Background: YidC is required for the insertion of membrane proteins.
Results: YidC meets its substrate Pf3 coat at multiple contact sites in four of six transmembrane helices, each on one helical face.
Conclusion: The contact sites are important for substrate binding and transmembrane insertion.
Significance: YidC provides a hydrophobic surface and a surrounding for the inserting substrate proteins.
Keywords: Bacteriophage, Cysteine-mediated Cross-linking, Membrane Biogenesis, Protein Cross-linking, Protein Translocation, Pf3 Coat Protein, YidC, Hydrophobic Platform, Membrane Insertion
Abstract
The membrane insertase YidC inserts newly synthesized proteins into the plasma membrane. While defects in YidC homologs in animals and plants cause diseases, YidC in bacteria is essential for life. Membrane insertion and assembly of ATP synthase and respiratory complexes is catalyzed by YidC. To investigate how YidC interacts with membrane-inserting proteins, we generated single cysteine mutants in YidC and in the model substrate Pf3 coat protein. The single cysteine mutants were expressed and analyzed for disulfide formation during 30 s of synthesis. The results show that the substrate contacts different YidC residues in four of the six transmembrane regions. The residues are located either in the region of the inner leaflet, in the center, as well as in the periplasmic leaflet, consistent with the hypothesis that YidC presents a hydrophobic platform for inserting membrane proteins. In a YidC mutant where most of the contacting residues were mutated to serines, YidC function was severely disturbed and no longer active in a complementation test, suggesting that the residues are important for function. In addition, a Pf3 mutant with a defect in membrane insertion was deficient to contact the periplasmic residues of YidC.
Introduction
The membrane insertase YidC of Escherichia coli is required for the biogenesis of respiratory complexes, ATP synthase, and other membrane proteins (1, 2). When YidC is depleted from bacterial cells, growth ceases, and the cells die, revealing that it is an essential protein (3, 4). In the absence of YidC, the subunit c protein of the Fo part of the ATP synthase has been shown to accumulate in the cytoplasm because its membrane insertion is blocked (5, 6). Also a number of other proteins, such as the CyoA protein of the terminal oxidase complex (7), the mechanosensitive channel protein MscL (8), and the coat proteins of filamentous phage Pf3 and M13 require YidC for their membrane insertion. All these substrate proteins comprise less than 200 amino acids and contain periplasmic regions of less than 30 residues. The best studied substrate is the 44 amino acid-long Pf3 coat protein. After its synthesis, it binds reversibly to YidC and is membrane-inserted within a few minutes. The binding of purified Pf3 coat protein has been investigated by its ability to quench the 1-anilinonaphthalene-8-sulfonate (ANS)-labeled YidC protein in detergent (9) and also to quench the membrane-integrated insertase (10). The insertion of the Pf3 coat protein had been studied in a reconstituted system with proteoliposomes that only contained YidC (11). When limiting amounts of YidC were present in the proteoliposomes, up to 150 Pf3 coat proteins were inserted per YidC molecule, suggesting that YidC is catalytically driving the membrane insertion of proteins.
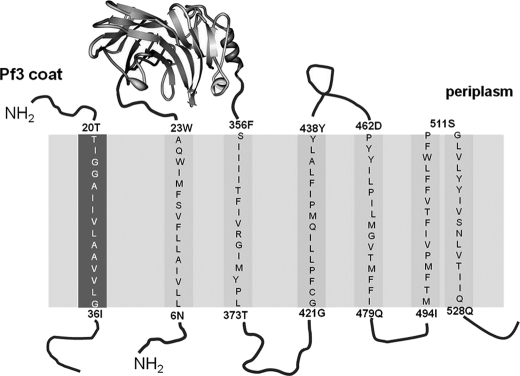
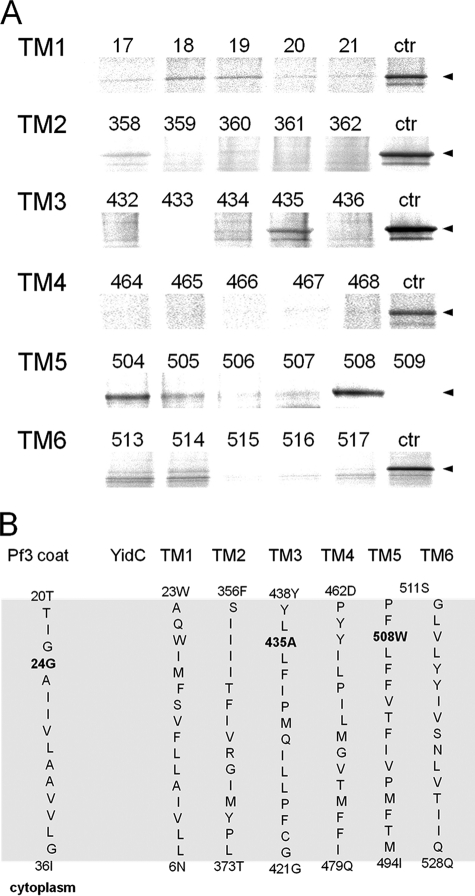
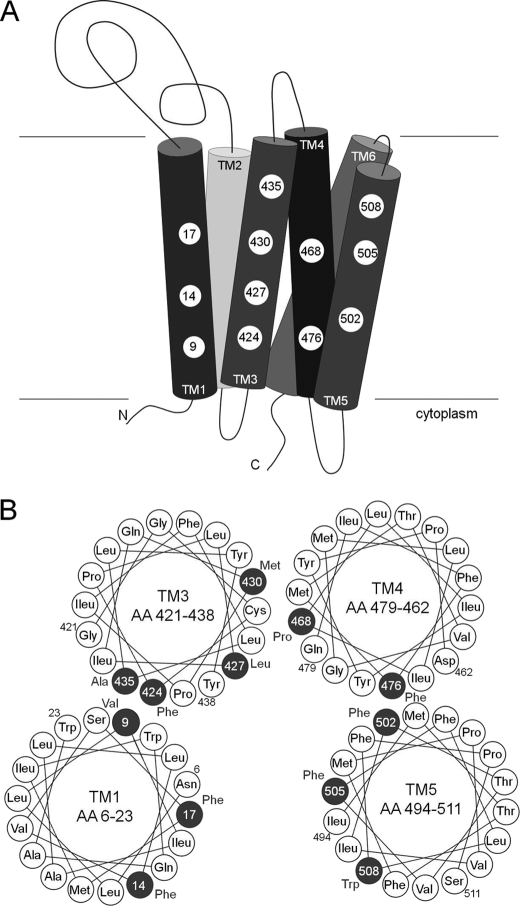
YidC is a six-spanning membrane protein of 548 amino acids (12). The structure of the large periplasmic domain between the first two transmembrane (TM)2 regions has recently been solved to high resolution (Fig. 1) (13, 14). The homologues found in mitochondria and chloroplasts lack the first transmembrane region and most of the first periplasmic domain of 330 amino acid residues. Accordingly, the deletion of 300 residues from the periplasmic domain of the YidC from E. coli retained a still functional YidC mutant deficient for the interaction with SecF (15). When the first transmembrane region of YidC was replaced by a cleavable signal peptide, it maintained activity (16). However, the transmembrane regions were shown to be essential for the function of YidC. Deletions of TM3/4 and TM5/6 resulted in inactive YidC (17). Also, the replacement of residues 418 to 425 by serines did not retain activity. Surprisingly, a number of substitutions of residues in the transmembrane regions with serines or alanines had no noticeable effect (17). Presumably, a number of residues in the hydrophobic transmembrane regions of YidC are involved in substrate binding and are required during the insertion process.
FIGURE 1.
Schematic representation of YidC and the Pf3 coat protein and their topology in the inner membrane of E. coli. All indicated residues (single letter abbreviation) in the predicted membrane spanning segments (TM) of the Pf3 coat protein (left column) and YidC protein (right columns) were replaced by single cysteine residues. The gray area represents the membrane region.
The transmembrane regions are the most conserved parts of the YidC protein. Particularly, this was observed for the sequence of TM2 and TM3 throughout, and for the periplasmic portions of TM5 and TM6 (12). Single cysteines in TM3 were shown to be involved in substrate contacts in vivo (18) and in vitro (19). Here, we have extended this study and systematically scanned all six transmembrane regions of YidC for contacts with a substrate by single cysteine mutants. The results show that TM1, TM3, and TM4 provide nine major contacting residues. Three additional contacts are located in the periplasmic half of TM5. A substrate mutant that is deficient for membrane insertion was able to contact the YidC residues close to the cytoplasm but failed to contact the residues in the periplasmic half of the transmembrane region.
EXPERIMENTAL PROCEDURES
Bacterial Growth, Pulse Labeling, and Disulfide Formation
E. coli MK6 (18) was transformed with pMS-Pf3 coat and pACYC-YidC and then grown in M9 minimal media in the presence of ampicillin (final concentration, 100 μg/ml) and chloramphenicol (final concentration, 25 μg/ml). In the MK6 strain, the promoter of the yidC gene had been exchanged with the araC-araBAD promoter cassette. Depletion of the chromosomally encoded YidC was achieved by growth in the presence of 0.2% glucose for 3 h at 37 °C. Unless indicated otherwise, the cells were treated with 1 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG) at A600 nm 0.2 for 10 min to induce expression of the Pf3 coat and pulse-labeled for the indicated time by the addition of [35S]methionine. The cells were chilled on ice and, if indicated, treated with 1 mm freshly prepared copper 1,10-phenanthroline for 3 min (20). The samples were precipitated with 20% TCA, washed with acetone, and resuspended in 2% SDS in Tris buffer (pH 8). Immunoprecipitation was done as described (21) using antiserum to Pf3 phage or to YidC. The samples were analyzed by SDS-PAGE and phosphor imaging.
Generation of Single Cysteine Mutants
The site-directed mutations in the Pf3 coat and YidC were generated by the QuikChange method with minor modifications (22). All mutations were verified by DNA sequencing of the entire gene.
Protease Mapping with Proteinase K
E. coli BL-21 (23) cells bearing the respective plasmids were grown overnight with 0.2% arabinose and diluted 1:100 into fresh minimal medium with 0.2% glucose and amino acids but lacking methionine. After 2 h of growth, the cells were washed and resuspended in M9 medium with 0.2% glucose. Expression of the Pf3 coat protein was induced with 1 mm IPTG for 10 min and pulse-labeled with [35S]methionine (10 μCi/ml) for 30 s. To generate spheroplasts, the pulse-labeled cells were collected by centrifugation at 4 °C and resuspended in 40% sucrose and 33 mm Tris-HCl (pH 8.0). Lysozyme (5 μg/ml) and 1 mm EDTA (pH 8.0) were added and kept on ice for 20 min. Where indicated, proteinase K was added (0.5 mg/ml) and incubated on ice for 1 h. Immunoprecipitation to Pf3, YidC, OmpA, and GroEL was performed as described previously (22). The samples were acid-precipitated and analyzed by SDS-PAGE.
Complementation with YidC-depleted Cells
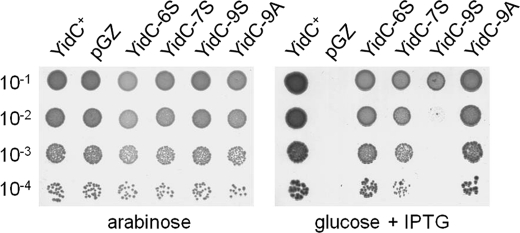
E. coli MK6 cells bearing the respective plasmids were grown overnight in Luria broth (LB) medium with 0.2% arabinose and 25 μg/ml chloramphenicol and then diluted 1:100 into LB medium without arabinose. After 2h of growth the cells were serially diluted in 1:10 steps and spotted on LB plates containing 0.2% arabinose or 0.2% glucose, respectively. Where indicated, 1 mm IPTG was present. The plates were incubated overnight at 37 °C.
RESULTS
The Pf3 Coat Protein Contacts YidC in the Center of TM1, TM3, TM4, and TM5
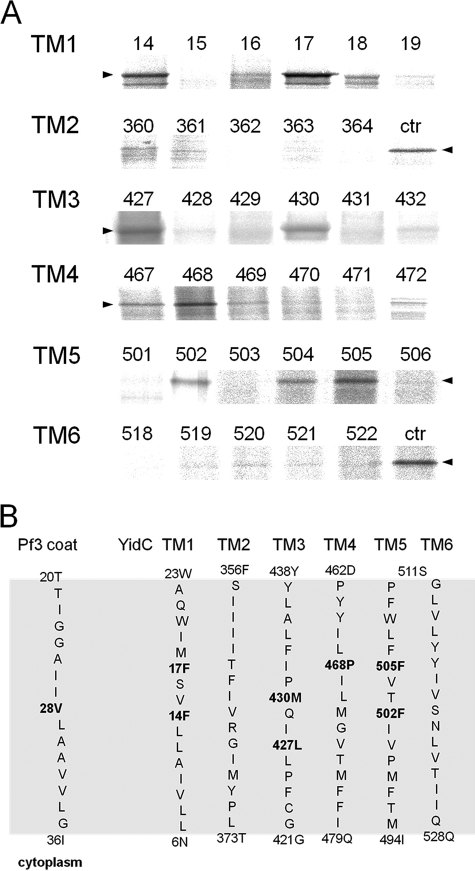
To follow the insertion process of the Pf3 coat protein, single cysteine mutants were generated in the Pf3 coat protein and in YidC (Fig. 1). These were placed into each of the transmembrane regions covering all the membrane-spanning positions. In total, more than 100 single cysteine mutants were collected. E. coli MK6 cells were transformed with a plasmid coding for one of the Pf3 coat protein single-cysteine mutants and with a second plasmid coding for one of the YidC single cysteine mutants. The MK6 cells that have the chromosomal yidC under control of the araBAD promoter (18) were grown in glucose media to deplete the YidC wild-type and allow the plasmid-borne expression of YidC. All mutant proteins showed a comparable expression level. To map the contact sites during the insertion event, the two proteins were coexpressed in E. coli MK6 cells and pulse-labeled with [35S]methionine for 30 s and treated with oxidant for 3 min. Then, the proteins were acid-precipitated and immunoprecipitated with an antibody to Pf3 coat protein. When the single cysteine residue was at position 28 of the Pf3 coat protein, which is in the center of the TM region, cross-linked YidC-Pf3 complexes were detected with certain cysteine mutants of YidC (Fig. 2). These mutants had their cysteine residue in the center of TM1 at position 14 and 17, in TM3 at position 427 and 430, in TM4 at position 468, and in TM5 at positions 502 and 504/505. The neighboring positions showed significantly weaker or no cross-links. No strong contacts were detected in TM2 and TM6. To evaluate exactly the level of cross-linking, a positive control with cells coexpressing YidC 430C and Pf3–28C was added and loaded on the same gel.
FIGURE 2.
Disulfide complexes of inserting Pf3-28C with YidC cysteine mutants in the center of TM1 to TM6. A, MK6 cells bearing the plasmid pMS-Pf3-28C and a second plasmid, pACYC, encoding the respective single cysteine YidC mutant, were grown in the presence of 0.2% glucose to deplete the cells from the chromosomally encoded YidC. The cells were induced with 1 mm IPTG for 10 min and pulse-labeled with [35S]methionine for 30 s. The cells were put on ice and treated with 1 mm copper phenanthroline. The cells were TCA-precipitated, immunoprecipitated with antibody to the Pf3 virus, and analyzed on non-reducing SDS-PAGE by phosphor imaging. For the control lanes, Pf3-28C and YidC430C (ctr) were coexpressed and treated as described above. The arrowheads depict the position of YidC-Pf3 complex. B, positions of the Pf3-28C contacts with YidC in the six transmembrane segments. The residues that provided stable disulfide cross-links when mutated to a cysteine residue are shown in boldface. The contacts were found in the center of TM1, 3, 4, and 5.
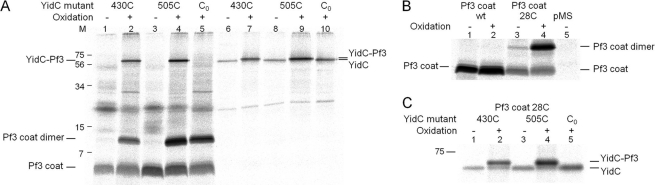
The Pf3 coat protein contacts with YidC430 and 505 were analyzed in more detail (Fig. 3). The pulse-labeled samples were immunoprecipitated either with Pf3 (Fig. 3A, lanes 1–5) or with YidC antiserum (lanes 6–10) and analyzed by polyacrylamide electrophoresis, respectively. The contacts only occurred under oxidative conditions and when a cysteine is present on both protein partners (Fig. 3, A and C). Under oxidative conditions, most of the Pf3 coat protein was present as dimers, but some monomers were also detectable. In addition, a significant proportion was found at about 65 kDa corresponding to a Pf3-YidC cross-link product. Clearly, the Pf3-YidC cross-link product was immunoprecipitated with both antibodies and migrated a little slower (Fig. 3C, lanes 2-4) than the cysteine-less YidC on SDS-PAGE (lane 5). When the cysteine-less YidC was coexpressed, no YidC-cross-linked product was detectable (Fig. 3A, lane 5). However, the formation of Pf3 coat protein dimers was unaffected. The contacts we observed with Pf3–28C were all at positions of the YidC protein that were predicted, on the basis of the hydrophobicity profile, to be in the center of the bilayer (Fig. 2, lower panel), in TM1, 3, 4, and 5.
FIGURE 3.
Pf3-YidC disulfide complexes are recognized by antibodies to Pf3 and to YidC. A, coexpression of Pf3-28C and YidC with a single cysteine at residue 430 (lanes 1, 2, 6, and 7) and 505 (lanes 3, 4, 8, and 9) were analyzed as described for Fig. 2, except that in lanes 1, 3, 6, and 8 no copper phenanthroline was added. Lanes 1-5 were immunoprecipitated with an antibody to Pf3. Lanes 6-10 were immunoprecipitated with an antibody to YidC. As a control, the cysteine-less YidC mutant (lanes 5 and 10) was expressed. The Pf3 coat protein was found as a monomer, dimer, and cross-linked with YidC. Molecular weight markers are indicated in kDa at the left margin (lane M). B, E. coli MK6 cells expressing the wild-type Pf3 coat protein (lanes 1 and 2) and Pf3–28C (lanes 3 and 4) were pulse-labeled as described above and treated with copper phenanthroline (lanes 2 and 4). The empty vector is shown as a control (lane 5). C, for better separation, lanes 6-10 of Fig. 3A were applied to a long PAGE.
Pf3 Contacts in the YidC Region Located in the Inner and Periplasmic Leaflet
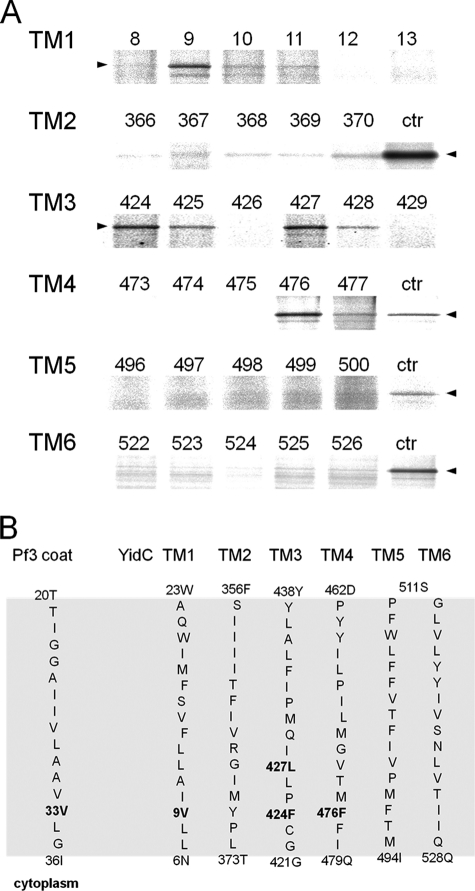
We then tested which YidC residues were contacted by a Pf3 coat protein that had its single cysteine at position 33, predicted to localize in the inner leaflet of the membrane (Fig. 4). MK6 cells bearing the plasmids for expressing a single cysteine mutant of YidC and a second plasmid expressing the Pf3 coat protein with a cysteine at position 33, respectively, were pulse-labeled for 30 s and analyzed for disulfide formation. YidC-Pf3 cross-link products were found to the YidC-TM1 (position 9), to TM3 (positions 424 and 427), and TM4 (position 476). Weak contacts were observed in TM3 at positions 425 and 428 that are next to the strong cross-linking signals and also in TM2 at positions 367 and 370. All these contacts are located in YidC at the predicted inner leaflet of the membrane bilayer (Fig. 4, lower panel). However, no strong contacts were observed with TM5 and TM6 in the region of the inner leaflet on the basis of the comparison of the background signals with a control culture (coexpression of Pf3–28C and YidC-505C).
FIGURE 4.
Inserting the Pf3-33C coat protein contacts YidC transmembrane residues located in the inner leaflet. A, E. coli MK6 cells bearing the plasmid pMS-Pf3-33C and a second plasmid encoding the respective single cysteine YidC mutant were grown and treated as described in the legend to Fig. 2. Cross-links were detected by immunoprecipitations with an antibody to Pf3. For the control lanes, Pf3-28C and YidC505C (ctr) were coexpressed and treated as described above. The arrowheads depict the position of the YidC-Pf3 complex. B, positions of the Pf3-33C contacts with YidC in the six transmembrane segments. The residues that provided stable disulfide cross-links when mutated to a cysteine residue are shown in boldface. The contacts were found in the inner half of TM1, 3, and 4.
Next, the YidC residues predicted to be located in the periplasmic leaflet of the transmembrane regions were investigated. When a Pf3 coat protein with a cysteine residue at position 24 was coexpressed, contacts were observed for YidC-TM3 at position 435 and for YidC-TM5 at 504 and 508 (Fig. 5A). No major contacts were found with TM2 and TM4, as compared with the background. Weak interactions were detected with the YidC residue 18, 19 (in TM1) and 514 (in TM6). We noticed that in general, the YidC contacts to Pf3–24C were weaker than to Pf3–33C and Pf3–28C, presumably because the periplasmic contacts may occur for a shorter time.
FIGURE 5.
Inserting the Pf3-24C coat protein contacts YidC in the transmembrane residues of the outer leaflet. A, E. coli MK6 cells bearing the plasmid pMS-Pf3-24C and a second plasmid encoding the respective single cysteine YidC mutant were grown and treated as described in the legend to Fig. 2. Cross-links were detected by immunoprecipitations with an antibody to Pf3. For the control lanes, Pf3-28C and YidC505C (ctr) were coexpressed and treated as described above. The arrowheads depict the position of the YidC-Pf3 complex. B, positions of the Pf3-24C contacts with YidC in the six transmembrane segments. The residues that provided stable disulfide cross-links when mutated to a cysteine residue are shown in boldface. The contacts were found in the outer half of TM 3 and 5.
Taken together, we have observed 12 strong contact sites in TM1, 3, 4, and 5. They were often flanked by residues that also showed a contact to the substrate but, mostly, much weaker. This was the case for the flanking residues of residues 9, 17, 424, 427, 468, 477, and 505. The residues 504 and 505 gave about the same signal to Pf3–24C and Pf3–28C (Figs. 2 and 5), suggesting that both residues are equally contacting the substrate.
Contacts of Insertion-deficient Pf3 Coat Protein Mutants with YidC
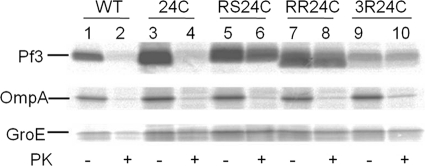
To study the different stages of YidC-substrate interaction, experiments were performed with Pf3 coat protein mutants that are deficient for membrane insertion (24). The Pf3-RR mutant has two arginines substituted for the amino acids at positions 17 and 18, whereas the Pf3-RS mutant has an arginine and a serine at these positions, respectively. A third mutant, Pf3-3R, is identical to Pf3-RR but has an additional arginine at position 7 replacing a glutamate. Single cysteines were introduced in all these mutants at position 24 to study their interaction with YidC and their insertion into the membrane. First, the membrane insertion of the mutants was studied in vivo by proteinase mapping (Fig. 6). E. coli BL-21 cells expressing the respective Pf3 coat protein mutant were grown in minimal medium lacking methionine to a density of 2 × 108 cells/ml. [35S]methionine was added for 3 min, and the cells were analyzed for Pf3 coat protein insertion. Although the wild-type Pf3 coat was accessible to the protease added to the periplasmic side (lanes 2 and 4), the mutants Pf3-RS, Pf3-RR, and Pf3-3R were not digested by the protease demonstrating that they are inhibited for membrane insertion. For a control, the cytoplasmic protein GroEL and OmpA were immunoprecipitated, and we verified that the cells remained intact and the periplasm was accessible to the outside-added protease.
FIGURE 6.
Membrane insertion of Pf3-RS, Pf3-RR, and Pf3-3R is inhibited. E. coli BL-21 cells expressing Pf3, Pf3-24C, Pf3-24C-RS, Pf3-24C-RR, or Pf3-24C-3R, respectively, were pulse-labeled for 3 min with [35S]methionine. The outer membranes of the cells were opened by osmotic shock, and proteinase K (+ PK lanes) was added to the outside of the cells, and they were incubated on ice for 1 h. The samples were immunoprecipitated with antiserum to Pf3, GroEL, or OmpA, respectively, and analyzed by PAGE and phosphor imaging.
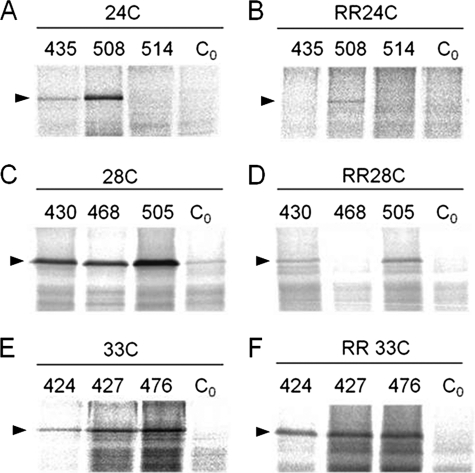
The Pf3-RR mutant was then coexpressed in E. coli MK6 with a single cysteine mutant of YidC and analyzed for contacts to YidC at the respective positions (Fig. 7). When the cysteine at position 24 in the Pf3 wild-type (Fig. 7A) was compared with Pf3-RR-24C (B), the contacts to the YidC residues at 435 and 508 were substantially reduced in the insertion-deficient Pf3-RR mutant. These residues are in the periplasmic membrane portion of YidC. Similarly, the residues in the center of the membrane at 430, 468, and 505 were not or only weakly cross-linked with Pf3-RR with a cysteine at position 28 (Fig. 7D). However, the YidC residues 424, 427, and 476 located in the inner leaflet were not inhibited for contacting Pf3-RR-33C (compare Fig. 7, E and F).
FIGURE 7.
Pf3-RR is blocked for contacting the YidC residues in the outer leaflet. E. coli MK6 cells expressing Pf3-24C (A), Pf3-24C-RR (B), Pf3-28C (C), Pf3-28C-RR (D), Pf3-33C (E), or Pf3-33C-RR (F) were coexpressed with the respective YidC mutants to analyze close contacts by disulfide cross-linking. The cells were labeled with [35S]methionine for 30 s and immunoprecipitated with antiserum to Pf3. As a control, the cysteine-less YidC (Co) was coexpressed. The samples were analyzed by PAGE and phosphor imaging.
The Pf3–3R mutant containing three arginine residues in the amino terminal domain was also tested for YidC interaction (Fig. 8). When this Pf3-3R mutant had a cysteine at position 24, 28, or 33, it was inhibited to interact with the residues 508, 505, and 476 of YidC located in the outer, central, and inner leaflet, respectively (Fig. 8A). When the samples were immunoprecipitated with antibodies to YidC, the shift of the band caused by a bound Pf3 protein was not observed when the 3R mutations were present in Pf3 (Fig. 8B), indicating that this Pf3 mutant is unable to interact with the YidC insertase.
FIGURE 8.
Pf3-3R is inhibited to contact YidC. E. coli MK6 cells expressing Pf3-24C (lane 1), Pf3-24C-3R (lane 2), Pf3-28C (lane 3), Pf3-28C-3R (lane 4), Pf3-33C (lane 5), or Pf3-33C-3R (lane 6) were coexpressed with YidC508C (lanes 1 and 2), YidC505C (lanes 3 and 4), or YidC476C (lanes 5 and 6), respectively. The cells were labeled with [35S]methionine for 30 s and immunoprecipitated with antiserum to Pf3 (A) or to YidC (B). The samples were analyzed by PAGE and phosphor imaging.
YidC Function Is Affected When Multiple Contact Sites Are Mutated
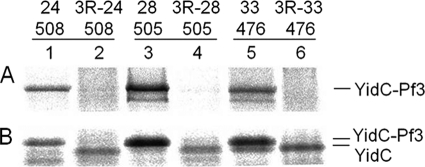
A further way to show that the contacting residues are important for function is to substitute all these residues into alanines or serines. We rationalized that for the YidC residues that are involved in substrate interactions are most likely not involved in helix-helix interactions important for the structure of the YidC protein. Hence, the structure of YidC should not be affected when only the contacting residues are mutated. However, it is expected that the substrate interaction is disturbed when the contacting YidC residues are changed to alanines and to serines, which are less hydrophobic than the corresponding residue in the wild-type. YidC function can be assessed in the MK6 strain where the chromosomal YidC is under control of the arabinose promoter. When YidC is depleted under glucose conditions, growth is inhibited unless a plasmid with a functional yidC gene is present and induced. In a mutant where the nine major contacting residues were substituted to serines, termed YidC-9S, YidC function was affected, and the plasmid-derived copy did no longer complement the chromosomally encoded gene (Fig. 9, right panel). In contrast, the YidC mutant with alanines at these positions still complemented the chromosomally encoded gene. Interestingly, cell growth was still observed when only the residues located at the central and periplasmic positions in the transmembrane segments were changed to serines (YidC-6S). Likewise, when the residue at the central and cytoplasmic positions were changed to serines (YidC-7S), growth was not affected. Taken together, we conclude that YidC function is only distorted when multiple contacting residues along the entire membrane region were changed.
FIGURE 9.
Cumulative mutations in the substrate contacting residues of YidC inhibit its function. The Pf3 contacting residues at 424, 427, 430, 435, 468, 476, 502, 505, and 508 were substituted to Ala (YidC-9A) and to Ser (YidC-9S) and expressed in MK6 under YidC-depleted conditions (in the presence of glucose, right panel) or non-depleted conditions (in the presence of arabinose, left panel). A mutant that has the contacting residues in the central and periplasmic leaflet (YidC-6S; 430, 435, 468, 502, 505, 508) and a mutant in the central and cytoplasmic positions mutated (YidC-7S; 424, 427, 430, 468, 476, 502, 505) were tested. 1 mm IPTG was added to induce the plasmid-directed synthesis of each YidC mutant. Serial dilutions of the cultures were spotted and incubated over night at 37 °C. For a control, cells transformed with the empty plasmid (pGZ) and cells encoding the wild-type YidC (YidC+) are shown.
DISCUSSION
The membrane insertase YidC catalyzes the insertion of the Pf3 coat protein into the inner membrane of E. coli (11). During this process, the Pf3 protein binds to YidC mainly by hydrophobic interactions (9) and induces a conformational change within YidC (10, 25). As a result, the Pf3 coat protein adopts a transmembrane conformation, exposing its N-terminal domain to the periplasm. Although the substrate binding step most likely involves the cytoplasmic residues of YidC, insertion of the TM region and translocation of the N-terminal region are likely to require other contacting residues of YidC. The molecular interactions of newly synthesized Pf3 coat protein were analyzed during 30 s after the onset of its synthesis. The data presented here show that multiple YidC-substrate contacts are observed involving the participation of most of the transmembrane segments of YidC. Only strong disulfide signals were considered. The weak signals were mostly observed at positions flanking the strong signals. Weak signals at other positions (e.g. at 358, 360, 370, and 514) may arise from a structural flexibility of YidC. Although TM3 showed contacts all across the predicted membrane-spanning α-helix, TM5 has three positions (502, 505, and 508), located in the central and outer leaflet, that make a contact with the newly expressed Pf3 coat protein. These contacts reveal a first three-dimensional picture of how the substrate is possibly binding YidC (Fig. 10). In the cytoplasmic leaflet, the substrate contacts TM1, 3, and 4 of YidC, whereas in the periplasmic leaflet, TM3 and 5 are contacted. This suggests that the residues at positions 424, 427, and 476 are primarily involved in the first substrate binding step, whereas residues 430, 435, 468, 502, 505, and 508 are required for translocation. The data that we have obtained with the substrate mutant Pf3-RR that is defective for membrane insertion are in support of this model. The insertion-deficient mutant RR-Pf3 was contacting 424, 427, and 476 in the cytoplasmic leaflet. However, the contacts to the central and periplasmic residues 430, 435, 468, 505 and 508 were substantially reduced (Fig. 7).
FIGURE 10.
Model of the YidC hydrophobic protein binding platform. A, the major contact sites in TM1, 3, 4, and 5 are depicted by a numbered circle referring to the amino acid residue position. B, helical wheel projection to illustrate the substrate contact sites of the YidC TM regions. The major contact sites are highlighted as dark circles with the respective position. Both ends of each TM segment are labeled with the position number outside the circle. Because TM2 and TM6 showed no strong contacts, they are not included in the model.
Intriguingly, the residues of YidC that are most conserved among the bacterial homologues are present in TM2, TM3, and in the C1 loop (12). Also, the periplasmic region of TM5 shows a high degree of homology (26). In these conserved TM regions of YidC we found seven major contacting residues. Surprisingly, several contacts were found within TM1, which is less conserved and is missing in Gram-positive bacteria as well as in the mitochondrial and thylakoidal homologues. This suggests that TM1 contributes to the YidC function, although it is not essential in the YidC family of proteins. The importance of the other TM regions for the function of YidC has been implicated from a number of site-directed mutants (17). The deletion of these TM regions inactivates the protein. Single residue substitutions had shown that only a few residues within the transmembrane regions are sensitive. Most of these locate in TM3, corroborating its central role for the insertase activity (17). Interestingly, the critical residues are not at the exact positions where our substrate contacts are localized but adjacent to them. Most likely, these residues are not involved in substrate interaction but rather in helix-helix contacts within YidC. One such contact was proposed for residue 423 in TM3 with TM2 (27). When the cysteine at 423 was substituted for an arginine, a cold-sensitive phenotype, was observed. Second-site suppressors identified residue T362E forming a possible charge pair with Arg-423, suggesting that TM2 and TM3 are close together. A further suppressor mutation of Arg-423 within TM3 was found at L426E in the same TM, indicating that residues 362, 423, and 426 are involved in helix-helix interaction and not in substrate binding. In accordance with this, we did not find Pf3-YidC contacts at 362, 423, or 426.
A previous study (19) had also identified TM3 of YidC as the substrate contact site. However, different residues were found to form disulfide contacts to the in vitro synthesized substrates, possibly because the cysteine mutants at positions 427 and 428 were not available. The Sec-dependent FtsQ protein was found to contact residues 425 and 426. With residue 425, we also found a weak interaction with Pf3, but residue 426 did not form a disulfide with Pf3. Another difference between that study and ours is that they had a nascent chain-arrested ribosome interacting with YidC in membrane vesicles in vitro. It is possible that the nascent chain could bind to the outside of YidC. Our experimental approach was in vivo, and to detect the formed disulfides we looked shortly (30-s pulse) after the addition of [35S]methionine. In this time span we expected to observe the naturally occurring contacts. In our results, the transmembrane segment TM3 showed four contacting residues at 424, 427, 430, and 435, corroborating that it is the most important YidC helix involved in substrate binding. When the residues are projected as a helical wheel, the contacting residues are on the same helical face (Fig. 10B). A similar result was obtained for TM1 and TM5, where more than one contact was observed. All substrate-contacting residues are hydrophobic. In particular, the extremely hydrophobic phenylalanine was found in six different contact sites. Because these phenylalanine residues are located in four different transmembrane helices, they together may constitute a stacking-like arrangement over the entire membrane-spanning region. In conclusion, the substrate-interacting surface of YidC is hydrophobic across the entire membrane span. We propose that the contacting residues form a hydrophobic platform that supports the transmembrane alignment of an incoming α-helix of the substrate. This proposed mechanism differs from the one of SecY, which is mostly hydrophilic with a small hydrophobic pore ring in its center (28).
One important question is whether the observed contacting residues are indeed of functional importance for membrane insertion. To answer this question, we studied Pf3 mutants that were deficient for membrane insertion. These mutants should fail to contact YidC at least at the periplasmic positions. The Pf3-RR mutant has an alanine residue at position 17 and an aspartic acid residue at position 18 substituted with arginines. Pf3-RR was not found to contact YidC at the periplasmic and central positions. For the Pf3-3R mutant, which had the aspartic acid at position 7 substituted to an arginine, in addition to the two arginines at 17 and 18, showed no contacts to YidC, not even to the cytoplasmic residue at 476 (Fig. 8). These data suggest that the Pf3-RR mutant protein still binds to the cytoplasmic side of YidC but cannot reach out to contact the more periplasmically located residues of YidC. Therefore, the translocation process of Pf3-RR across the membrane was perturbed, and the Pf3-3R mutant appeared to be blocked for both binding and translocation.
Previously, the TM regions of YidC had been analyzed by serine scanning, and thereby most of the contacting residues had been mutated to serine individually (17). All of these mutants turned out to be functional. However, in our study we found that when we combined the serine mutations of most major contacting sites, YidC function was severely affected (Fig. 9).
Taken together, our data show that YidC contacts its substrate Pf3 at multiple sites within the transmembrane domain (Fig. 10). This is in line with the proposal that YidC provides a hydrophobic platform supporting membrane insertion of proteins (29). The contacts are required for the insertase activity because the Pf3 cysteine mutants that are defective for membrane insertion were impaired to contact YidC.
This work was supported by Deutsche Forschungsgemeinschaft grant Ku749/6-1 (to A. K.).
- TM
- transmembrane
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- PK
- proteinase K.
REFERENCES
- 1. Facey S. J., Kuhn A. (2010) Biogenesis of bacterial inner-membrane proteins. Cell. Mol. Life Sci. 67, 2343–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Price C. E., Driessen A. J. (2010) Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim. Biophys. Acta 1803, 748–766 [DOI] [PubMed] [Google Scholar]
- 3. Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 4. Wang P., Kuhn A., Dalbey R. E. (2010) Global change of gene expression and cell physiology in YidC-depleted Escherichia coli. J. Bacteriol. 192, 2193–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yi L., Celebi N., Chen M., Dalbey R. E. (2004) Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279, 39260–39267 [DOI] [PubMed] [Google Scholar]
- 6. van der Laan M., Bechtluft P., Kol S., Nouwen N., Driessen A. J. (2004) F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Celebi N., Yi L., Facey S. J., Kuhn A., Dalbey R. E. (2006) Membrane biogenesis of subunit II of cytochrome bo oxidase. Contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357, 1428–1436 [DOI] [PubMed] [Google Scholar]
- 8. Facey S. J., Neugebauer S. A., Krauss S., Kuhn A. (2007) The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J. Mol. Biol. 365, 995–1004 [DOI] [PubMed] [Google Scholar]
- 9. Gerken U., Erhardt D., Schubert A., Bär G., Ghosh R., Kuhn A. (2008) Initial binding process of the membrane insertase YidC with its substrate Pf3 coat protein is reversible. Biochemistry 47, 6052–6058 [DOI] [PubMed] [Google Scholar]
- 10. Winterfeld S., Imhof N., Roos T., Bär G., Kuhn A., Gerken U. (2009) Substrate-induced conformational change of the Escherichia coli membrane insertase YidC. Biochemistry 48, 6684–6691 [DOI] [PubMed] [Google Scholar]
- 11. Serek J., Bauer-Manz G., Struhalla G., van den Berg L., Kiefer D., Dalbey R., Kuhn A. (2004) Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23, 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kiefer D., Kuhn A. (2007) YidC as an essential and multifunctional component in membrane protein assembly. Int. Rev. Cytology 259, 113–138 [DOI] [PubMed] [Google Scholar]
- 13. Oliver D. C., Paetzel M. (2008) Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J. Biol. Chem. 283, 5208–5216 [DOI] [PubMed] [Google Scholar]
- 14. Ravaud S., Stjepanovic G., Wild K., Sinning I. (2008) The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J. Biol. Chem. 283, 9350–9358 [DOI] [PubMed] [Google Scholar]
- 15. Xie K., Kiefer D., Nagler G., Dalbey R. E., Kuhn A. (2006) Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45, 13401–13408 [DOI] [PubMed] [Google Scholar]
- 16. Jiang F., Yi L., Moore M., Chen M., Rohl T., Van Wijk K. J., De Gier J. W., Henry R., Dalbey R. E. (2002) Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J. Biol. Chem. 277, 19281–19288 [DOI] [PubMed] [Google Scholar]
- 17. Jiang F., Chen M., Yi L., de Gier J. W., Kuhn A., Dalbey R. E. (2003) Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 278, 48965–48972 [DOI] [PubMed] [Google Scholar]
- 18. Klenner C., Yuan J., Dalbey R. E., Kuhn A. (2008) The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS Letters 582, 3967–3972 [DOI] [PubMed] [Google Scholar]
- 19. Yu Z., Koningstein G., Pop A., Luirink J. (2008) The conserved third transmembrane segment of YidC contacts nascent Escherichia coli inner membrane proteins. J. Biol. Chem. 283, 34635–34642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagler C., Nagler G., Kuhn A. (2007) Cysteine residues in the transmembrane regions of M13 procoat protein suggest that oligomeric coat proteins assemble onto phage progeny. J. Bacteriol. 189, 2897–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhn A., Wickner W. (1985) Isolation of mutants in M13 coat protein that affect its synthesis, processing, and assembly into phage. J. Biol. Chem. 260, 15907–15913 [PubMed] [Google Scholar]
- 22. Kiefer D., Kuhn A. (1999) Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 18, 6299–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Studier F. W., Moffatt B. A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 24. Kiefer D., Hu X., Dalbey R., Kuhn A. (1997) Negatively charged amino acid residues play an active role in orienting the Sec-independent Pf3 coat protein in the Escherichia coli inner membrane. EMBO J. 16, 2197–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imhof N., Kuhn A., Gerken U. (2011) Substrate-dependent conformational dynamics of the Escherichia coli membrane insertase YidC. Biochemistry 50, 3229–3239 [DOI] [PubMed] [Google Scholar]
- 26. Kuhn A., Stuart R., Henry R., Dalbey R. E. (2003) The Alb3/Oxa1/YidC protein family. Membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13, 510–516 [DOI] [PubMed] [Google Scholar]
- 27. Yuan J., Phillips G. J., Dalbey R. E. (2007) Isolation of cold-sensitive yidC mutants provides insights into the substrate profile of the YidC insertase and the importance of transmembrane 3 in YidC function. J. Bacteriol. 189, 8961–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 29. Dalbey R. E., Kuhn A. (2004) YidC family members are involved in the membrane insertion, lateral integration, folding, and assembly of membrane proteins. J. Cell Biol. 166, 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]