Background: Cofilin rods are associated with Alzheimer disease, but their pathological significance is unclear.
Results: Time-lapse imaging revealed that cofilin rods inhibit the movement of mitochondria and early endosomes. Cofilin rods reduce dendritic spines and impair synaptic transmission. Cofilin rods are discovered in senile rat brains.
Conclusion: Cofilin rods block intracellular transport and induce synaptic loss.
Significance: Our work identifies a signaling pathway underlying neurodegeneration and brain aging.
Keywords: Actin, Alzheimer Disease, Cofilin, Neurodegeneration, Synapses, Trafficking
Abstract
Cofilin is an actin-binding protein and a major actin depolymerization factor in the central nervous system (CNS). Cofilin-actin aggregates are associated with neurodegenerative disorders, but how cofilin-actin aggregation induces pathological effects in the CNS remains unclear. Here, we demonstrated that cofilin rods disrupted dendritic microtubule integrity in rat hippocampal cultures. Long term time-lapse imaging revealed that cofilin rods block intracellular trafficking of both mitochondria and early endosomes. Importantly, cofilin rod formation induced a significant loss of SV2 and PSD-95 puncta as well as dendritic spines. Cofilin rods also impaired local glutamate receptor responses. We discovered an inverse relationship between the number of synaptic events and the accumulation of cofilin rods in dendrites. We also detected cofilin rods in aging rat brains in vivo. These results suggest that cofilin aggregation may contribute to neurodegeneration and brain aging by blocking intracellular trafficking and inducing synaptic loss.
Introduction
Actin dynamics is tightly regulated by a variety of actin-binding proteins (1–3). Cofilin is one of the major actin-binding proteins in the brain and severs filamentous actin (F-actin) into short segments and creates free barbed ends for actin elongation (4–7). Cofilin is inactivated after phosphorylation of Ser3 by LIM kinase (8, 9). Phosphorylated cofilin can be reactivated through dephosphorylation by phosphatase slingshot or chronophin (10–13). Together with LIM kinase, slingshot, and chronophin, cofilin plays a critical role in neurite growth and growth cone turning (14–18), receptor trafficking (19–21), and dendritic spinogenesis and synaptic plasticity (20, 22–26).
Recent studies have identified cofilin-actin rods in the brains of postmortem Alzheimer disease (AD)4 patients and in transgenic animal models for AD (27–30). Rods were originally identified as actin-immunopositive inclusions (31), but subsequent studies found that cofilin was also abundant in rods (27, 32). Interestingly, cofilin-actin rods can be induced by amyloid β oligomers and oxidative stress (13, 27, 28, 33–35). Cofilin rod formation may also be regulated by microRNAs (36). In the hippocampus and cortex of AD patients or mouse models, abnormal cofilin aggregation has been detected in amyloid β plaques and possibly tau tangles (27, 29, 37). In a Drosophila AD model expressing human mutant tauR406W, cofilin-actin rods are found colocalizing with hyperphosphorylated tau (30). These studies suggest a potential role of cofilin pathology during neurodegeneration. However, how cofilin aggregation impairs neuronal functions in mammalian central nervous system has not been well understood.
In this study, we employed live time-lapse imaging to demonstrate that the formation of cofilin rods block intracellular organelle trafficking, including early endosomes and mitochondria. Rod formation also induces synaptic loss, as revealed by the lack of pre- and postsynaptic markers and reduction of dendritic spines in rod areas. When the number of cofilin rods increases, the frequency of synaptic events is progressively reduced. Interestingly, we have identified cofilin rods in senile but not young adult rat brains, suggesting that cofilin rods may be associated with brain aging.
EXPERIMENTAL PROCEDURES
Cell Culture
Hippocampal cultures were prepared similarly to our previous reports (38, 39). Hippocampal CA1–CA3 region was dissected out from 18-day rat embryos of either sex, dissociated in 0.05% trypsin-EDTA solution, and plated on a monolayer of cortical astrocytes. The culture medium contained 500 ml of MEM (Invitrogen), 5% FBS (HyClone, Logan, UT), 10 ml of B-27 supplement (Invitrogen), 100 mg of NaHCO3, 20 mm d-glucose, 0.5 mm l-glutamine, and 25 units/ml penicillin/streptomycin. Cultured hippocampal neurons were maintained at 37 °C in a 5% CO2-humidified incubator.
Transfection
Calcium-phosphate transfection was performed similar to the protocol previously described (40). Transfection efficiency was optimized through short incubation of neurons with Ca2+/phosphate/DNA mixture (25 min) and a washing phase with 10% CO2 preequilibrated transfection medium. Both improvements increased transfection efficiency and reduced cell toxicity by minimizing the formation of large calcium-phosphate particles. After incubation, the precipitate was washed out with 10% CO2 transfection medium three times and then incubated for 6 min before being transferred back to the original wells. Typically, each set of experiments was repeated by at least three independent transfections.
cDNA Constructs
Human full-length cofilin constructs (peGFP-N1-cofilin-WT/S3E/S3A and pmRFP-N1-cofilin-WT/S3A) were generously provided by Dr. James Bamburg (Colorado State University, Fort Collins, CO). GFP-Rab5 was a gift from Dr. Yong-Jian Liu (University of Pittsburgh, Pittsburgh, PA). GFP-mito was provided by Dr. M. Bienengraeber (Medical College of Wisconsin, Milwaukee, WI).
Imaging
Phase-contrast and fluorescence images were obtained on an inverted Nikon (Tokyo, Japan) TE 2000-S microscope using a 40× objective. Images were captured with a Hamamatsu (Hamamatsu City, Japan) ORCA 100-cooled CCD camera driven by Simple PCI software (Hamamatsu). Confocal images were collected on an Olympus FV1000 confocal microscope. Fluorescent images were processed with Adobe Photoshop software.
Cells were quantified as having rods if they possessed at least three rods, and each with a length ≥ 3 μm. Rod length varied from 3 to 70 μm with an average between 5 and 10 μm. The percentage of rod coverage of dendrites was measured by ImageJ with NeuronJ plugin, calculated by the total length of rods divided by the total length of dendrites.
Long term time-lapse imaging was performed using an inverted Olympus IX81 microscope using a 20× objective equipped with an Ultraview VoX live cell scanning unit (PerkinElmer Life Sciences). Cell culture dishes containing live neurons and glial cells growing in culture medium were placed in a chamber that was mounted on the XY stage of the microscope. The chamber had an automatic heating control unit to maintain the temperature at 37 °C and an airflow system to keep the chamber at 5% CO2. Image acquisition, analysis, and movie construction were carried out with Volocity Software 5.0 (Improvision; PerkinElmer Life Sciences). The kymograph was constructed using ImageJ software. The height of kymographs represents recording time, and the width represents the length of dendrites. To estimate the velocity of early endosome and mitochondria movement quantitatively, we quantified moving organelles in dendrites with or without cofilin rods. Time-lapse images from 3 h before to 2 h after rod formation were included for data analysis. The built-in functional module of Volocity Software 5.4 was used to find and track objects before calculating the velocity of different organelles.
Immunostaining
Cultured Cells
Cells were rinsed with PBS twice and fixed for 45 min in a solution of 4% paraformaldehyde and 0.1% glutaraldehyde, pH 7.4. Fixation for 15 min was attempted but could not fix cofilin rods. Coverslips were then rinsed three times in PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 min or 98% methanol for 3 min. Permeabilization solution was removed and washed three times with PBS. Primary antibodies were added together with a donkey serum blocking solution in PBS and set to incubate overnight at 4 °C. The following day, coverslips were rinsed three times in PBS for 15 min. Subsequently, samples were incubated for 45 min in anti-mouse Alexa Fluor 647-conjugated or anti-rabbit Alexa Fluor 546-conjugated secondary antibodies (Invitrogen). Cofilin was immunostained with rabbit polyclonal antibody (1:300; Cytoskeleton). Presynaptic nerve terminals were identified with a mouse monoclonal antibody specific for SV2 (1:2000; Developmental Studies Hybridoma Bank, University of Iowa, IA), and glutamatergic postsynaptic puncta were identified using mouse monoclonal antibody specific for PSD-95 (1:2000; Affinity BioReagents, IL). Early endosomes were labeled with a mouse monoclonal antibody specific for early endosome-associated antigen 1 (EEA1) (1:200; BD Transduction Laboratories). Coverslips were then rinsed six times in PBS for 15 min and then mounted with mounting solution (50% glycerol, 50% 0.1 m NaHCO3, pH 7.4) on glass slides.
Brain Sections
Brains were dissected out from six young adult (3–4 months) Sprague-Dawley rats and six old rats (∼2 years old) and fixed in 4% paraformaldehyde at 4 °C for 3 days and then dehydrated with 20% sucrose overnight at 4 °C. Coronal sections (30 μm) of cortex were made on a freezing microtome (Microm, HM 400; Thermo Scientific) cooled by dry ice, and tissue was mounted using a drop of PBS. Sections were stored in cryoprotectant solution (20% sucrose, 30% ethylene glycol, 0.02% sodium azide, in sodium phosphate buffer, pH 7.4) until immunostaining (less than a week). Free-floating sections were washed 3 times in PBS for 10 min. Sections were permeabilized in 98% methanol for 20 min at −20 °C. Sections were washed three times in PBS for 10 min and further permeabilized and blocked with an antibody dilution buffer (normal donkey serum, 0.5% Triton X-100, and PBS) for 30 min at room temperature. After blocking, sections were incubated overnight at 4 °C in rabbit polyclonal antibody to human cofilin (2 μg/ml; Cytoskeleton). Another cofilin antibody was a gift kindly provided by Dr. James Bamburg (27, 28). Both cofilin antibodies gave similar results. Sections were washed three times in PBS for 10 min and incubated with secondary antibody for 2 h (goat anti-rabbit Alexa Fluor 546, 1:300, Invitrogen) in a dark box. Sections were again washed three times in PBS for 10 min each and then mounted onto glass slides with aqueous nonfluorescing mounting medium (Shandon Immu-Mount; Thermo Scientific). Cofilin rods in brain sections were visualized using an Olympus FV1000 confocal microscope. Z-stacks of 0.3–0.4-μm slices were merged into a composite image.
In Vivo Quantification
For calculating SV2 fluorescent signal, oval regions of interest (ROIs) were drawn around rods of varying sizes (5–15 μm) after compressing a series of confocal images (Z-stack). Oval ROIs completely encompassed each single rod and left ∼3 μm spacing around each side of the rod. ROIs for non-rod areas and young cultures/cortices were selected from the four corners of the image. ROIs were measured using the Olympus Fluoview 1000 image processing software. Both cultured neurons and cortical slices used 60× with digital zoom (1–2×).
Electrophysiology
Whole-cell patch clamp recordings were performed using Multiclamp 700A amplifier (Axon Instruments) as described before (38, 39). Patch pipettes were pulled from borosilicate glass and fire-polished (4–6 megohms). The recording chamber was perfused continuously with a bath solution containing 128 mm NaCl, 30 mm glucose, 25 mm HEPES, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, pH 7.3 adjusted with NaOH. The pipette solution contained 147 mm KCl, 5 mm Tris phosphocreatine, 2 mm EGTA, 10 mm HEPES, 4 mm MgATP, 0.5 mm Na2GTP, pH 7.3 adjusted with KOH. The membrane potential was clamped at −70 mV. Data were acquired using pClamp 9 software, sampled at 5 kHz, and filtered at 1 kHz. For local ejection of glutamate, a fine pipette (1–2-μm tip) filled with glutamate (500 μm), 0.5 μm TTX, and 20 μm bicuculline was positioned very close to soma or dendrites (1–2 μm). A Picospritzer (Parker Instrumentation) was used to eject glutamate with air pressure of 10 p.s.i. and duration of 15 ms. Coverslips were oriented so that the recorded dendrites or rods were perpendicular to the ejecting pipette. Rods and control dendrites at similar distal locations were selected for experiments. Bath solution with 0.5 μm TTX and 20 μm bicuculline was perfused before and during the ejection to block action potential and GABAA receptor responses. Data were expressed as mean ± S.E., and an unpaired Student's t test was used for statistical analysis unless otherwise noted.
Drugs
TTX was purchased from Sigma and bicuculline from Tocris (Ellisville, MO). All drugs were freshly diluted to final concentrations right before experiments.
RESULTS
Cofilin Rod Formation in Dendrites
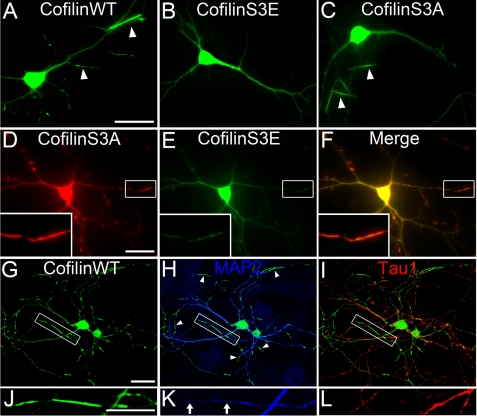
Cofilin has high affinity for actin and rarely forms aggregation in neuronal cells under normal physiological conditions. However, under neuropathological conditions such as AD or overexpressing cofilin in cells, cofilin rod formation has been reported (26, 27, 41). Here, we found that transfecting hippocampal neurons with wild-type (WT) GFP-cofilin resulted in numerous cofilin rods (Fig. 1A, 90% of neurons showed rods, n = 76). In contrast, overexpression of phosphomimetic GFP-cofilin mutant (S3E) resulted in no cofilin rod formation (Fig. 1B, 0%, n = 36), whereas overexpression of constitutively active GFP-cofilin mutant (S3A) resulted in reduced cofilin rod formation (Fig. 1C, 48% of neurons showed rods, n = 21), suggesting that both cofilin phosphorylation and dephosphorylation are required for rod formation. Furthermore, coexpression of RFP-cofilin S3A and GFP-cofilin S3E formed fewer rods compared with GFP-cofilin S3A alone (Fig. 1, D–F, 33%, n = 66). Note that RFP-cofilin S3A and GFP-cofilin S3E localized throughout the cytoplasm, but only RFP-cofilin S3A concentrated in rods. Overexpression of GFP or RFP alone never induced rod formation (data not shown). These experiments suggest that the serine 3 residue (S3) of cofilin is a critical regulation site for rod formation.
FIGURE 1.
Cofilin rods form in distal dendrites. A, representative image shows cofilin rods (indicated by white arrowheads) in neurons transfected with wild-type GFP-cofilin (7 days in vitro transfection and 9 days in vitro imaging). B and C, overexpression of GFP-cofilin S3E did not induce rod formation whereas transfection of GFP-cofilin S3A did induce rod formation. D–F, coexpression of RFP-cofilin S3A and GFP-cofilin S3E resulted in rod formation with RFP-cofilin S3A concentrating in rods and GFP-cofilin S3E largely absent in rods. Inset in left corner is the enlarged view of a cofilin rod. Note the presence of RFP-cofilin S3A but not GFP-cofilin S3E in rod region. G–I, immunostaining with dendritic marker MAP2 (blue) and axon marker tau1 (red) showed that the majority of cofilin rods were formed in distal dendrites and less in axons. J–L, enlarged view shows cofilin rods in dendrites versus axons. Note that MAP2 staining is significantly reduced in rod areas, suggesting a possible destruction of microtubule integrity in rod regions. Scale bars, 20 μm.
To better understand where cofilin rods are distributed in neurons, we performed immunostaining with antibodies specific for the dendritic marker MAP2 and axon marker tau1 (Fig. 1, G–I). The majority of cofilin rods were found in distal (but not apical) dendrites and only occasionally in axons (n = 11). It is worth of noting that whereas most cofilin rods were in dendrites labeled by MAP2, the actual intensity of MAP2 signal in rod areas was greatly reduced compared with neighboring non-rod dendritic branches (Fig. 1, J–L), suggesting that microtubule integrity was impaired by cofilin rods.
Although high levels of active cofilin are sufficient to induce rod formation, we tested whether a low level of cofilin signal in dendrites may also be targeted for immediate rod formation. To address this question, neurons were treated with glutamate (200 μm) or neurotoxic ATP depletion solution (Fig. 2). Immunostaining of endogenous cofilin detected linear rod-like structures in distal dendrites after ATP depletion (Fig. 2, A and B) or glutamate stimulation (Fig. 2, E and F), similar to previous reports (13, 27, 41). Interestingly, overexpression of GFP-cofilin plus glutamate or ATP depletion stimulation resulted in immediate cofilin aggregation within minutes, not only in distal dendrites but also in apical dendrites (Fig. 2, C and D, and G and H, 100% of neurons showed rods; ATP depletion, n = 23; glutamate, n = 19). Nontransfected control neurons without any stimulation showed no rods after immunostaining of endogenous cofilin (Fig. 2I). Thus, neurons with an increased level of active cofilin are prone to form cofilin rods and that rod formation can be exacerbated by neurotoxic stimulations to affect all regions of dendrites.
FIGURE 2.
Neurotoxic stimulation induces cofilin rod formation. A, hippocampal neurons were immunostained with cofilin-specific antibodies after being stimulated with ATP depletion medium (10 mm NaN3 and 6 mm 2-deoxyglucose, 30 min). After stimulation, endogenous cofilin formed rod structures. B, enlarged view of rods in A are shown. C and D, neurons were first transfected with cofilin-GFP and then stimulated with ATP depletion medium. Note severe rod formation in these cofilin-transfected neurons. E and F, glutamate (200 μm, 30 min) stimulation induced endogenous cofilin rod formation. G and H, neurons first transfected with cofilin-GFP and then stimulated with glutamate also showed severe rods. I, cofilin immunostaining affected nontransfected and nonstimulated control neurons. Arrows point to growth cones with accumulated cofilin signal. Scale bar, 10 μm.
Cofilin Rods Block Intracellular Transport
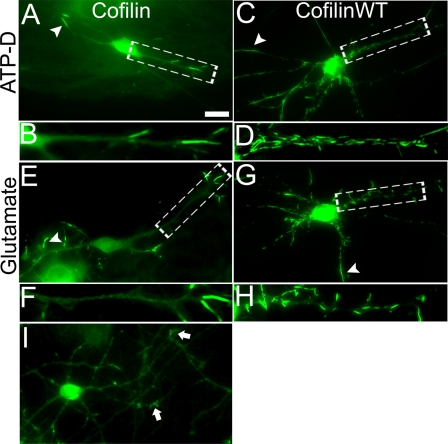
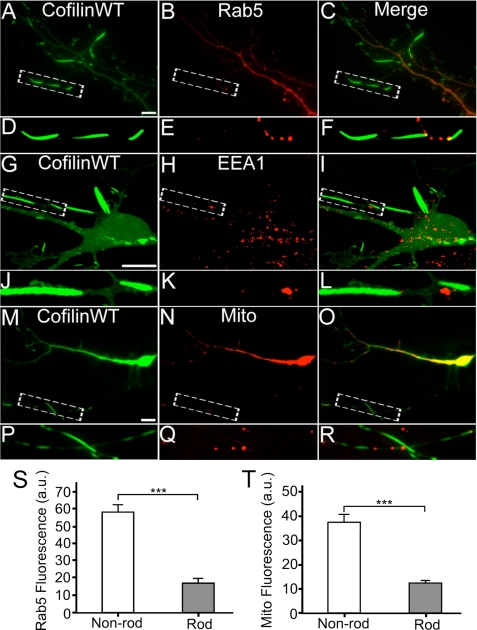
To investigate the functional consequences of cofilin rods, we first examined the effect of cofilin rods on the localization of intracellular organelles. We coexpressed RFP-tagged cofilin and GFP-tagged Rab5, a small GTPase localized specifically to early endosomes (42, 43), in cultured hippocampal neurons. We found that Rab5-labeled early endosomes were largely absent in rod areas or were stuck at either distal or proximal end of the rods (Fig. 3, A–F, quantification shown in S). Similarly, immunostaining with early endosome marker EEA1 confirmed the lack of early endosomes in rod areas (Fig. 3, G–L). Next, we coexpressed RFP-cofilin with GFP-Mito to monitor mitochondria distribution. GFP-Mito signal was also largely absent in rod regions or trapped in-between rods (Fig. 3, M–R, quantification shown in T). Therefore, cofilin rod formation disrupted normal distribution of intracellular organelles.
FIGURE 3.
Lack of intracellular organelles in cofilin rod areas. A and B, coexpression of RFP-cofilin WT (colored green, A) and GFP-Rab5 (colored red, B) in cultured hippocampal neurons. C, merged image. D–F, enlarged view revealing a lack of Rab5 puncta in rod areas. G and H, cofilin WT transfected neurons (G) and EEA1 immunostaining (H). I, merged image. J–L, enlarged view confirming the lack of early endosomes in rod areas. M–O, coexpression of RFP-cofilin WT (M) and GFP-Mito (N) to assess mitochondria distribution. P–R, enlarged view revealing a lack of mitochondria in rod areas. S, quantitative analysis of the average fluorescence intensity of GFP-Rab5 in rod regions (17.0 ± 2.7 a.u., n = 9, total rod length = 82 μm) versus non-rod areas (58.2 ± 4.4 a.u., n = 9, total non-rod dendritic length = 85 μm; ***, p < 0.001). T, quantitative analysis of the average fluorescence intensity of GFP-Mito in rod regions (13.0 ± 1.3 a.u., n = 8, total rod length = 76 μm) versus non-rod areas (37.1 ± 4.2 a.u., n = 8, total non-rod dendritic length = 101 μm; ***, p < 0.001). Scale bars, 10 μm.
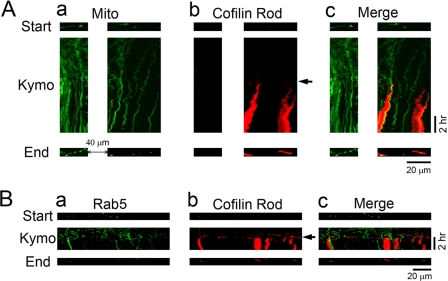
We further employed long term time-lapse imaging to monitor intracellular organelle trafficking before, during, and after rod formation (Fig. 4). Using GFP-Mito to label mitochondria, we found that mitochondria moved freely before rod formation, as shown by the kymograph illustrating their trafficking trajectory (Fig. 4Aa, green signal). However, coinciding with rod formation (Fig. 4Ab, red signal, arrow indicates rod starting point), the movement of mitochondria in rod areas was largely arrested, represented by vertical lines in the kymograph (Fig. 4Ac). We quantified the velocity of mitochondria movement in transfected cells before and after rod formation. Before rod formation (3-h time period), mitochondria moved at a velocity of 0.57 ± 0.04 μm/min (n = 132), whereas the velocity decreased to 0.36 ± 0.05 μm/min (n = 40; p < 0.01, Student's t test) within the 1st h of rod formation and further decreased to 0.21 ± 0.03 μm/min (n = 29; p < 0.001) within the 2nd h after rod formation. Control dendrites without rod formation did not show significant changes in mitochondria movement in the same time window (initial 3 h, 0.67 ± 0.08 μm/min, n = 43; later 2 h, 0.70 ± 0.11 μm/min, n = 25; p > 0.7). Interestingly, even in a non-rod area proximal to the soma and ∼40 μm away from a rod region (Fig. 4Aa, left), mitochondria movement was partially inhibited, suggesting that the formation of cofilin rods can affect the intracellular trafficking beyond rod regions. It is also worth of pointing out that mitochondria were normally in elongated shape before rod formation (Fig. 4Aa, top) but became round after rod formation (Fig. 4Aa, bottom), suggesting that mitochondrial function may also be compromised. Similarly, Rab5-labeled early endosomes also moved freely before rod formation (0.81 ± 0.04 μm/min, n = 207), but significantly reduced the velocity after rod formation (0.44 ± 0.05 μm/min, n = 87; p < 0.001) (Fig. 4B). Therefore, we conclude that cofilin rod formation blocks intracellular transport, possibly through the disruption of microtubule integrity shown by MAP2 staining.
FIGURE 4.
Time-lapse imaging showing the block of intracellular trafficking by cofilin rods. A, changes of mitochondrial trafficking before and after cofilin rod formation. a, kymograph illustrating the spatial-temporal trajectory of GFP-Mito-labeled mitochondria (green) before and after rod formation. b, time-lapse imaging of the formation of cofilin rods labeled by RFP-cofilin (red). Arrow indicates the start of rod formation. c, merged view of mitochondria and cofilin rods. Note that the vertical lines of GFP-Mito signal in the kymograph represent arrested mitochondria, which coincides with the appearance of cofilin rods. Top and bottom rows, distribution of mitochondria in dendrites at the beginning and end of time-lapse imaging. Note that besides rod formation, the shape of mitochondria changed from elongated form into small round ones, suggesting possible functional change of mitochondria after rod formation. B, dendritic trafficking of early endosomes also blocked after cofilin rod formation. a, kymograph illustrating the moving trajectory of GFP-Rab5-labeled early endosomes (green) before and after cofilin rod formation. b, rod formation (indicated by arrow) labeled by RFP-cofilin (red). c, merged view of early endosomes and cofilin rods. Note that early endosomes ceased to move upon the appearance of cofilin rods. In addition, the number of early endosomes in the end of imaging (bottom row) was greatly reduced compared with the starting point before any rod formation (top row).
Cofilin Rods Induce Synaptic Loss
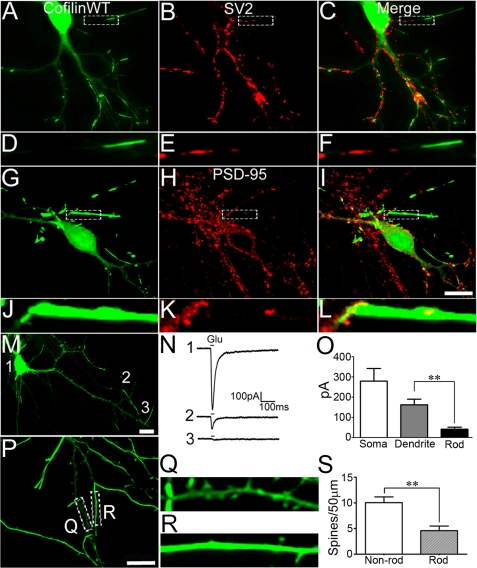
We further examined the effects of cofilin rods on synaptic structures and functions. To examine synaptic changes, we first performed immunostaining with antibodies specific for presynaptic marker SV2 and postsynaptic marker PSD-95. Interestingly, there appeared to be a significant reduction of both SV2 (Fig. 5, A–F) and PSD-95 puncta (Fig. 5, G–L) in rod areas. Quantitatively, the average density of SV2 puncta in non-rod areas was 7.4 ± 0.2/10 μm (n = 15) and significantly reduced to 1.4 ± 0.4/10 μm in rod areas (n = 15; p < 0.001). Similarly, the average density of PSD-95 puncta in non-rod areas was 7.2 ± 0.2/10 μm (n = 11) and decreased to 0.9 ± 0.3/10 μm in rod areas (n = 11, p < 0.001). We next employed patch clamp recordings to test glutamate responses directly in the rod versus non-rod dendritic areas. A short pulse of glutamate (500 μm, 15 ms) was ejected through a micropipette (1–2 μm) to soma, distal dendrites, and rod areas (Fig. 5M). Glutamate application induced a large current at soma, a significant current at distal dendrites, but a small current at rod areas (Fig. 5N). The average current amplitude at rod areas was 42 ± 10 pA (n = 7), significantly smaller than that at non-rod dendrites (162 ± 28 pA, n = 7, p < 0.01) (Fig. 5O). Furthermore, we found that dendritic spines were reduced in cofilin rod areas. Control dendrites without rods showed a significant number of spines (Fig. 5, P and Q, 10.1 ± 1.1 spines/50 μm, n = 56), whereas rod regions showed fewer spines (Fig. 5, R and S, 4.6 ± 0.9 spines/50 μm, n = 18; p < 0.001). Together, these results indicate that rod formation results in a significant decrease of synapses.
FIGURE 5.
Cofilin rods induce local synaptic loss. A–C, immunostaining of synaptic vesicle protein 2 (SV2), a presynaptic marker, in neurons transfected with GFP-cofilin WT revealing lack of SV2 puncta in rod areas. D–F, enlarged view of cofilin rods and SV2 staining. G–I, immunostaining for PSD-95, a postsynaptic marker, in GFP-cofilin WT transfected neurons showing loss of PSD-95 puncta within rod areas. J–L, enlarged view of cofilin rods and PSD-95 puncta. M, example of a cultured hippocampal neuron transfected with GFP-cofilin WT used for whole cell patch clamp recording. Local glutamate ejection (500 μm, 15 ms) was applied at soma (1), distal dendrite (2), and rod area (3). N, glutamate responses from the same neuron at locations shown in M. O, quantitative analysis of glutamate responses showing the average peak current from soma (279 ± 63 pA), dendrite (162 ± 28 pA), and rod areas (42 ± 10 pA) (p < 0.01; n = 7). Scale bars, 10 μm. P, GFP-cofilin-transfected neuron with substantial rod coverage (21 days in vitro). Q, dendritic segment without much rod coverage showing many spines. R, rod area with very few spines. S, quantitative analysis showing a significant reduction of spines in rod-containing dendrites (p < 0.001, n = 56 for non-rod branch, n = 18 for rod branch).
Accumulation of Cofilin Rods Gradually Decreases Synaptic Transmission
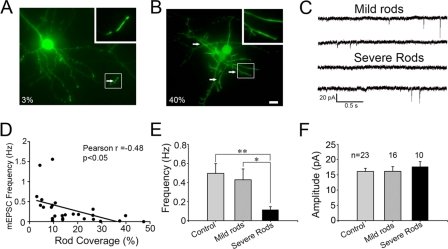
In accordance with the morphological reduction of synapses, electrophysiological analysis also revealed functional impairment associated with cofilin rods. Although rods occurred in the majority of cofilin-transfected neurons, the dendritic coverage by rods varied greatly among transfected neurons, ranging from 1 to 50%. To analyze cofilin rod effects on synaptic transmission accurately, we recorded on cofilin-transfected neurons with a wide range of rod coverage, from mild to severe. Nontransfected neurons served as controls. The miniature excitatory postsynaptic currents (mEPSCs) were recorded in the presence of TTX (0.5 μm) and GABAA receptor blocker bicuculline (20 μm). Cofilin-transfected neurons with mild rods (<15% rod coverage) showed frequency and amplitude of similar mEPSCs to the nontransfected controls (Fig. 6, A, C, E, and F). However, neurons with severe rods (>15% rod coverage) showed a significant reduction of mEPSC frequency, but not amplitude (Fig. 6, B, C, E, and F). When plotting the mEPSC frequency against the percentage of rod coverage, it showed a clear inverse relationship, that is, the more rods the less synaptic events (Fig. 6D). Neurons with severe rods did fire action potentials, suggesting that they were still viable (data not shown). Together, these results suggest that a progressive accumulation of cofilin rods will gradually induce synaptic loss and eventually impair neuronal functions.
FIGURE 6.
Effect of cofilin rods on mEPSCs. A and B, examples of cofilin-transfected neurons with mild rods (A, 3% rod coverage) and severe rods (B, 40% rod coverage). Arrows point to typical rods. Scale bar, 10 μm. C, representative traces of mEPSCs recorded in neurons with mild or severe rods. D, Pearson's correlation test revealing the inverse correlation between rod coverage and mEPSC frequency (n = 26, r = −0.48, p < 0.05). E and F, bar graphs showing that severe rods resulted in a significant decrease of mEPSC frequency (E), but not amplitude (F). Neurons were transfected at 6–9 days in vitro and recorded at 10–12 days in vitro. mEPSC frequency: control 0.5 ± 0.1 Hz (n = 23), mild rods 0.43 ± 0.11 Hz (n = 16), severe rods 0.12 ± 0.03 Hz (n = 10); mEPSC amplitude: control 16.1 ± 1.0 pA, mild rods 16.1 ± 1.6 pA, severe rods 17.7 ± 1.7 pA. *, p < 0.05; **, p < 0.01.
Cofilin Rods Discovered in Aging Brains
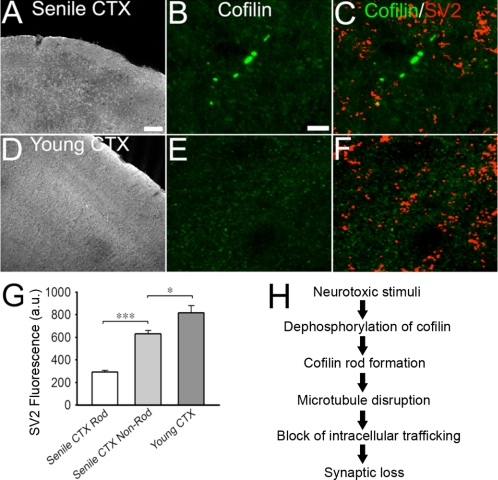
Cofilin rods have been found previously in AD brains (27–30). Here, we discovered that cofilin rods were also detectable in the brains of normal aging rats. Cofilin immunostaining were performed on coronal sections of brain tissue dissected out from senile (23 months old) or young adult (3–4 months old) rats. We detected cofilin rods in the cortices of all six old rats examined (Fig. 7, A and B). The rod size ranged from 3 to 20 μm with the majority near 5 μm. Rods were scattered in all regions of the brain and different layers of cortex, with 2–10 rods/30-μm coronal section (>20 sections/animal analyzed). In contrast, young adult rat brains rarely showed cofilin rods (Fig. 7, D and E). Because cofilin rods induced synaptic loss in vitro, we further investigated whether cofilin rods in vivo also have such similar functional consequence. Coimmunostaining of cofilin and presynaptic marker SV2 revealed sparse SV2 puncta around the rod areas in cortices of senile rat brains (Fig. 7C, quantified in G; SV2 intensity in rod areas was 291 ± 16 a.u., n = 122 rods; in non-rod areas 630 ± 31 a.u.; p < 0.001). In young adult brains, the SV2 intensity (817 ± 64 a.u.) was significantly higher than that of senile rat brains (Fig. 7F, quantified in G), consistent with a decline of synaptic density during brain aging (44). Therefore, cofilin rod formation may contribute to synaptic loss during normal brain aging.
FIGURE 7.
Cofilin rods detected in aging rat brains. A, immunostaining of cofilin in coronal sections (30 μm) of cortex from senile rat (23 months). B, enlarged local cortical region from (A) showing cofilin rods. C, coimmunostaining of cofilin and SV2 in senile cortex. Note the lack of SV2-positivie puncta in rod areas. Average fluorescence intensity of SV2 in rod regions was 291 ± 15.6 a.u. (***, p < 0.001; 122 rods; ∼1,500 μm2) whereas non-rod areas was 630 ± 30.9 a.u. (∼4,777 μm2). Quantification is shown in G. D, immunostaining of young adult (3–4 months) rat cortex showed no cofilin rods. Cofilin signal was diffuse throughout cortex. E and F, coimmunostaining for cofilin and SV2 in young adult cortex. Average fluorescence intensity of SV2 in young cortex was 817 ± 64.1 a.u. (∼16,020 μm2). G, quantitative analysis of SV2 fluorescence intensity under various conditions. H, proposed model of cellular events from rod formation induced by neurotoxic stimulation to synaptic loss. Scale bar for A and D, 100 μm; for other panels, 10 μm.
DISCUSSION
In this study, we demonstrated that cofilin rods impair neuronal functions through two major mechanisms: (i) blocking intracellular trafficking; and (ii) inhibiting synaptic functions, including reducing synaptic proteins, receptor responses, dendritic spines, and impairing neurotransmission. Moreover, cofilin rods are discovered in the brains of senile rats but not young adults, indicating that cofilin aggregation may be associated with normal brain aging. Our studies suggest a cellular mechanism of neurodegeneration in which cofilin rod formation disrupts microtubule integrity, blocks intracellular transport, and induces synaptic loss (illustrated in Fig. 7H).
Cofilin Rods Block Intracellular Trafficking
Cofilin has been shown to regulate secretory cargo sorting at the trans-Golgi network, possibly through Ca2+ ATPase SPCA1 (45, 46). Such a role of cofilin in Golgi-related trafficking is likely regulated by LIM kinase (47). Here, we further demonstrate that cofilin regulates the trafficking of early endosomes and mitochondria. Our time-lapse imaging illustrated that the movement of early endosomes and mitochondria was arrested within minutes of the formation of cofilin rods. The rods formed during our long term imaging process may be caused by both cofilin accumulation and phototoxicity. Intracellular organelle trafficking may be physically blocked by cofilin rods, as indicated by frequent observations of early endosomes and mitochondria stalled at either proximal or distal end of rods. Occasionally, these organelles can also be found in the middle of rod areas, suggesting that rods may not completely block the passage of organelles in dendrites. Another mechanism for trafficking block is the disruption of the transportation route. We demonstrate that microtubule integrity may be impaired as shown by MAP2 dissociation from rod areas. Thus, in addition to physical presence of rod structures, cell signaling pathways may be altered during rod formation to affect intracellular trafficking. In support of this notion, we found that even dendritic areas not in the rod regions (at least 40 μm away from rods, proximal to soma) were affected in terms of intracellular trafficking.
Early endosomes are critical for recycling and trafficking of membrane proteins such as membrane receptors. Our live imaging with GFP-Rab5 and immunostaining of EEA1 both showed occlusion of early endosomes in rod areas. Therefore, the lack of glutamate receptor responses at rod areas is likely due to the block of glutamate receptor trafficking through recycling endosomes. Indeed, AMPA receptor insertion into the postsynaptic sites has been tightly regulated by cofilin (20) and slingshot (21). The vesicular trafficking of acetylcholine receptors in neuromuscular junctions is also highly dependent on cofilin regulation (19). When cofilin rods formed in dendrites, the receptor trafficking is hampered, and postsynaptic apparatus may be disassembled without continuing supply of receptors and postsynaptic proteins.
Mitochondria are important energy source for cellular activities. The block of mitochondria transport by cofilin rods may cut off the ATP supply necessary for the maintenance of synaptic structures. The ATP level in old rod areas has been reported to be lower than non-rod areas (41). Conversely, ATP depletion or oxidative stress also induces cofilin rods (13, 27, 35). Our own study found that when the cofilin level is increased in neurons, ATP depletion greatly enhances cofilin rod formation. In addition to the block of mitochondria movement, we also demonstrated that mitochondria undergo morphological changes from elongated to round-shaped structures after rod formation, an indication of a functional alteration. In addition, cofilin has been shown to be translocated to mitochondria outer membranes and induce apoptosis under stimulation of staurosporine or oxidative stress (48, 49). Mitochondria dysfunction has been closely linked to brain aging and neurodegeneration (50, 51). Therefore, the impairment of mitochondrial trafficking by cofilin rods may have broad impact on neuronal functions, including causing synaptic loss and inhibiting new synapse formation.
Cofilin Rods and Synaptic Loss
As one of the major actin-binding proteins, cofilin is enriched in growth cones (16, 18, 52) and dendritic spines (20, 53–55). The formation of cofilin-actin rods may sequester local cofilin and actin in and around spines. Because actin dynamics is crucial for spine structure and function (56–58), cofilin rods may directly affect dendritic spines by impairing actin dynamics. Indeed, cofilin has been suggested to be an important regulator of spine morphology and plasticity (20, 23, 53, 59, 60). We compared rod-rich dendrites with non-rod dendrites from the same neurons and found that cofilin rods significantly reduced spine numbers in rod areas. This is consistent with the reduction of postsynaptic puncta labeled by PSD-95. The decrease of presynaptic puncta may be secondary to postsynaptic loss, possibly due to the lack of retrograde signal (61). The second mechanism for synaptic loss induced by cofilin rods is due to the block of intracellular trafficking and hence the lack of replenishment of synaptic proteins as discussed above. Our findings are consistent with previous work showing synaptic defects induced by cofilin rods in Aplysia neurons (26). Importantly, in our electrophysiology studies, we quantitatively analyzed the effect of cofilin rods on synaptic transmission according to the relative rod coverage of the total dendrites in each neuron. We found that mild rod formation had no effect on synaptic transmission, and only when substantial rod formation occurs will neurotransmission be impaired. Thus, cofilin rods affect neuronal functions in an accumulative manner, consistent with a possible role during progressive neurodegenerative disorders.
Cofilin rods or similar forms of inclusions have been identified in a number of degenerative disorders including AD (27, 62–64). Since the identification of cofilin rods in the postmortem brains of human AD patients (27), cofilin rods have been reported in several AD mouse models (28, 29, 37) as well as a Drosophila AD model (30). Our identification of cofilin rods in senile rat brains further suggests that in addition to AD, cofilin rods may also contribute to synaptic loss during normal brain aging. We demonstrated that neurons with increased cofilin level form many more rods even within apical dendrites upon neurotoxic stimulation such as glutamate or ATP depletion, both of which are contributing factors toward brain aging (65, 66). We propose that cofilin rods gradually accumulate over an extended period during brain aging, possibly through an imbalanced activity among actin-regulatory proteins such as LIM kinase and slingshot/chronophin. Cofilin rod formation will be exacerbated by glutamate excitotoxicity and oxidative stress during brain aging. When the accumulation of cofilin rods reaches a threshold level in the brain, neuronal functions and synaptic connections will be impaired.
CONCLUSION
We demonstrate that the major pathological consequences of cofilin rod are the block of intracellular trafficking and synaptic loss. Synaptic deficits induced by cofilin rods may be cumulative during normal brain aging or during the progression of neurodegenerative disorders. Because synaptic loss is a hallmark of neurodegeneration (44, 67), we propose that cofilin rods may be a common pathological marker for early stages of neurodegeneration.
Acknowledgments
We thank Dr. Wenbiao Gan and Dr. James Bamburg for thorough discussion about our data and careful editing of the manuscript with many insightful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants NS054858 and MH083911. This work was also supported by National Science Foundation Grant 0821955 and American Heart Association Grant 0765258U (to G. C.), Natural Science Foundation of China Grants 30828014 (to G. C. and Y. W.) and 31129003 and 81171224 (to Y. W.), and Ministry of Science and Technology of China Grant 2009CB522000 and Science and Technology Commission of Shanghai Municipality Grant 09JC1401900 (to Y. W.).
- AD
- Alzheimer disease
- a.u.
- arbitrary units
- EEA1
- early endosome-associated antigen 1
- mEPSC
- miniature excitatory postsynaptic current
- ROI
- region of interest
- TTX
- tetrodotoxin.
REFERENCES
- 1. Dent E. W., Gertler F. B. (2003) Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40, 209–227 [DOI] [PubMed] [Google Scholar]
- 2. Sarmiere P. D., Bamburg J. R. (2004) Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J. Neurobiol. 58, 103–117 [DOI] [PubMed] [Google Scholar]
- 3. Bernard O. (2007) Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 39, 1071–1076 [DOI] [PubMed] [Google Scholar]
- 4. Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D. S., Condeelis J. S. (2004) Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746 [DOI] [PubMed] [Google Scholar]
- 5. Andrianantoandro E., Pollard T. D. (2006) Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 [DOI] [PubMed] [Google Scholar]
- 6. Chan C., Beltzner C. C., Pollard T. D. (2009) Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. (1997) Actin-depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arber S., Barbayannis F. A., Hanser H., Schneider C., Stanyon C. A., Bernard O., Caroni P. (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805–809 [DOI] [PubMed] [Google Scholar]
- 9. Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809–812 [DOI] [PubMed] [Google Scholar]
- 10. Gohla A., Birkenfeld J., Bokoch G. M. (2005) Chronophin, a novel HAD-type serine-protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7, 21–29 [DOI] [PubMed] [Google Scholar]
- 11. Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. (2002) Control of actin reorganization by slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108, 233–246 [DOI] [PubMed] [Google Scholar]
- 12. Huang T. Y., DerMardirossian C., Bokoch G. M. (2006) Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18, 26–31 [DOI] [PubMed] [Google Scholar]
- 13. Huang T. Y., Minamide L. S., Bamburg J. R., Bokoch G. M. (2008) Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev. Cell 15, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Montani L., Gerrits B., Gehrig P., Kempf A., Dimou L., Wollscheid B., Schwab M. E. (2009) Neuronal Nogo-A modulates growth cone motility via Rho-GTP/LIMK1/cofilin in the unlesioned adult nervous system. J. Biol. Chem. 284, 10793–10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Endo M., Ohashi K., Mizuno K. (2007) LIM kinase and slingshot are critical for neurite extension. J. Biol. Chem. 282, 13692–13702 [DOI] [PubMed] [Google Scholar]
- 16. Meberg P. J., Bamburg J. R. (2000) Increase in neurite outgrowth mediated by overexpression of actin-depolymerizing factor. J. Neurosci. 20, 2459–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuhn T. B., Meberg P. J., Brown M. D., Bernstein B. W., Minamide L. S., Jensen J. R., Okada K., Soda E. A., Bamburg J. R. (2000) Regulating actin dynamics in neuronal growth cones by ADF/cofilin and Rho family GTPases. J. Neurobiol. 44, 126–144 [PubMed] [Google Scholar]
- 18. Wen Z., Han L., Bamburg J. R., Shim S., Ming G. L., Zheng J. Q. (2007) BMP gradients steer nerve growth cones by a balancing act of LIM kinase and slingshot phosphatase on ADF/cofilin. J. Cell Biol. 178, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee C. W., Han J., Bamburg J. R., Han L., Lynn R., Zheng J. Q. (2009) Regulation of acetylcholine receptor clustering by ADF/cofilin-directed vesicular trafficking. Nat. Neurosci. 12, 848–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu J., Lee C. W., Fan Y., Komlos D., Tang X., Sun C., Yu K., Hartzell H. C., Chen G., Bamburg J. R., Zheng J. Q. (2010) ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat. Neurosci. 13, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuen E. Y., Liu W., Kafri T., van Praag H., Yan Z. (2010) Regulation of AMPA receptor channels and synaptic plasticity by cofilin phosphatase slingshot in cortical neurons. J. Physiol. 588, 2361–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meng Y., Zhang Y., Tregoubov V., Janus C., Cruz L., Jackson M., Lu W. Y., MacDonald J. F., Wang J. Y., Falls D. L., Jia Z. (2002) Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 35, 121–133 [DOI] [PubMed] [Google Scholar]
- 23. Zhou Q., Homma K. J., Poo M. M. (2004) Shrinkage of dendritic spines associated with long term depression of hippocampal synapses. Neuron 44, 749–757 [DOI] [PubMed] [Google Scholar]
- 24. Tavazoie S. F., Alvarez V. A., Ridenour D. A., Kwiatkowski D. J., Sabatini B. L. (2005) Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat. Neurosci. 8, 1727–1734 [DOI] [PubMed] [Google Scholar]
- 25. Hotulainen P., Llano O., Smirnov S., Tanhuanpää K., Faix J., Rivera C., Lappalainen P. (2009) Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J. Cell Biol. 185, 323–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jang D. H., Han J. H., Lee S. H., Lee Y. S., Park H., Lee S. H., Kim H., Kaang B. K. (2005) Cofilin expression induces cofilin-actin rod formation and disrupts synaptic structure and function in Aplysia synapses. Proc. Natl. Acad. Sci. U.S.A. 102, 16072–16077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Minamide L. S., Striegl A. M., Boyle J. A., Meberg P. J., Bamburg J. R. (2000) Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2, 628–636 [DOI] [PubMed] [Google Scholar]
- 28. Maloney M. T., Minamide L. S., Kinley A. W., Boyle J. A., Bamburg J. R. (2005) β-Secretase-cleaved amyloid precursor protein accumulates at actin inclusions induced in neurons by stress or amyloid β: a feedforward mechanism for Alzheimer disease. J. Neurosci. 25, 11313–11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao L., Ma Q. L., Calon F., Harris-White M. E., Yang F., Lim G. P., Morihara T., Ubeda O. J., Ambegaokar S., Hansen J. E., Weisbart R. H., Teter B., Frautschy S. A., Cole G. M. (2006) Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat. Neurosci. 9, 234–242 [DOI] [PubMed] [Google Scholar]
- 30. Fulga T. A., Elson-Schwab I., Khurana V., Steinhilb M. L., Spires T. L., Hyman B. T., Feany M. B. (2007) Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol. 9, 139–148 [DOI] [PubMed] [Google Scholar]
- 31. Rathke P. C., Seib E., Weber K., Osborn M., Franke W. W. (1977) Rod-like elements from actin-containing microfilament bundles observed in cultured cells after treatment with cytochalasin A (CA). Exp. Cell Res. 105, 253–262 [DOI] [PubMed] [Google Scholar]
- 32. Nishida E., Iida K., Yonezawa N., Koyasu S., Yahara I., Sakai H. (1987) Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 84, 5262–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davis R. C., Maloney M. T., Minamide L. S., Flynn K. C., Stonebraker M. A., Bamburg J. R. (2009) Mapping cofilin-actin rods in stressed hippocampal slices and the role of cdc42 in amyloid-β-induced rods. J. Alzheimers Dis. 18, 35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis R. C., Marsden I. T., Maloney M. T., Minamide L. S., Podlisny M., Selkoe D. J., Bamburg J. R. (2011) Amyloid β dimers/trimers potently induce cofilin-actin rods that are inhibited by maintaining cofilin-phosphorylation. Mol. Neurodegener. 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim J. S., Huang T. Y., Bokoch G. M. (2009) Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol. Biol. Cell 20, 2650–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao J., Hennessey T., Flynt A., Lai E., Beal M. F., Lin M. T. (2010) MicroRNA-related cofilin abnormality in Alzheimer disease. PLoS ONE 5, e15546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whiteman I. T., Gervasio O. L., Cullen K. M., Guillemin G. J., Jeong E. V., Witting P. K., Antao S. T., Minamide L. S., Bamburg J. R., Goldsbury C. (2009) Activated actin-depolymerizing factor/cofilin sequesters phosphorylated microtubule-associated protein during the assembly of Alzheimer-like neuritic cytoskeletal striations. J. Neurosci. 29, 12994–13005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao J., Qi J., Chen G. (2006) Actin-dependent activation of presynaptic silent synapses contributes to long term synaptic plasticity in developing hippocampal neurons. J. Neurosci. 26, 8137–8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deng L., Yao J., Fang C., Dong N., Luscher B., Chen G. (2007) Sequential postsynaptic maturation governs the temporal order of GABAergic and glutamatergic synaptogenesis in rat embryonic cultures. J. Neurosci. 27, 10860–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang M., Chen G. (2006) High Ca2+-phosphate transfection efficiency in low density neuronal cultures. Nat. Protocols 1, 695–700 [DOI] [PubMed] [Google Scholar]
- 41. Bernstein B. W., Chen H., Boyle J. A., Bamburg J. R. (2006) Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am. J. Physiol. Cell Physiol. 291, C828–839 [DOI] [PubMed] [Google Scholar]
- 42. Wilson J. M., de Hoop M., Zorzi N., Toh B. H., Dotti C. G., Parton R. G. (2000) EEA1, a tethering protein of the early sorting endosome, shows a polarized distribution in hippocampal neurons, epithelial cells, and fibroblasts. Mol. Biol. Cell 11, 2657–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bucci C., Lütcke A., Steele-Mortimer O., Olkkonen V. M., Dupree P., Chiariello M., Bruni C. B., Simons K., Zerial M. (1995) Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 366, 65–71 [DOI] [PubMed] [Google Scholar]
- 44. Masliah E., Mallory M., Hansen L., DeTeresa R., Terry R. D. (1993) Quantitative synaptic alterations in the human neocortex during normal aging. Neurology 43, 192–197 [DOI] [PubMed] [Google Scholar]
- 45. von Blume J., Duran J. M., Forlanelli E., Alleaume A. M., Egorov M., Polishchuk R., Molina H., Malhotra V. (2009) Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J. Cell Biol. 187, 1055–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. von Blume J., Alleaume A. M., Cantero-Recasens G., Curwin A., Carreras-Sureda A., Zimmermann T., van Galen J., Wakana Y., Valverde M. A., Malhotra V. (2011) ADF/cofilin regulates secretory cargo sorting at the TGN via the Ca2+ ATPase SPCA1. Dev. Cell 20, 652–662 [DOI] [PubMed] [Google Scholar]
- 47. Rosso S., Bollati F., Bisbal M., Peretti D., Sumi T., Nakamura T., Quiroga S., Ferreira A., Cáceres A. (2004) LIMK1 regulates Golgi dynamics, traffic of Golgi-derived vesicles, and process extension in primary cultured neurons. Mol. Biol. Cell 15, 3433–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chua B. T., Volbracht C., Tan K. O., Li R., Yu V. C., Li P. (2003) Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 5, 1083–1089 [DOI] [PubMed] [Google Scholar]
- 49. Klamt F., Zdanov S., Levine R. L., Pariser A., Zhang Y., Zhang B., Yu L. R., Veenstra T. D., Shacter E. (2009) Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol. 11, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schon E. A., Przedborski S. (2011) Mitochondria: the next (neurode) generation. Neuron 70, 1033–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Müller W. E., Eckert A., Kurz C., Eckert G. P., Leuner K. (2010) Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer's disease–therapeutic aspects. Mol. Neurobiol. 41, 159–171 [DOI] [PubMed] [Google Scholar]
- 52. Garvalov B. K., Flynn K. C., Neukirchen D., Meyn L., Teusch N., Wu X., Brakebusch C., Bamburg J. R., Bradke F. (2007) Cdc42 regulates cofilin during the establishment of neuronal polarity. J. Neurosci. 27, 13117–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen L. Y., Rex C. S., Casale M. S., Gall C. M., Lynch G. (2007) Changes in synaptic morphology accompany actin signaling during LTP. J. Neurosci. 27, 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi Y., Pontrello C. G., DeFea K. A., Reichardt L. F., Ethell I. M. (2009) Focal adhesion kinase acts downstream of EphB receptors to maintain mature dendritic spines by regulating cofilin activity. J. Neurosci. 29, 8129–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Racz B., Weinberg R. J. (2006) Spatial organization of cofilin in dendritic spines. Neuroscience 138, 447–456 [DOI] [PubMed] [Google Scholar]
- 56. Matus A. (2005) Growth of dendritic spines: a continuing story. Curr. Opin. Neurobiol. 15, 67–72 [DOI] [PubMed] [Google Scholar]
- 57. Hotulainen P., Hoogenraad C. C. (2010) Actin in dendritic spines: connecting dynamics to function. J. Cell Biol. 189, 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Okamoto K., Nagai T., Miyawaki A., Hayashi Y. (2004) Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7, 1104–1112 [DOI] [PubMed] [Google Scholar]
- 59. Rex C. S., Lin C. Y., Kramár E. A., Chen L. Y., Gall C. M., Lynch G. (2007) Brain-derived neurotrophic factor promotes long term potentiation-related cytoskeletal changes in adult hippocampus. J. Neurosci. 27, 3017–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rust M. B., Gurniak C. B., Renner M., Vara H., Morando L., Görlich A., Sassoè-Pognetto M., Banchaabouchi M. A., Giustetto M., Triller A., Choquet D., Witke W. (2010) Learning, AMPA receptor mobility and synaptic plasticity depend on n-cofilin-mediated actin dynamics. EMBO J. 29, 1889–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Futai K., Kim M. J., Hashikawa T., Scheiffele P., Sheng M., Hayashi Y. (2007) Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat. Neurosci. 10, 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gearing M., Juncos J. L., Procaccio V., Gutekunst C. A., Marino-Rodriguez E. M., Gyure K. A., Ono S., Santoianni R., Krawiecki N. S., Wallace D. C., Wainer B. H. (2002) Aggregation of actin and cofilin in identical twins with juvenile-onset dystonia. Ann. Neurol. 52, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Culebras A., Feldman R. G., Merk F. B. (1973) Cytoplasmic inclusion bodies within neurons of the thalamus in myotonic dystrophy: a light and electron microscope study. J. Neurol. Sci. 19, 319–329 [DOI] [PubMed] [Google Scholar]
- 64. Ono S., Inoue K., Mannen T., Kanda F., Jinnai K., Takahashi K. (1987) Neuropathological changes of the brain in myotonic dystrophy: some new observations. J. Neurol. Sci. 81, 301–320 [DOI] [PubMed] [Google Scholar]
- 65. Mattson M. P. (2003) Excitotoxic and excitoprotective mechanisms: abundant targets for the prevention and treatment of neurodegenerative disorders. Neuromolecular Med. 3, 65–94 [DOI] [PubMed] [Google Scholar]
- 66. Navarro A., Boveris A. (2007) The mitochondrial energy transduction system and the aging process. Am. J. Physiol. Cell Physiol. 292, C670–686 [DOI] [PubMed] [Google Scholar]
- 67. Morrison J. H., Hof P. R. (1997) Life and death of neurons in the aging brain. Science 278, 412–419 [DOI] [PubMed] [Google Scholar]