP as substrate (Fig. 4, D and E). A small amount of deoxynucleotide diphosphate was generated from the appropriate triphosphate with no evidence for the production of deoxynucleoside (Fig. 4, F and G). This activity was, however, very weak compared with the utilization of the corresponding nucleotides.
FIGURE 4.
R1-NB generates nucleoside from nucleotides. 5.2 μm R1-NB was assayed in 100 μl reactions in the presence of 100 μm (A) GTP, (B) GDP, (C) GMP, (D) CTP, (E) UTP, (F) dATP, and (G) dGTP. Reactions were analyzed by HPLC.
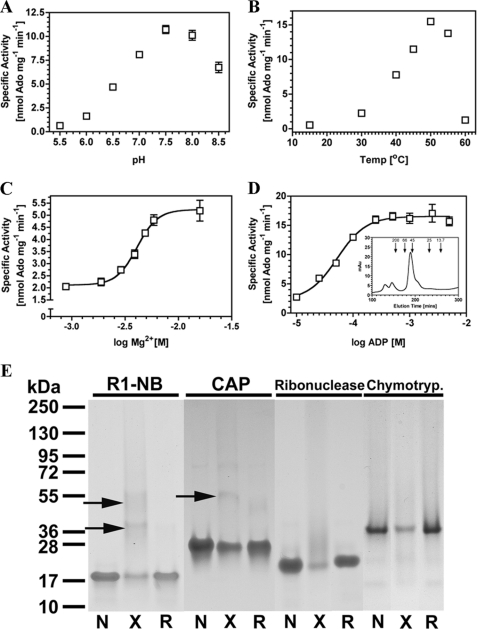
To further characterize the R1-NB protein we examined the response of R1-NB to temperature and pH. A pH optimum of 7.5 and temperature optimum of 50 °C was found using ADP as substrate (Fig. 5, A and B). Further assays demonstrated that divalent metal was required for R1-NB activity as would be expected for a P-loop family protein with a nucleotidase activity and an EC50 for Mg2+ of 4.1 mm is consistent with related molecules (Fig. 5C) (8). Kinetic analysis of R1-NB using ADP as substrate gave a Km(app) of 53 ± 7 μm and Vmax(app) of 16.6 ± 0.4 nmol Ado mg−1 min−1 (corresponding to a kcat of 0.28 min−1) (Fig. 5D). Although the affinity of R1-NB for the nucleotide is lower than that of the affinity of the I-2 CC-NB-ARC domains for ATP (Kd = 2.2 μm; (8)), an affinity in the μm range demonstrates that the nucleoside generating activity is a likely biologically relevant activity and not an artifact of assay in vitro under extreme conditions. A sigmoid for the substrate response gives a Hill Slope of 1.7 consistent with R1-NB acting as a multisubunit protein. Analytical gel filtration revealed a major peak at just over 45 kDa that contained only the R1-NB protein (17 kDa) as assessed by SDS-PAGE (Fig. 5D). It is tempting to assign this peak as a R1-NB dimer structurally analogous to the asymmetric dimer formed by the CED-4 nucleotide binding α/β fold in octameric apoptosome assembly (18). To provide additional evidence for dimer formation we performed chemical cross-linking of R1-NB using the E. coli catabolite-activated protein (CAP) as positive control and ribonuclease A1 and chymotrypsinogen A1 as negative controls (Fig. 5E). Cross-linking of R1-NB and CAP gave clear bands corresponding to a protein dimer that could be eliminated by reduction of the cross-link (lower arrow for R1-NB). No such bands were evident for ribonuclease A1 or chymotrypsinogen A. The protein bands for the negative controls are reduced through smearing up the gel by nonspecific modification by the cross-linking agent but no clear multimer bands are observed. A faint higher molecular weight band in the CAP lanes (at around 72 kDa) is evident but was a minor contaminant from protein purification and not a band corresponding to cross-linking. A higher molecular weight band was consistently observed on cross-linking of R1-NB (upper arrow for R1-NB). This band could be speculatively assigned as a trimer, consistent with analytical gel filtration and asymmetric multimerization, but an unambiguous assignment of the multimer status will require a full structural analysis. R1-NB was assayed in the presence of the reducing agents dithiothreitol or monothioglycerol (supplemental Fig. S1) to examine whether a redox sensitive linkage might contribute to enzyme activity, for example through an intermolecular bond. Increasing concentrations of dithiothreitol had no influence on specific activity. Elevated concentrations of monothioglycerol gave a very small but statistically relevant reduction in specific activity. The lack of any effect of dithiothreitol and the small effect of monothioglycerol does not support a role for an intra- or intermolecular redox sensitive bond essential for activity.
FIGURE 5.
Influence of assay variables on R1-NB activity assayed by TLC. A, 3.76 μm R1-NB was assayed in the presence of 1 mm ADP at varying pH (n = 8). B, 3.76 μm R1-NB was assayed in the presence of 1 mm ADP at varying temperature (n = 8). C, 5.28 μm R1-NB was assayed in the presence of 1 mm ADP at varying MgCl2 (n = 8). D, 5.24 μm R1-NB was assayed in the presence of varying ADP substrate (n = 8). Independent assays confirmed reactions were linear over the timescale of the assay. The inset shows an analytical gel filtration trace with molecular weight standards in kDa indicated. E, 5.8 μg of protein was analyzed (SDS/PAGE analysis and Coomassie Blue staining) after no treatment (N), chemical cross-linking (X), or cross-linking and subsequent reduction (R). Arrows indicate protein multimers.
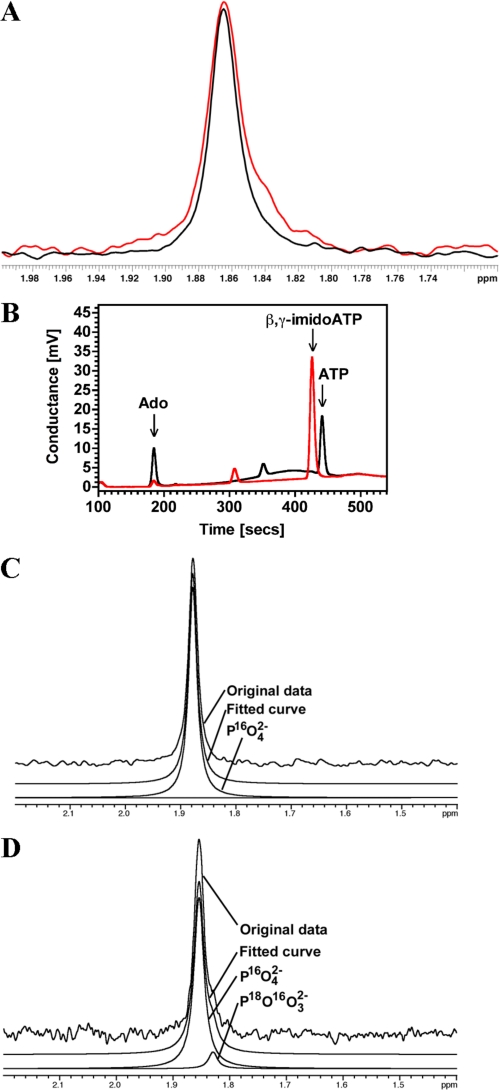
The generation of nucleoside from nucleotide could occur by two distinct mechanisms. The first possibility is that R1-NB is a nucleotide monoester hydrolase that cleaves nucleotides at the nucleoside/α-PO42− bond. The second possibility is that R1-NB is a nucleotide phosphatase that sequentially cleaves terminal phosphates. 31P NMR analysis of R1-NB using ADP as substrate was used to select between these two alternatives. A single peak corresponding to PO42− was observed (Fig. 6A, black trace) and no peak corresponding to P2O82− was observed indicating that R1-NB is a nucleotide phosphatase. A formal possibility is that P2O82− is generated and subsequently cleaved by R1-NB, however, R1-NB was unable to convert the non-hydrolyzable ATP analog adenosine 5′-(β,γ-imido)triphosphate demonstrating that the terminal phosphate bond is the target (Fig. 6B). NMR in the presence of 40% (v/v) H2O18 revealed an asymmetric PO42− peak with a shoulder, indicating a second signal in this peak that is not present when the assay is performed in H2O16 (Fig. 6A, red trace). Deconvolution of the PO42− peaks demonstrated the presence of a minor peak with a small chemical shift in the presence of H2O18 (Fig. 6D) but not with H2O16 (Fig. 6C). This finding is consistent with the production of P16O318O2− in the assay performed with H2O18 (19, 20). Taken together all the data are consistent with a mechanism whereby divalent metal activated water mounts a nucleophilic attack on the terminal ester bond, releasing the phosphate. The substrate preference order demonstrates that all phosphates are sequentially cleaved with the eventual release of the nucleoside.
FIGURE 6.
R1-NB is a nucleotide phosphatase. A, 31P-NMR PO42− peak obtained from an assay in which R1-NB was provided with ADP as substrate in the presence of H2O16 (black line) or 60:40 H2O16: H2O18 (red line). B, 5.2 μm R1-NB was assayed in 100 μl reactions in the presence of 100 μm ATP (black trace) or β,γ-imidoATP (red trace). Reactions were analyzed by HPLC. C, deconvolution of 31P-NMR spectrum obtained in reaction containing only H2O16. Only a single peak corresponding to P16O42− can be extracted from the signal. D, deconvolution of 31P-NMR spectrum obtained in reaction containing 40:60 H2O18:H2O16. Two signals are obtained from the fitting routine with the P18O16O32− peak at around 10% of the P16O42− peak independent of fitting parameters.
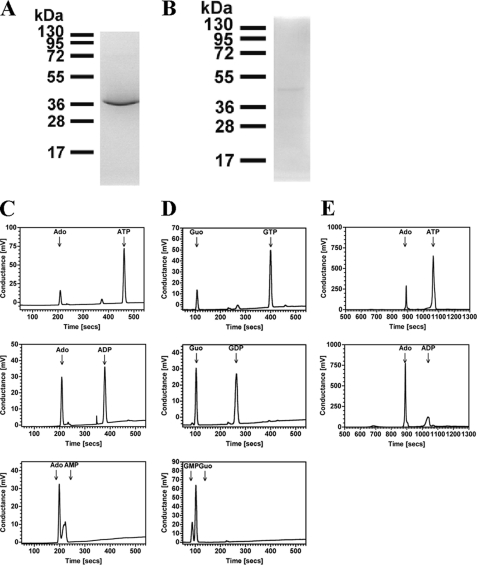
We chose to establish that the nucleotide phosphatase activity of R1-NB was not an artifact of the truncated nature of the protein. Previous work with refolded preparations of Mi-1 and I-2 R-proteins utilized constructs that encompassed the entire NB-ARC domain (8). Residues within the tandem ARC domains may confer an additional specificity on catalytic activity and nucleotide binding. Unfortunately we were unable to recover a soluble recombinant protein corresponding to the complete NB-ARC domain for Os02g_25900 and therefore turned our attention to other R-proteins. We specifically investigated the expression of the NB-ARC domains of PSiP and the well-characterized R-protein Rpm1 (hereafter called PSiP-NBARC and Rpm1-NBARC, respectively). Both Rpm1 and PSiP are derived from the CNL clade of NB-ARC family proteins and enable us to investigate whether the nucleotide phosphatase activity of R1-NB is present in other R-proteins. PSiP was an attractive experimental target for protein expression as this would enable us to validate whether an AC activity is associated with this ORF as hypothesized. PSiP-NBARC expression was comparable to R1-NB and it could be purified to homogeneity using similar methodology (Fig. 7A). Rpm1 was an attractive experimental target for protein expression as this has an established role in the innate immune response. Rpm1-NBARC was expressed at extremely low levels and was often lost on purification but a single pure preparation of protein was obtained on which multiple experiments could be performed (Fig. 7B). Analysis of PSiP-NBARC nucleotide utilization by HPLC showed the generation of the nucleoside with an identical substrate preference to R1-NB of AMP>ADP>ATP (Fig. 7C) and GMP>GDP>GTP (Fig. 7D). Specific activity using ADP as substrate was 6.0 ± 0.5 nmol Ado mg−1 min−1 (3 μm protein; SD) equivalent to values obtained for R1-NB. PSiP-NBARC contains all residues and protein subdomains required for nucleotide binding/catalysis in P-loop ATPases based on bioinformatic and structural analysis (2, 13). PSiP is therefore not an AC. Using the limited Rpm1-NBARC material available we demonstrated that Rpm1-NBARC catalyzed the production of Ado from ATP and ADP demonstrating that the nucleotide phosphatase activity observed in R1-NB and PSiP-NBARC is also present in an established R-protein (Fig. 7E). Specific activity using ADP as substrate was 38.5 ± 2.3 nmol Ado mg−1 min−1 (0.38 μm protein; SD). Together these data demonstrate that the substrate specificity in the NB-ARC domain proteins is inherent to the NBD and that our data is unlikely to be due to an altered activity caused by protein truncation.
FIGURE 7.
Purification and assay of recombinant NB-ARC domains (SDS/PAGE analysis and Coomassie Blue staining). A, purification of recombinant PSiP-NBARC. A 1.5 μg portion of protein was applied and molecular mass standards (in kDa) are indicated. B, purification of recombinant Rpm1-NBARC. A 0.5 μg portion of protein was applied and molecular mass standards (in kDa) are indicated. C, 1.3 μm PSiP-NBARC was assayed in 100 μl reactions in the presence of 100 μm ATP (upper panel), ADP (middle panel), or AMP (lower panel). Reactions were analyzed by HPLC. D, 1.3 μm PSiP-NBARC was assayed in 100 μl reactions in the presence of 100 μm GTP (upper panel), GDP (middle panel), or GMP (lower panel). Reactions were analyzed by HPLC. E, 10 nm Rpm1-NBARC was assayed in 100 μl reactions in the presence of 100 μm ATP (upper panel) or ADP (lower panel). Reactions were analyzed by HPLC.
DISCUSSION
The description of biochemically active soluble recombinant plant NBDs and NB-ARC domains represents a significant advance in our analysis of plant R-protein biochemistry. It is, however, somewhat of a surprise that the proteins examined possessed a nucleotide phosphatase activity. The phosphatase activity was not an artifact of the methodology used as it was observed with two independent methods of analysis (TLC and HPLC). The activity is also not an artifact of protein expression as the same catalytic activity is observed in three independent proteins over multiple preparations. The fact that the nucleotide phosphatase activity is intrinsic to the NBD, as opposed to a contaminating protein, is unambiguously demonstrated by the fact that multiple mutations widely accepted as reducing the activity of P-loop containing enzymes (2) significantly reduced the specific activity of R1-NB. Mutations in residues required for phosphate co-ordination (K211A) and co-factor binding (D294A D295A) diminished enzyme activity by >80%. The percentage reduction in activity of the R1-NB K211A mutant is identical in extent to that reported for the equivalent K207R mutant of I-2 (8) and K222N mutation of N (21). Despite the differences in biochemistry, therefore, between R1-NB and I-2, the behavior of the active site mutants is identical. The nucleotide phosphatase activity observed is not an artifact of co-purification with DnaK. The E. coli chaperones including the DnaK system (DnaK with its DnaJ and GrpE co-chaperones) are a normal part of the cellular machinery and are widely used to optimize recombinant protein folding (22). R1-NB biochemistry is identical both in the presence of DnaK and when purified away by anion exchange indicating that the activity of the purified protein is not an artifact of misfolding. While a model of R1-NB proved useful in identifying likely important catalytic residues, it is of limited use in determining the structural basis for the nucleotide phosphatase activity, and this awaits structural analysis through x-ray crystallography.
Characterization of R1-NB revealed pH and temperature optima of ∼7.5 and 50 °C, respectively. The pH optimum is not unusual and approximates cytosolic pH. The temperature optimum is high but not unusual for rice enzymes (for example tryptophan decarboxylase and glyceraldehyde-3-phosphate dehydrogenase have temperature optima of 45 and 50 °C, respectively (23, 24)). While the temperature optimum is certainly beyond normal growth temperatures, the high optimum might ensure that R-protein responses in the wild can be maintained over widely fluctuating daily temperatures rather than be compromised during the hottest hours of the day.
The nucleotide phosphatase activity observed is intrinsic to the NBD. Although a contribution of the tandem ARCs domains to catalysis cannot be ruled out (e.g. in determining substrate affinity) it is important to note that PSiP is a nucleotide phosphatase and not an AC as previously proposed (17). Previous analysis has asserted that PSiP does not encode an AC (2). This study assigns a nucleotide phosphatase activity to this protein and demonstrates conclusively that it is not an AC. Of central significance is the observation of a nucleotide phosphatase activity in the well-studied Rpm1 R-protein. The finding of a nucleotide phosphatase activity in Rpm1 is not inconsistent with the behavior of genetically analyzed mutants in vivo (25). Unfortunately, Rpm1 proved so refractory to expression and purification that a highly desirable study correlating the biochemistry of Rpm1 to its activity in vivo is not possible without further major advances in methodology.
The identification of a nucleotide phosphatase activity that can utilize any nucleotide to generate nucleoside in an R-protein has implications for our working models for signaling in response to pathogens. Current models envisage the NBD as a switch domain; “off” in the ADP bound state and “on” in the ATP bound state. Hydrolysis of ATP to ADP is required to switch “off” a signaling event. The nucleotide phosphatase activity of R1-NB, PSiP-NBARC, and Rpm1-NBARC is entirely consistent with this model except that hydrolysis continues to the nucleoside rather than stopping at the diphosphate. Sustained hydrolysis to the nucleoside might permit a sustained signaling event without the need to exchange ADP for ATP. Another intriguing observation for R1-NB is its multimer status. Multimer formation is supported by three independent lines of experiment (kinetic evidence, analytical gel filtration, chemical cross-linking) and has never been observed in a R-protein. Many P-loop-containing proteins function as multimers, the significance of which is not always clear. The role of multimer formation in R-protein function will require future structural and biochemical analysis. Such analysis will enable the identification of key regions of a R-protein likely to be involved in dimer formation that can then be mutated and analyzed for their effect on enzyme activity and the immune response in plants.
As the P-loop residues analyzed required for the strict ATPase activity of I-2 and nucleotide phosphatase activity of R1-NB are equivalent, straightforward in vivo analysis of these mutants is unlikely to reveal anything of substance regarding the function of the two activities in plant defense. Future experiments should be directed toward a structural analysis of R1-NB to uncover likely determinants of its nucleotidase activity and the dimer interface. This will enable the rationale design of the appropriate mutants to constrain activity and dimer formation for further analysis in vivo.
Supplementary Material
Acknowledgments
We thank David Dixon, Alan Kenwright, and Aileen Congreve for outstanding technical support and many helpful scientific discussions.
This work was supported by Leverhulme Trust Grant F/00 128/AU and BBSRC Grant BB/I011994/1 (to M. J. C.).

This article contains supplemental Table S1 and Fig. S1.
- MAMP
- microbe-associated molecular pattern
- NTPase
- nucleotide triphosphatase
- Avr
- Avirulence
- NB-ARC
- nucleotide binding, Apaf-1, R-protein, and CED-4
- NBD
- nucleotide binding domain
- CC
- coiled coil
- LRR
- leucine-rich repeat
- Ado
- adenosine
- TOF
- time of flight
- AC
- adenylyl cyclase.
REFERENCES
- 1. Dangl J. L., Jones J. D. (2001) Plant pathogens and integrated defense responses to infection. Nature 411, 826–833 [DOI] [PubMed] [Google Scholar]
- 2. Leipe D. D., Koonin E. V., Aravind L. (2004) STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343, 1–28 [DOI] [PubMed] [Google Scholar]
- 3. van der Biezen E. A., Jones J. D. (1998) The NB-ARC domain: a novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R227 [DOI] [PubMed] [Google Scholar]
- 4. Jones J. D., Dangl J. L. (2006) The plant immune system. Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- 5. Takken F. L., Tameling W. I. (2009) To nibble at plant resistance proteins. Science 324, 744–746 [DOI] [PubMed] [Google Scholar]
- 6. Allen R. L., Bittner-Eddy P. D., Grenville-Briggs L. J., Meitz J. C., Rehmany A. P., Rose L. E., Beynon J. L. (2004) Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960 [DOI] [PubMed] [Google Scholar]
- 7. Kim H. E., Du F., Fang M., Wang X. (2005) Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. U.S.A. 102, 17545–17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tameling W. I., Elzinga S. D., Darmin P. S., Vossen J. H., Takken F. L., Haring M. A., Cornelissen B. J. (2002) The tomato R gene products I-2 and MI-1 are functional ATP-binding proteins with ATPase activity. Plant Cell 14, 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tameling W. I., Vossen J. H., Albrecht M., Lengauer T., Berden J. A., Haring M. A., Cornelissen B. J., Takken F. L. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moffett P., Farnham G., Peart J., Baulcombe D. C. (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rairdan G. J., Moffett P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18, 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bao Q., Lu W., Rabinowitz J. D., Shi Y. (2007) Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol. Cell 25, 181–192 [DOI] [PubMed] [Google Scholar]
- 13. Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y. (2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926–933 [DOI] [PubMed] [Google Scholar]
- 14. Dixon D. P., Hawkins T., Hussey P. J., Edwards R. (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 60, 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) Comparative protein structure modeling with MODELLER. Current Protocols in Bioinformatics, Supplement 15, 5.6.1–5.6.30, John Wiley & Sons, Inc., New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moutinho A., Hussey P. J., Trewavas A. J., Malhó R. (2001) cAMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. U.S.A. 98, 10481–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi S., Pang Y., Hu Q., Liu Q., Li H., Zhou Y., He T., Liang Q., Liu Y., Yuan X., Luo G., Li H., Wang J., Yan N., Shi Y. (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141, 446–457 [DOI] [PubMed] [Google Scholar]
- 19. Cohn M., Hu A. (1978) Isotopic (18O) shift in 31P nuclear magnetic resonance applied to a study of enzyme-catalyzed phosphate-phosphate exchange and phosphate (oxygen)-water exchange reactions. Proc. Natl. Acad. Sci. U.S.A. 75, 200–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Etten R. L., Risley J. M. (1978) Phosphate (oxygen)-water exchange reaction catalyzed by human prostatic acid phosphatase. Proc. Natl. Acad. Sci. U.S.A. 75, 4784–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ueda H., Yamaguchi Y., Sano H. (2006) Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61, 31–45 [DOI] [PubMed] [Google Scholar]
- 22. de Marco A., Deuerling E., Mogk A., Tomoyasu T., Bukau B. (2007) Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang K., Kang S., Lee K., Park M., Back K. (2008) Enzymatic features of serotonin biosynthetic enzymes and serotonin biosynthesis in plants. Plant Signal Behav. 3, 389–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arumugam Pillai M., Lihuang Z., Akiyama T. (2002) Molecular cloning, characterization, expression, and chromosomal location of OsGAPDH, a submergence responsive gene in rice (Oryza sativa L.). Theor. Appl. Genet. 105, 34–42 [DOI] [PubMed] [Google Scholar]
- 25. Takken F. L., Albrecht M., Tameling W. I. (2006) Resistance proteins: molecular switches of plant defense. Curr. Opin. Plant Biol. 9, 383–390 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.