Background: p300 and CBP have overlapping functions, but p300 is required for advanced prostate cancer cell proliferation.
Results: p300, not CBP, is widely required for androgen-induced gene regulation and histone acetylation and methylation.
Conclusion: p300 is dominant over CBP in advanced prostate cancer cells and required for early steps in transcriptional activation.
Significance: Genes controlled by p300 are critical for prostate cancer proliferation.
Keywords: Androgen Receptor, Histone Modification, p300, Prostate Cancer, Transcription Coactivators, Transcription Regulation
Abstract
The protein acetyltransferases p300 and cAMP response element-binding protein binding protein (CBP) are homologous, ubiquitously expressed proteins that interact with hundreds of proteins involved in transcriptional regulation and are involved globally as transcriptional coregulators. Although these two proteins acetylate and interact with overlapping sets of proteins, we found that p300 and CBP contribute to androgen-induced regulation of distinct sets of genes in C4-2B prostate cancer cells, a model of advanced prostate cancer. CBP cannot compensate for the loss of p300 to support androgen-induced expression of many genes, such as TMPRSS2 and PSA. Global gene expression analysis indicated that 47% of androgen-regulated genes are p300-dependent in these cells, whereas, surprisingly, only 0.3% of them are CBP-dependent. Chromatin immunoprecipitation analysis after depletion of cellular p300 indicated that p300 is required for androgen-induced acetylation of histones H3 and H4, methylation of histone H3 at Lys-4, and recruitment of TATA box binding protein (TBP) and RNA polymerase II, but not recruitment of the androgen receptor, on the TMPRSS2 gene in response to androgen. Thus, p300 is the dominant coregulator of the CBP/p300 pair for androgen-regulated gene expression in C4-2B cells. p300 is required at an early stage of chromatin remodeling and transcription complex assembly after binding of androgen receptor to the gene but before many critical histone modifications occur.
Introduction
The androgen receptor (AR)2 is a ligand-activated transcription factor belonging to the nuclear receptor superfamily which regulates the expression of hundreds of different genes in any given cell type in response to its cognate hormone, dihydrotestosterone (DHT). Upon binding of the ligand, the AR translocates to the nucleus where it binds directly to DNA sequences known as androgen-responsive elements (AREs), which serve as regulatory (enhancer and silencer) elements for associated genes (1, 2). The DNA-bound AR recruits coregulators (coactivators and/or corepressors) that remodel chromatin conformation around the ARE and the associated transcription start site (TSS) and regulate the assembly or disassembly of an active transcription complex on the TSS.
AR plays important roles in male reproductive system development, including the development of the prostate gland and in the onset and progression of prostate cancer (3). Initially, prostate adenocarcinoma cells depend on androgens to drive their proliferation by regulating genes that control that process. This dependence on the hormone-stimulated actions of AR makes therapy possible by interfering with the physiological levels or actions of androgens either by chemical or physical castration. Androgen ablation therapy of early-stage tumors is generally successful, leading to a decrease in tumor size. However, in many cases the tumors begin to grow again, and they no longer rely on high levels of androgen for their growth or survival, although they do still depend on a functional AR. The disease is now regarded as androgen depletion-independent (ADI), and a key difference in this phenotype is hyperactivation of AR through various mechanisms. AR amplification or gain of function mutations can allow AR activation by castration levels of androgens or even by non-androgenic steroids and antiandrogens, which do not activate the wild-type AR. Enhanced activity of other signaling pathways or altered androgen metabolism can also enhance AR activity (4). Increased levels or activity of coregulators or posttranslational modifications to AR or coregulators as a result of deregulation of other signaling pathways can also contribute to the ADI phenotype (5, 6).
Although dozens or even hundreds of coregulators are generally recruited by DNA-bound transcription factors, including AR, to carry out the regulation of chromatin conformation and transcription complex assembly required to control transcription, increasing evidence suggests that different subsets of coregulators may be required for the transcription of different target genes (7–9). In this study, we explored the differential roles of two specific coregulators, CBP and p300, in androgen-regulated gene expression in the C4-2B cell line, a model for ADI prostate cancer. p300 and CBP are known coregulators for the nuclear hormone receptors as well as many other transcription factors and are known to enhance transcriptional activity through either their protein acetyltransferase activity or by acting as scaffold proteins to recruit other coregulators or components of the basal transcription machinery (10–13). Because of their high degree of homology (about 63% at the amino acid level) and significant overlap of their acetylation substrate specificity, they are sometimes regarded as interchangeable. In fact, it has been shown that they do have similar substrates in vitro and may indeed perform some redundant functions. However, it is important to note that the two share regions of lesser homology as well as greater homology. Considerable evidence has suggested that they are not functionally redundant and that expression of a specific gene may preferentially require the action of one over the other (7, 8, 14, 15). In vivo studies show that not only homozygous null mutations of p300 or CBP but also double heterozygosity in mice is embryonic lethal, indicating that they have unique individual functions and highlighting their importance early on in development (16). Also, although both are essential in hematopoiesis, CBP but not p300 is important for stem cell proliferation, whereas p300 but not CBP is essential for stem cell differentiation (17). Genetic mutations in one copy of the CBP gene lead to a congenital developmental disorder known as Rubinstein-Taybi syndrome, but no genetic diseases have yet been associated with p300 abnormalities (16, 18). Mice lacking one functional copy of the CBP allele, as opposed to those lacking one functional copy of p300, exhibit growth retardation and craniofacial abnormalities (19). Mice lacking one functional copy of p300 exhibit significant embryonic lethality, and those that are p300−/− show severe abnormalities in heart development as compared with their CBP-deficient counterparts (20).
Recently, roles of coregulators in disease progression have been increasingly recognized. Coregulators are known to be important for the proper assembly of a transcriptional complex, and a subset of these coregulators, including the SRCs, p300 and CBP, are overexpressed during prostate cancer progression (21–23). Of particular relevance to this study, expression of p300 correlates positively with prostate tumor grade, tumor volume, proliferation markers, and other features of aggressive tumors (24). Furthermore, over-expression of p300 is required for the transition from androgen dependent to ADI prostate cancer (6). In this study, we explored the reasons for the importance of p300 versus CBP in advanced prostate cancer. We investigated the individual contributions that CBP and p300 make to androgen-regulated gene expression in the ADI cell line C4-2B. Our study defined the specific sets of genes that require CBP or p300 for their androgen-regulated expression. Through microarray analysis we show that there are indeed separate groups of genes that preferentially require p300 or CBP in ADI prostate cancer cells. In addition, we explored the promoter-specific mechanisms of androgen-regulated gene expression by defining the specific stage of the androgen-induced chromatin remodeling and transcription complex assembly processes at which p300, in particular, contributes to these processes. These novel findings are critical for understanding the hypersensitive actions of AR in ADI prostate cancer and lay the foundation for further study of the mechanism of progression to advanced prostate cancer and for subsequently designing novel avenues for therapeutic intervention.
EXPERIMENTAL PROCEDURES
Cell Culture
C4-2B cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS in a 37 °C and 5% CO2 incubator. Prior to any hormone treatment, cells were plated and maintained for 3 days in RPMI 1640 phenol red-free medium supplemented with 5% charcoal-stripped serum, to which we will subsequently refer as treatment medium. All hormone treatments were with 10 nm DHT unless noted otherwise, and an equivalent volume of ethanol was used as the vehicle control.
siRNA Transfections
C4-2B cells were plated in 12-well plates in treatment medium at 8 × 104 cells per well. The following day they were transfected with 60 pmol of On-Targetplus SMARTpool siRNA (Dharmacon) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. After transfection, cells were maintained in treatment medium for 3 days before harvest, including treatment with 10 nm DHT for the specified times. Either human EP300, human CREBBP (CBP) On-Targetplus SMARTpool (Dharmacon), or both in combination were used. Total RNA was harvested using the TRIzol method (Invitrogen), and whole cell protein extracts were obtained using radioimmune precipitation assay buffer (50 mm Tris HCl (pH 8), 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40).
Illumina Microarray Analysis
Global gene expression analysis was performed at the Southern California Genotyping Consortium at University of California Los Angeles with an Illumina Human HT12v4 microarray using randomized placement of four biological replicates generated on different days. Using Illumina Genomestudio software, output of summarized probe information was done with minimal preprocessing and removal of imputed genes. These data were imported into Bioconductor (25), where it was transformed, normalized, and corrected for background and batch, before being used to determine significant differential gene expression. Background correction, normalization, and transformation, in that order, were done by the limma package using neqc (26). Combat was used to remove batch effects because of experimental day bias (27). Differentially expressed genes were identified using a two-sample modified Student's t test from eBayes in the limma package (28). False discovery was accounted for by using q values (29). Original microarray data sets have been submitted to NCBI GEO (accession number GSE31873).
Quantitative RT-PCR (qRT-PCR)
Total RNA was harvested using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 10–25 μg total RNA was treated with DNase I (Roche) prior to reverse transcription. For measuring mRNA, cDNA was synthesized by reverse transcribing 500 ng total RNA using the iScript cDNA synthesis kit (Bio-Rad). For measuring pre-mRNA, 1 μg of total RNA was reverse-transcribed using the standard RT-PCR procedure with random hexamers of the Transcriptor First Strand cDNA synthesis kit (Roche). In each case, the resulting cDNA was diluted 5-fold, and 1.5 μl of diluted cDNA was amplified by qPCR using the LightCycler 480 SYBR Green I master mix (Roche) on the LightCycler 480 system (Roche) and primers described in supplemental Table 1.
ChIP Assay
C4–2B cells were plated at 3 × 106 cells per 150-mm dish in treatment medium and maintained for 3 days prior to hormone treatment and cross-linking. ChIP assays were performed as described previously (30) with the following modifications. Cells were grown to confluence and treated with 10 nm DHT for 4 h and subsequently incubated for 10 min with a final concentration of 1% formaldehyde. Reactions were quenched for 5 min with 125 mm glycine. Cells were harvested and sonicated for 15 to 30 min (in repeated cycles of 30 s on and 30 s off) using a BioRuptor sonicator (Diagenode) kept at 4 °C with a circulating water bath to achieve fragments 200–800 bp in size. After a 1-h preclearing step using protein A/G Plus-agarose (Santa Cruz Biotechnology), chromatin was immunoprecipitated by end-over-end rotation overnight at 4 °C with 40 μl of protein G-Sepharose beads (GE Healthcare) and the following antibodies: 3 μg of AR PG-21 (Millipore), 3 μg of p300 (Bethyl Laboratories), 4 μg of CBP (A-22, Santa Cruz Biotechnology), 1 μg RNA polymerase II clone CTD4H8 (Millipore), 3 μg TATA box binding protein (TBP) (N-12, Santa Cruz Biotechnology), 0.5 μg of H3K18Ac (Abcam), 1 μg of H3K27Ac (Abcam), 1 μg of H4Ac (Millipore), 1 μg of H3K4me1 (Abcam), and 3 μg of H3K4me3 (Active Motif). Immunoprecipitates were washed and eluted, and purified DNA was obtained as described previously. Purified immunoprecipitated DNA and input DNA (preimmunoprecipitation) were used as template for subsequent quantitative real-time PCR. Briefly, 1.5 μl of DNA was amplified with 2.5 pmol each of forward and reverse primer (supplemental Table 2) in a final reaction volume of 10 μl using LightCycler 480 SYBRGreen I Master mix (Roche) on a LightCylcer 480 system (Roche). Percent input was calculated using the percent input method (Invitrogen).
RESULTS
Requirement for p300 and CBP in Androgen-regulated Gene Expression
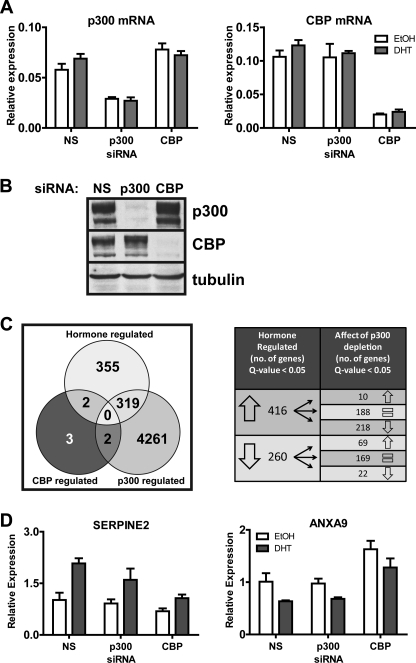
To identify genes that are specifically regulated by p300 or CBP, we used C4-2B prostate cancer cells, a model of advanced ADI prostate cancer. These cells express AR and exhibit androgen-regulated gene expression. Transfection of cells with siRNA against p300 (sip300), CBP (siCBP), or a nonspecific sequence (siNS) specifically depleted the intended target mRNA and protein, and cellular levels of p300 and CBP mRNA and protein were not altered by DHT treatment (Fig. 1, A and B, and data not shown). For global gene expression analysis, C4-2B cells were transfected with siRNA and, after 3 days to allow depletion of the target protein, treated with DHT or vehicle for 16 h before preparing total RNA for microarray analysis. Four independent experiments were conducted on different days. Each experiment consisted of six samples: siNS vehicle, siNS DHT, si p300 vehicle, si p300 DHT, siCBP vehicle, and siCBP DHT. Of the greater than 40,000 probes present on the Illumina BeadChip, we found that a total of 676 probes were hormone-regulated (q value threshold ≤ 0.05), i.e. when we compared siNS vehicle-treated versus siNS DHT-treated samples (Fig. 1C, hormone-regulated set). Each gene or putative gene is represented by a single probe on the BeadChip with only a small number of exceptions where one gene is represented by multiple probes. Of these 676 DHT-regulated genes, 416 were up-regulated and 260 were down-regulated in response to DHT. With depletion of p300 a total of 4582 genes (q value threshold ≤ 0.05) had altered expression in the sip300 DHT sample as compared with the siNS DHT sample (Fig. 1C, p300-regulated set). One of the down-regulated genes was p300, thus validating our initial qRT-PCR and Western blot observations. Of those 4582 p300-dependent genes, 319 were also members of the 676-member DHT-regulated gene set. Thus, 47% of the DHT-regulated genes required p300 to achieve their DHT-regulated expression level. For the 416 genes that were up-regulated by DHT, depletion of p300 interfered with the DHT-induced expression levels of 218 of those (52.4%) (q value threshold ≤ 0.05). This includes the known AR target genes PSA/KLK3, KLK2, and TMPRSS2. A small number (10 genes) were hyperactivated by DHT after p300 depletion. When we look at the 260 genes down-regulated by DHT, p300 depletion interfered with the DHT-stimulated down-regulation of 69 genes (26.5%). Depletion of p300 also caused increased repression of 22 genes by DHT. The large number of DHT-regulated genes affected by p300 depletion indicates that p300 plays a major role in gene regulation by DHT, a role for which CBP cannot compensate.
FIGURE 1.
Effects of p300 and CBP depletion on global and DHT-induced gene expression in C4-2B cells. A, qRT-PCR was performed on RNA extracted from cells transfected with siRNA against p300, CBP, or a nonspecific (NS) sequence. Cells were treated with ethanol (EtOH) or 10 nm DHT for 16 h before harvest. Levels of mRNA are shown relative to 18 S rRNA and represent the mean ± S.D. of three technical PCR replicates. B, whole cell extracts from cells transfected with siRNA were analyzed by immunoblot using antibodies against p300, CBP, or tubulin as a control. C, total RNA was extracted from cells transfected with siRNA and treated with DHT or ethanol as in A. Four independent experiments of this type were conducted on different days, and the 24 resulting RNA samples were analyzed on Illumina microarrays. The Venn diagram for the hormone-regulated gene set shows the number of genes with significantly different expression (q ≤ 0.05) for RNA from cells transfected with nonspecific siRNA and then treated with ethanol versus DHT. The Venn diagram for the p300-regulated gene set shows the number of genes with significantly different expression (q ≤ 0.05) for RNA from DHT-treated cells that had been transfected with nonspecific siRNA versus siRNA against p300. The Venn diagram for the CBP-regulated gene set shows the number of genes with significantly different expression (q ≤ 0.1) for RNA from DHT-treated cells that had been transfected with nonspecific siRNA versus siRNA against CBP. Overlap areas indicate the number of genes belonging to two or all three of the three gene sets. The table on the right depicts the number of genes up- or down-regulated with DHT and the number that are either up-regulated (↑), down-regulated (↓), or unchanged ( =) with p300 knockdown. The complete lists of genes for the three major sets in the Venn diagram are found in supplemental Tables 3–5. D, Illumina microarray results for the genes which were DHT-regulated and CBP-dependent. For each gene, expression is given relative to cells transfected with siNS and treated with ethanol and is plotted as the mean ± S.D. of four independent experiments.
Surprisingly, when we analyzed the data set for CBP-dependent genes, we found that only seven genes were affected by CBP depletion (q-value threshold ≤ 0.1) (Fig. 1C, CBP-regulated set). It is important to note that we had to raise the q value cut-off to obtain these candidate genes, as a q value cut-off ≤ 0.05 did not yield any results of genes affected by CBP depletion, except for CBP itself. Of those seven genes, only two (SERPINE2 and ANXA9) also belonged to the 676-member DHT-regulated gene set. DHT normally up-regulates the SERPINE2 mRNA level, but the DHT-induced level of this mRNA was significantly reduced in the microarray data when CBP (but not p300) was depleted (Fig. 1D, left panel). ANXA9 mRNA was reduced by DHT treatment, but CBP depletion caused a significant increase in the DHT-treated level of mRNA (Fig. 1D, right panel). From the data, it appears that the basal as well as the DHT-treated levels of the mRNA were affected by CBP depletion, but the significance of these differences was not evaluated in this analysis. In the microarray data, neither of the two DHT-regulated, CBP-dependent genes was significantly altered in its DHT-regulated expression level by depletion of p300.
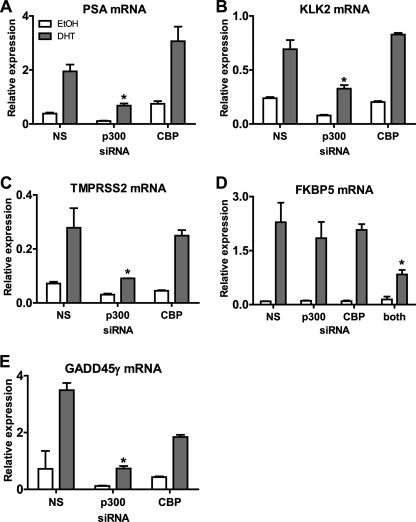
Expression of some of the androgen-regulated, p300-dependent genes identified from the microarray analysis was examined further using qRT-PCR. Because the biological phenotype of DHT-treated cells is dependent on the level of gene expression achieved after DHT treatment, we focused our attention on the effects of p300 depletion on mRNA levels in DHT-treated cells rather than the fold induction by DHT. PSA, probably the most well characterized androgen-regulated gene, is used as a biomarker of prostate cancer onset and progression. The qRT-PCR analysis indicated that the PSA mRNA level was up-regulated in C4-2B cells after 16 h of DHT treatment (Fig. 2A). Depletion of p300 caused a decrease in both the basal and DHT-induced levels of PSA mRNA. In contrast, CBP depletion did not decrease PSA expression (the increase observed in this experiment was not reproducible). Similar results were obtained with two other DHT-regulated genes identified as p300-dependent in the microarray analysis, KLK2 and TMPRSS2. Depletion of p300, but not CBP, caused statistically significant (p ≤ 0.05) decreases in the DHT induced levels of these mRNAs compared with RNA from cells treated with siNS and DHT (Fig. 2, B and C).
FIGURE 2.
Effect of p300 and CBP depletion on androgen-regulated expression of specific genes. A–E, qRT-PCR was performed on total RNA extracted from C4-2B cells transfected with siRNA against p300, CBP, both p300 and CBP, or a nonspecific (NS) sequence. Cells were treated either with ethanol (EtOH) or 10 nm DHT for 16 h. Levels of mRNA are shown relative to 18 S rRNA. Results shown are mean ± S.D. of three replicate PCR analyses of RNA samples from a single experiment. *, p < 0.05 compared with the siNS/DHT sample in the same graph, calculated using a paired Student's t test from at least three biological replicates.
As also predicted by the microarray results, DHT-induced levels of mRNAs for some genes were not affected by depletion of either p300 or CBP. FKBP5 is an example of such a gene (Fig. 2D). This indicates either a possible compensatory role for these cofactors on genes such as FKBP5 or no involvement of either coactivator in the expression of this gene. To discriminate between these two possibilities, siRNAs specific for p300 and CBP were transfected in combination. The DHT-induced FKBP5 mRNA level was significantly reduced only when both p300 and CBP were depleted, indicating that either p300 or CBP is required and that the function of these proteins is redundant for DHT induction of FKBP5 gene expression (Fig. 2D). We also observed other DHT-regulated genes that were modestly affected or unaffected by single coactivator depletion, whereas dual siRNA treatment caused a significant effect (data not shown). Results similar to those observed for FKBP5 were also observed for genes that were repressed by DHT. For example, the level of LMO4 mRNA decreased in response to DHT. Individual depletion of either p300 or CBP had little or no effect on the level of mRNA observed after DHT treatment, but depletion of both p300 and CBP caused a significant elevation of the DHT-depressed level of mRNA (data not shown). In general, results from the microarray and qRT-PCR analyses were in good agreement. However, there were some cases where differences that did not reach the q value cut-off in the microarray results still turned out to be significant in the qRT-PCR results. One of those differences defined a new class of genes with respect to the requirements for p300 and CBP. The microarray results for the DHT-regulated gene GADD45γ indicated a requirement for p300 (q value ≤ 0.05) but not for CBP (q value > 0.1), even though the CBP depletion appeared to reduce the level of mRNA in the presence of DHT. Likewise, in qRT-PCR analysis, depletion of only p300 but not CBP caused a dramatic and significant (p < 0.05) reduction in the DHT-induced mRNA (Fig. 2E). However, although the decrease in GADD45γ expression caused by CBP depletion did not reach significance, there was a reproducible trend indicating a requirement for CBP.
Thus, from microarray and qRT-PCR analyses, we identified four different categories of DHT-regulated genes on the basis of their requirements of p300 and CBP for their DHT-regulated expression. Category 1 genes (e.g. PSA, KLK2, and TMPRSS2) are solely dependent on p300 for DHT-regulated gene expression levels. Category 2 genes (SERPINE2 and ANXA9) are solely dependent on CBP. Category 3 genes (e.g. FKBP5 and LMO4) require either p300 or CBP but not both of them; i.e. p300 and CBP have important but redundant functions for these genes. Category 4 (e.g. GADD45γ) require the actions of both p300 and CBP. Presumably there are also DHT-regulated genes that require neither p300 nor CBP (category 5), but we did not identify these genes because our microarray analyses did not include RNA samples from cells depleted of both p300 and CBP. In addition, results from the microarray analysis indicate that p300 is the dominant member of this homologous protein pair in these cells.
p300 Acts at the Transcriptional Level
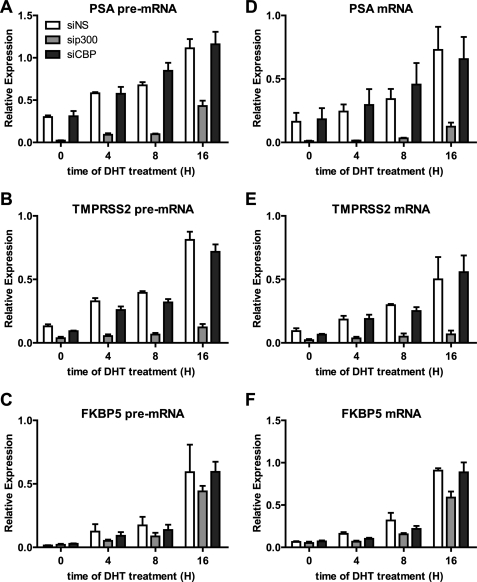
Our ultimate goal was to investigate what p300 contributes to the transcriptional activation process by using ChIP analysis in conjunction with p300 depletion. However, because ChIP provides a snapshot of promoter occupancy at a given time point, we first determined an appropriate time point for ChIP analysis after DHT administration by examining mRNA and pre-mRNA levels of AR target genes at various times after beginning DHT treatment. The mRNA levels provide a cumulative assessment of transcriptional activity since the beginning of DHT treatment. Because pre-mRNAs are rapidly converted to mRNA by RNA processing machinery, pre-mRNA levels provide a more accurate assessment of the instantaneous transcriptional activity on the gene promoter at any given time. To detect pre-mRNA levels, we designed primers for qRT-PCR that specifically spanned an early exon/intron boundary for each gene of interest.
We chose PSA and TMPRSS2 as examples of genes that require p300 but not CBP for DHT-induced expression, and as a control we examined FKBP5 as a gene that is induced normally by DHT when either p300 or CBP is depleted. Previous studies in LNCaP cells (a model for hormone-dependent prostate cancer) indicated that maximum AR recruitment and transcription rates occur between 4 and 16 h after initiating DHT treatment (31). Although C4-2B cells are derived from LNCaP cells, less is known about the time course for AR recruitment and for AR target gene expression in C4-2B cells. Total RNA was collected from C4-2B cells treated with 10 nm DHT for 0, 4, 8, and 16 h. Expression of PSA, TMPRSS2, and FKBP5 pre-mRNAs increased after 4 h of DHT treatment and increased further after 8 and 16 h of DHT treatment (Fig. 3, A–C). Likewise, there was a concomitant gradual increase in mRNA expression for those three genes over the same time period (Fig. 3, D–F). In a subsequent experiment we observed increased levels of pre-mRNA for these genes after 2 h of DHT treatment (data not shown).
FIGURE 3.
Effect of p300 and CBP depletion on the time course of DHT-induced increases in pre-mRNA and mRNA from AR target genes. A–C, qRT-PCR with primers specific for pre-mRNA was performed on total RNA extracted from C4-2B cells transfected with siRNA against nonspecific, p300, or CBP and then treated for 0, 4, 8, or 16 h with 10 nm DHT before harvesting. cDNA was made with random hexamers as primers. The pre-mRNA levels are calculated relative to 18 S rRNA, and each value is the mean ± S.E. for three biological replicates. D–F, the same cDNA samples described in A–C were analyzed by qPCR with primers specific for mature mRNA. The mRNA levels are calculated relative to 18 S rRNA, and each value is the mean ± S.E. for three biological replicates.
The hormone-induced levels of mRNA and pre-mRNA for PSA and TMPRSS2 were substantially reduced at all time points examined by depletion of p300 but not by depletion of CBP (Fig. 3, A, B, D and E). In contrast, pre-mRNA and mRNA levels for FKBP5 were affected very little or not at all by depletion of p300 or CBP (Fig. 3, C and F). The effect of p300 depletion on the DHT-induced levels of pre-mRNA for PSA and TMPRSS2 indicates that p300 is acting at the level of transcription rather than some posttranscriptional process.
Defining the Position of p300 in the DHT-induced Sequence of Events Leading to Transcriptional Activation
Assembly of an active transcription complex on the promoter of an AR target gene in response to DHT requires an ordered sequence of events, including the recruitment of specific coregulators, histone modifications, and recruitment of the basal transcription factors and RNA polymerase II. The role and position of p300 in this process can be defined partially by determining which events depend on p300 and which events do not depend on p300. To initiate this type of analysis, we first defined some of the histone modifications and the recruitment of coregulators and basal transcription factors that occur on AR target genes in response to DHT. On the basis of our time course analysis of pre-mRNA levels (Fig. 3), we chose 0 and 4 h of DHT treatment as the time points for ChIP analysis.
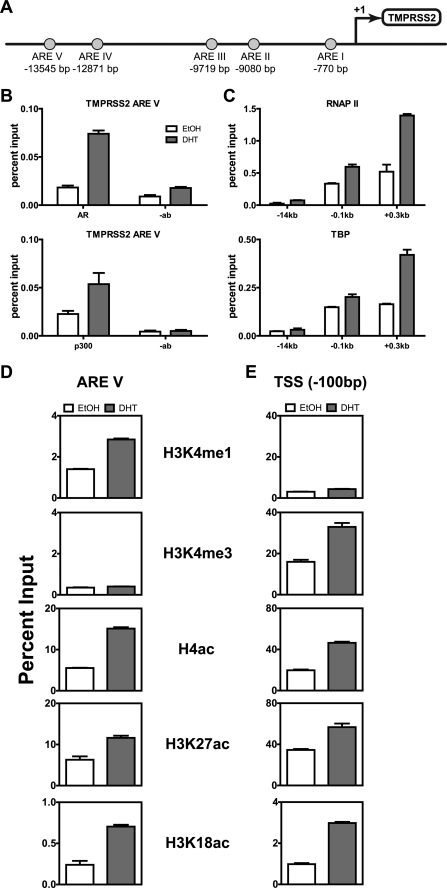
As our primary model of an AR target gene that requires p300, we chose TMPRSS2, and as a control AR target gene which does not require p300 we chose FKBP5. TMPRSS2 is an androgen-regulated transmembrane serine protease primarily expressed in prostate epithelial cells. The protein is highly expressed in primary and metastatic prostate cancers and known to mislocalize to the cytoplasm in high-grade prostate cancers (32, 33). A highly frequent occurrence in prostate cancer is the fusion of the 5′ untranslated region of the TMPRSS2 gene to the genes encoding the ETS transcription factors, primarily ERG, allowing for their androgen regulated overexpression (34). Although TMPRSS2 is a prominent AR target gene, it is far less characterized than the widely known prostate-specific antigen (PSA/KLK3) gene. TMPRSS2 has five motifs resembling AREs, all located upstream of the transcription start site (Fig. 4A). The most distal upstream motif (ARE V), located ∼14 kb upstream of the TSS, is the main site of AR recruitment upon DHT treatment and is well characterized in LNCaP cells (31). The four remaining AREs do not recruit AR upon DHT treatment. For our ChIP studies, we examined the ARE V region and two other regions located 100 bp upstream and 300 bp downstream of the TSS.
FIGURE 4.
DHT-induced histone modifications and transcription complex assembly on the TMPRSS2 gene. A, diagram of the regulatory region of the TMPRSS2 gene showing five putative AREs (circles) and their locations measured in bp relative to the TSS (arrow). ARE V is the main site of AR recruitment. B–E, after growth in hormone-free media for 3 days, C4-2B cells were treated with ethanol (EtOH) or 10 nm DHT for 4 h, and ChIP was performed with the indicated antibodies or no antibody (-ab) as a control, using primers for the indicated region of the TMPRSS2 gene. Each graph shows a single biological experiment that is representative of at least two independent biological experiments. Immunoprecipitated DNA was analyzed relative to input DNA (before immunoprecipitation), and values shown are mean ± S.D. of three technical qPCR replicates, using primers specific for the indicated region.
When C4-2B cells treated with 10 nm DHT for 4 h were compared with vehicle-treated cells, we observed DHT-induced recruitment of AR and p300 to TMPRSS2 enhancer ARE V (Fig. 4B). We also observed a very slight recruitment of RNA polymerase II and TBP to ARE V, but as expected, far more recruitment of these two proteins was observed near the TSS (Fig. 4C). DHT treatment increased the level of histone H3 Lys-4 monomethylation (H3K4me1), a known marker of active and poised enhancer elements (35, 36), at ARE V of the TMPRSS2 gene (Fig. 4D) and also increased histone H3 Lys 4 trimethylation (H3K4me3), a general marker of active and poised TSS regions, just upstream of the TMPRSS2 TSS (E). Acetylation of histone H4 (H4ac) and of histone H3 at Lys-18 (H3K18ac) and Lys-27 (H3K27ac), which generally mark active genes, were increased by DHT treatment at both the ARE V and the TSS regions (Fig. 4, D and E).
Similar DHT-induced events were observed on the regions surrounding the major enhancer and the TSS for the PSA gene (data not shown) and the FKBP5 gene (Fig. 5). AREs for the FKBP5 gene have been identified previously (37). The gene contains 13 in silico-predicted AREs, but ChIP in LNCaP and VCaP cells identified the major AR binding site at an intronic region located ∼86 kb downstream of the TSS, designated as ARE VIII/IX because it contains two AREs spaced 31 nucleotides apart (Fig. 5A). Our ChIP analysis focused on this enhancer region and a site 100 bp upstream from the TSS. As observed for the TMPRSS2 gene, DHT caused increased recruitment of AR and p300 primarily at the enhancer region, whereas RNA polymerase II and TBP were primarily recruited to the TSS (Fig. 5B).
FIGURE 5.
DHT-induced histone modifications and transcription complex assembly on the FKBP5 gene. A, diagram of the regulatory region of the FKBP5 gene showing putative AREs (circles) and their locations measured in bp relative to the TSS (arrow). ARE VIII/IX is the main site of AR recruitment. B–C, ChIP was performed with C4-2B cells treated with ethanol or DHT for 4 h as in Fig. 4.
DHT also caused enhancement of the same histone modifications on the FKBP5 enhancer and TSS regions (Fig. 5C), as were observed at the analogous sites of the TMPRSS2 gene (Fig. 4, D and E). The DHT-induced recruitment of p300 to the enhancer regions of both genes indicates that the lack of requirement for p300 by the FKBP5 gene is not because p300 is not recruited to this gene.
Having defined a number of DHT-induced events at the enhancer and TSS regions of the TMPRSS2 and FKBP5 genes, we examined the effects of p300 depletion on these hormone-induced events to determine the position at which p300 acts in the sequence of events leading to transcriptional activation. The results observed by ChIP for the DHT-regulated events after p300 depletion (Fig. 6A) were compared with the results seen in the absence of siRNA transfection (Figs. 4 and 5) and with the results obtained by depletion of CBP (which had no effect on the DHT-induced expression of TMPRSS2 or FKBP5) (Fig. 6B). After depletion of p300, there was still a robust DHT-induced increase in AR occupancy at ARE V of the TMPRSS2 gene (Fig. 6A), comparable with that observed after CBP depletion (B) or in the absence of any siRNA transfection (Fig. 4B). However, p300 depletion prevented or dramatically reduced DHT-induced increases in occupancy of TBP and RNA polymerase II at the TSS (Fig. 6A), increases in acetylation of histones H3 and H4 at ARE V and the TSS (C), and increases in H3K4me1 at ARE V and H3K4me3 at the TSS (C). This is in stark contrast to the robust DHT-induction of these events in the absence of siRNA transfection (Fig. 4, C–E). Furthermore, when CBP was depleted in parallel with the p300 depletion, robust DHT-induced increases were still observed for recruitment of TBP and RNA polymerase II at the TSS (Fig. 6B), for acetylation of histone H4 at ARE V and the TSS, and for H3K18 acetylation at the TSS (D). Thus, p300 depletion had no effect on DHT-induced binding of AR to the ARE of TMPRSS2 but interfered with all of the other DHT-induced events examined here. In contrast, all of the hormone-induced events were still observed after depletion of CBP.
FIGURE 6.
Effect of p300 and CBP depletion on DHT-induced histone modifications and transcription complex assembly on the TMPRSS2 gene. A–D, C4-2B cells were transfected with siRNA against p300 (A and C) or CBP (B and D), and cells were treated with ethanol or DHT for 4 h, prior to ChIP analysis performed as in Fig. 4. Each graph shows a single biological experiment that is representative of at least two independent biological experiments. Immunoprecipitated DNA was analyzed relative to input DNA (before immunoprecipitation), and values shown are mean ± S.D. of three technical qPCR replicates, using primers specific for the indicated region.
In parallel with the examination of TMPRSS2, we examined DHT-induced events at the enhancer and TSS regions of the FKBP5 gene after depletion of p300. We observed a robust DHT-induced increase in occupancy of ARE VIII/IX by AR, and a robust occupancy of TBP and RNA polymerase II at the TSS region, even though DHT-induced occupancy of p300 on the enhancer region was eliminated by p300 depletion (Fig. 7A). Thus, although p300 recruitment was prevented, there was no effect on subsequent recruitment of TBP and RNA polymerase II, in keeping with the lack of any effect on FKBP5 expression when p300 was depleted (Fig. 2). Surprisingly, we found that p300 depletion eliminated all of the DHT-induced histone modifications on the FKBP5 gene (Fig. 7B), similar to the effects seen for the TMPRSS2 gene (Fig. 6C). Thus, the DHT-induced histone modifications normally observed on the FKBP5 gene are not required for the DHT-induced occupancy of the TSS by TBP and RNA polymerase II or for the DHT-induced transcription of this gene.
FIGURE 7.
Effect of p300 depletion on DHT-induced histone modifications and transcription complex assembly on the FKBP5 gene. A–B, C4-2B cells were transfected with siRNA against p300, and cells were treated with ethanol or DHT for 4 h, prior to ChIP analysis performed as in Fig. 4. Each graph shows a single biological experiment that is representative of at least two independent biological experiments. Immunoprecipitated DNA was analyzed relative to input DNA (before immunoprecipitation), and values shown are mean ± S.D. of three technical qPCR replicates, using primers specific for the indicated region.
DISCUSSION
Role of p300 in Advanced Prostate Cancer
Recent evidence has strongly implicated p300 in the progression of prostate cancer. Clinically, p300 overexpression correlates positively with proliferation, stage, and other aggressive characteristics of prostate tumors, and p300 overexpression correlates negatively with progression-free survival of patients (24). Furthermore, androgen deprivation of cell lines that serve as prostate cancer models enhances p300 expression, and depletion of p300 from cell line models of ADI prostate cancer inhibits cell proliferation (6). These findings suggest an important role for p300 in the progression of prostate cancer to ADI status and maintenance of ADI prostate cancer. Why is it specifically p300 rather than CBP that is important in prostate cancer cells? Our results demonstrate that p300 is required for the expression of thousands of genes in an advanced prostate cancer cell line, including about half of the androgen-regulated genes. In contrast, we found that CBP was necessary for expression of only a very small number of genes, thus providing an explanation as to why p300 and not CBP is important for these cells.
p300 could represent a potential therapeutic target for prostate cancer. However, p300 and CBP are thought to serve as coregulators for hundreds of different transcription factors, raising the issue of many possible side effects if p300 is targeted therapeutically. Although p300 may be involved in the regulation of thousands of genes, the requirement of p300 for proliferation of ADI prostate cancer cells is presumably due to its requirement for expression of a much smaller subset and perhaps only a few of its total complement of target genes. Once identified, the p300 target genes responsible for ADI prostate cancer cell proliferation and tumor progression may represent much better therapeutic targets than p300 itself. Here we have identified the complete set of p300 target genes in an ADI prostate cancer cell line and the subset of those genes that are also regulated by androgen. These gene sets can now serve as a starting point for identification of the specific genes required for p300-dependent proliferation of these cells.
Major Differences in the Roles of p300 and CBP in C4-2B Cells
By global gene expression analysis, we found major differences in the roles of p300 and CBP in C4-2B cells. Depletion of p300 affected the expression of over 4000 genes in DHT-treated cells, whereas depletion of CBP affected fewer than 10. For the 676 DHT-regulated genes, the level of expression after DHT treatment was significantly altered (q ≤ 0.05) for approximately half of the genes when p300 was depleted, but we only found two that were significantly altered (q ≤ 0.1) by CBP depletion. A trivial explanation for this difference could be that our depletion of CBP was not efficient enough. However, we observed efficient depletion of each protein and the corresponding mRNA by siRNA transfection (Fig. 1, A and B). In addition, both p300 and CBP were found to be significantly reduced (q ≤ 0.05) by the appropriate siRNA treatment in our unbiased statistical analysis of the microarray data. Alternatively, we sometimes observed a small increase in p300 levels (which appeared to be less than 50%) after depletion of CBP, and this could in theory compensate for the loss of CBP. However, our microarray and qRT-PCR data confirmed that depletion of CBP caused a substantial and reproducible decrease in the DHT-induced expression of SERPINE2 and in the basal and DHT-induced expression of GADD45γ mRNA (Fig. 2E), thus demonstrating the functional efficacy of CBP depletion. In addition, previous studies which measured the function of p300 and CBP concluded that these proteins are generally present in limiting concentrations, such that there is competition for them among the hundreds of proteins that interact with them (19, 20, 38). We therefore conclude that p300 has a dominant role compared with CBP in global gene regulation in C4-2B cells. Other investigators have also observed a critical role for p300 in ADI prostate cancer cells (6). In addition, a genome-wide ChIP analysis of p300 and CBP in glioblastoma cells revealed that the majority of genes that were regulated by serum addition were bound preferentially by p300 rather than CBP (39).
Other differences in the functions of p300 and CBP have also been well documented, including their distinct actions in Wnt signaling, where preferential interaction of β-catenin with CBP or p300 leads to different gene expression patterns. In colon cancer cells, survivin gene expression was blocked by selectively inhibiting the interaction of β-catenin with CBP. This allowed β-catenin to interact with p300, leading to inhibition of survivin gene expression (7). In another study, murine embryonic stem cell pluripotency was maintained by inhibiting interaction of β-catenin with p300 and allowing for an increased β-catenin-CBP association. A switch back to conditions allowing β-catenin-p300 mediated transcription promoted differentiation of the embryonic stem cells (8).
The mechanisms that are responsible for gene-specific requirements of CBP versus p300 are still poorly understood, but a few previous studies, as well as our current investigation, offer some initial insights. Ramos et al. (39) found that genes which were preferentially bound by either CBP or p300 also exhibited differences in the enrichment of other transcription factor binding sites associated with either p300-specific or CBP-specific targets (39). Thus, the presence of specific transcription factors in the promoter environment may favor recruitment of either p300 or CBP, perhaps because of preferential interactions of various transcription factors with either p300 or CBP. In contrast, in our study, p300 was required for DHT-induced expression of the TMPRSS2 gene but not the DHT-induced expression of the FKBP5 gene. Nevertheless, p300 was recruited to the ARE of both genes in response to DHT (Figs. 4B and 5B). Although the differential requirement for p300 in this case was not due to differential recruitment of p300, the mechanism responsible for the differential requirement presumably lies in some different aspect of promoter environment at the two genes. As suggested by Ramos et al. (39), differences in binding sites for other transcription factors near the ARE could influence whether p300 or CBP is recruited. In addition, differences in the DNA sequence at steroid receptor binding sites can lead to altered conformations of the DNA-bound steroid receptor, resulting in recruitment of different sets of coactivators (40–43). Thus, although DHT induces recruitment of p300 to both the TMPRSS2 and FKBP5 genes, differential presence of other coregulators on these two promoters may cause p300 to be required on one gene but redundant on the other.
p300 Action Is Required Early in the Process of Chromatin Remodeling and Transcription Complex Assembly
ChIP analysis indicated that p300 is recruited to the promoter in response to DHT, and analysis of mRNA levels of AR target genes after p300 depletion demonstrates that p300 has an important function in DHT-regulated expression of many target genes of AR. However, neither ChIP analysis nor p300 depletion used alone provide any mechanistic information about what p300 contributes to the process of chromatin remodeling and transcription complex assembly. However, by performing ChIP analysis of p300-requiring genes in DHT-treated cells after p300 depletion, we have been able to specify which DHT-induced events on the target genes require p300 and which events do not require p300. This information provides insight into the specific position of p300 function within the ordered sequence of events that constitutes hormone-induced chromatin remodeling and transcription complex assembly. Thus, we observed that on the TMPRSS2 gene p300 is not required for the DHT-induced binding of AR to the ARE, but every other DHT-induced event we monitored was dependent on p300. This includes increased acetylation of histones H3 and H4, methylation of H3K4, and recruitment of TBP and RNA polymerase II. It is possible that increased histone acetylation may be required for recruitment or activity of specific H3K4 methyltransferases, or p300 may recruit such enzymes through protein-protein interactions. Thus, p300 is required for many aspects of DHT-induced chromatin remodeling as well as recruitment of the basal transcription machinery, specifically TBP and RNA polymerase II. TBP is an integral part of the Transcription Factor IID (TFIID) transcription complex and is believed to be required for all RNA polymerase II transcription. Because p300 is known to interact with TBP (44, 45), it is possible that p300 directly recruits TBP. Alternatively, histone acetylation or recruitment of other coregulators by p300 could lead indirectly to recruitment of TBP.
Histone H3 Lys-18 and Lys-27 are in vitro acetylation targets of p300 and CBP (46), and it has been suggested that CBP and p300 are responsible for over 90% of the H3K18ac and H3K27ac in MEFs (47). During ligand-mediated gene activation by nuclear receptor peroxisome proliferator activated receptor δ (PPARδ), three distinct phases of ligand-induced histone modifications have been identified. Acetylation of H3K18, H3K27, and H4 occurs in the early phase prior to RNA polymerase II recruitment and gene expression, whereas H3K4me3, H3K36me3, and H3K79me2 modifications occur after RNA polymerase II recruitment (47).
Assuming that p300 is responsible for at least some of the DHT-induced histone acetylation increases we observed, our results are also consistent with this model, where H3K4 methylation and recruitment of TBP and RNA polymerase II follow and depend on the actions of p300, which presumably include histone acetylation. We propose a model, on the basis of our results, to illustrate the events occurring on an androgen-regulated gene that is dependent on p300 (Fig. 8A). After DHT-activated AR binds to the ARE, p300 is among a group of coregulators recruited to the ARE. Other recruited coregulators include p160 coactivators which serve as scaffolds for recruitment of many other coregulators (48). Our results show that recruitment of p300 leads to increases in H3K18ac, H3K27ac, and H4ac in the region of the ARE and the TSS. Increased H3K4me1 is observed in the region of the ARE and increased H3K4me3 near the TSS. TBP and RNA polymerase II are recruited to the TSS, and active transcription ensues. In contrast, when p300 is depleted, DHT-induced transcription is inhibited on genes that require p300 for their DHT-induced expression. Although basal histone acetylation and H3K4me1 at the ARE and basal histone acetylation and H3K4me3 at the TSS are still observed, they are not increased by DHT. DHT also fails to induced recruitment of TBP and RNA polymerase II (Fig. 8B).
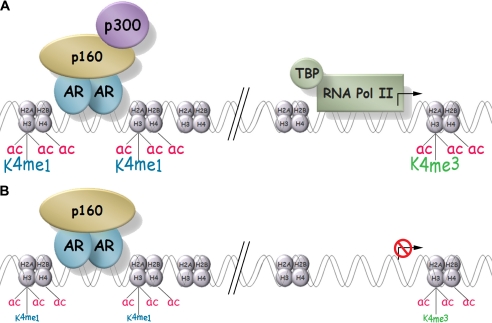
FIGURE 8.
Model of a p300-dependent AR target gene in intact and p300-depleted conditions. A, in intact cells DHT causes binding of AR to the ARE, which results in recruitment of p300 and a variety of other coactivators including p160 (or SRC) coactivators. p300 and other recruited histone acetyltransferases increase the acetylation levels of histones H3 and H4 (ac) near the AR binding site and the TSS. Subsequent events include increases in the level of H3K4me1 (K4me1) near the AR binding site and in the level of H3K4me3 (K4me3) near the TSS, recruitment of the basal transcription machinery including TBP and RNA polymerase II (RNA pol II), and enhanced transcription from the TSS (arrow). B, in the absence of p300, DHT still induces binding of AR to the ARE, and some coactivators (represented here by the p160 coactivator) may still be recruited normally to the promoter. However, acetylation of histones H3 and H4 and methylation of H3K4 are not enhanced by DHT; TBP and RNA polymerase II are not recruited to the TSS; and transcription enhancement by DHT is inhibited or eliminated (red barred circle covering the transcription arrow at the TSS).
In contrast, our results for FKBP5 show that different mechanisms of transcriptional regulation apply to different target genes of AR. p300 depletion prevented the normal pattern of androgen-induced increases in histone acetylation and H3K4 methylation on both the TMPRSS2 and FKBP5 genes. But although p300 depletion prevented the androgen-induced recruitment of TBP and RNA polymerase II on the TMPRSS2 gene (Fig. 6), these events occurred normally on the FKBP5 gene after p300 depletion (Fig. 7). Thus, neither p300 nor the androgen-induced increases in histone modifications are required for recruitment of TBP and RNA polymerase II to the FKBP5 gene or for androgen induced expression of FKBP5. These results suggest that the regulatory environment (binding of other transcription factors and/or chromatin conformation) associated with these two genes cause different coactivator requirements for the androgen regulated expression of the two genes.
By specifically placing the role of p300 within the ordered sequence of events that comprise chromatin remodeling and transcription complex assembly, our results have begun to elucidate how the actions of many different coregulators are coordinated to accomplish the complex task of DHT-induced transcriptional regulation.
Supplementary Material
Acknowledgments
We thank Dr. Gerhard Coetzee (University of Southern California) for the C4-2B cells and all the members of the Stallcup Laboratory for critical discussions. We also thank the Molecular Genomics Core and the Cancer Research Informatics Core of the University of Southern California Norris Comprehensive Cancer Center and the University of California Epigenome Center for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK043093 and R01DK055274 (to M. R. S.) and Training Grant T32GM067587 (for D. Y. W.). This work was also supported by Cancer Center award number P30CA014089 from the National Cancer Institute (for K. S.).
Microarray data sets can be accessed through the NCBI GEO Database under NCBI accession number GSE31873.

This article contains supplemental Tables 1–5 and references.
- AR
- androgen receptor
- DHT
- dihydrotestosterone
- ARE
- androgen response element
- TSS
- transcription start site
- ADI
- androgen depletion independent
- CBP
- cAMP response element-binding protein binding protein or CREB BP
- SRC
- steroid receptor coactivator
- qRT-PCR
- quantitative RT-PCR
- TBP
- TATA box binding protein
- sip300
- siRNA against p300
- siCBP
- siRNA against CBP
- siNS
- siRNA against a nonspecific sequence
- PSA
- prostate specific antigen
- H3K4me1
- histone H3 monomethylated at Lys-4
- H3K4me3
- histone H3 trimethylated at Lys-4
- H4ac
- acetylated histone H4
- H3K18ac
- histone H3 acetylated at Lys-18
- H3K27ac
- histone H3 acetylated at Lys-27.
REFERENCES
- 1. Claessens F., Denayer S., Van Tilborgh N., Kerkhofs S., Helsen C., Haelens A. (2008) Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl. Recept. Signal 6, e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heinlein C. A., Chang C. (2002) Androgen receptor (AR) coregulators. An overview. Endocr. Rev. 23, 175–200 [DOI] [PubMed] [Google Scholar]
- 3. Lamont K. R., Tindall D. J. (2010) Androgen regulation of gene expression. Adv. Cancer Res. 107, 137–162 [DOI] [PubMed] [Google Scholar]
- 4. Dutt S. S., Gao A. C. (2009) Molecular mechanisms of castration-resistant prostate cancer progression. Future Oncol. 5, 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heemers H. V., Tindall D. J. (2005) Androgen receptor coregulatory proteins as potential therapeutic targets in the treatment of prostate cancer. Curr. Cancer Ther. Rev. 1, 175–186 [Google Scholar]
- 6. Heemers H. V., Sebo T. J., Debes J. D., Regan K. M., Raclaw K. A., Murphy L. M., Hobisch A., Culig Z., Tindall D. J. (2007) Androgen deprivation increases p300 expression in prostate cancer cells. Cancer Res. 67, 3422–3430 [DOI] [PubMed] [Google Scholar]
- 7. Ma H., Nguyen C., Lee K. S., Kahn M. (2005) Differential roles for the coactivators CBP and p300 on TCF/β-catenin-mediated survivin gene expression. Oncogene 24, 3619–3631 [DOI] [PubMed] [Google Scholar]
- 8. Miyabayashi T., Teo J. L., Yamamoto M., McMillan M., Nguyen C., Kahn M. (2007) Wnt/β-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. U.S.A. 104, 5668–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeong K. W., Kim K., Situ A. J., Ulmer T. S., An W., Stallcup M. R. (2011) Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat. Struct. Mol. Biol. 18, 1358–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakravarti D., LaMorte V. J., Nelson M. C., Nakajima T., Schulman I. G., Juguilon H., Montminy M., Evans R. M. (1996) Role of CBP/P300 in nuclear receptor signalling. Nature 383, 99–103 [DOI] [PubMed] [Google Scholar]
- 11. Glass C. K., Rosenfeld M. G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 12. Bannister A. J., Kouzarides T. (1996) The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643 [DOI] [PubMed] [Google Scholar]
- 13. Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 14. Kawasaki H., Eckner R., Yao T. P., Taira K., Chiu R., Livingston D. M., Yokoyama K. K. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393, 284–289 [DOI] [PubMed] [Google Scholar]
- 15. Yuan Z. M., Huang Y., Ishiko T., Nakada S., Utsugisawa T., Shioya H., Utsugisawa Y., Shi Y., Weichselbaum R., Kufe D. (1999) Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene 18, 5714–5717 [DOI] [PubMed] [Google Scholar]
- 16. Kalkhoven E. (2004) CBP and p300. HATs for different occasions. Biochem. Pharmacol. 68, 1145–1155 [DOI] [PubMed] [Google Scholar]
- 17. Rebel V. I., Kung A. L., Tanner E. A., Yang H., Bronson R. T., Livingston D. M. (2002) Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. U.S.A. 99, 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McManus K. J., Hendzel M. J. (2001) CBP, a transcriptional coactivator and acetyltransferase. Biochem. Cell Biol. 79, 253–266 [PubMed] [Google Scholar]
- 19. Tanaka Y., Naruse I., Maekawa T., Masuya H., Shiroishi T., Ishii S. (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele. A partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. U.S.A. 94, 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yao T. P., Oh S. P., Fuchs M., Zhou N. D., Ch'ng L. E., Newsome D., Bronson R. T., Li E., Livingston D. M., Eckner R. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93, 361–372 [DOI] [PubMed] [Google Scholar]
- 21. Xu J., Wu R. C., O'Malley B. W. (2009) Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer 9, 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iyer N. G., Ozdag H., Caldas C. (2004) p300/CBP and cancer. Oncogene 23, 4225–4231 [DOI] [PubMed] [Google Scholar]
- 23. Culig Z., Bartsch G. (2006) Androgen axis in prostate cancer. J. Cell. Biochem. 99, 373–381 [DOI] [PubMed] [Google Scholar]
- 24. Debes J. D., Sebo T. J., Lohse C. M., Murphy L. M., Haugen D. A., Tindall D. J. (2003) p300 in prostate cancer proliferation and progression. Cancer Res. 63, 7638–7640 [PubMed] [Google Scholar]
- 25. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor. Open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi W., Oshlack A., Smyth G. K. (2010) Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38, e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson W. E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 [DOI] [PubMed] [Google Scholar]
- 28. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 29. Storey J. D. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 [Google Scholar]
- 30. Kim J. H., Li H., Stallcup M. R. (2003) CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell 12, 1537–1549 [DOI] [PubMed] [Google Scholar]
- 31. Wang Q., Li W., Liu X. S., Carroll J. S., Jänne O. A., Keeton E. K., Chinnaiyan A. M., Pienta K. J., Brown M. (2007) A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lucas J. M., True L., Hawley S., Matsumura M., Morrissey C., Vessella R., Nelson P. S. (2008) The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol. 215, 118–125 [DOI] [PubMed] [Google Scholar]
- 33. Lin B., Ferguson C., White J. T., Wang S., Vessella R., True L. D., Hood L., Nelson P. S. (1999) Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 59, 4180–4184 [PubMed] [Google Scholar]
- 34. Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 [DOI] [PubMed] [Google Scholar]
- 35. Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 37. Makkonen H., Kauhanen M., Paakinaho V., Jääskeläinen T., Palvimo J. J. (2009) Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 37, 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., Lin S. C., Heyman R. A., Rose D. W., Glass C. K., Rosenfeld M. G. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85, 403–414 [DOI] [PubMed] [Google Scholar]
- 39. Ramos Y. F., Hestand M. S., Verlaan M., Krabbendam E., Ariyurek Y., van Galen M., van Dam H., van Ommen G. J., den Dunnen J. T., Zantema A., 't Hoen P. A. (2010) Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 38, 5396–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., Yamamoto K. R. (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hall J. M., McDonnell D. P., Korach K. S. (2002) Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol. Endocrinol. 16, 469–486 [DOI] [PubMed] [Google Scholar]
- 42. Lefstin J. A., Yamamoto K. R. (1998) Allosteric effects of DNA on transcriptional regulators. Nature 392, 885–888 [DOI] [PubMed] [Google Scholar]
- 43. Schultz J. R., Loven M. A., Melvin V. M., Edwards D. P., Nardulli A. M. (2002) Differential modulation of DNA conformation by estrogen receptors α and β. J. Biol. Chem. 277, 8702–8707 [DOI] [PubMed] [Google Scholar]
- 44. Dallas P. B., Yaciuk P., Moran E. (1997) Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J. Virol. 71, 1726–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abraham S. E., Lobo S., Yaciuk P., Wang H. G., Moran E. (1993) p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene 8, 1639–1647 [PubMed] [Google Scholar]
- 46. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 47. Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y., Ge K. (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Q., Carroll J. S., Brown M. (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19, 631–642 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.