Background: ADP-ribosylation factor 6 is critical to a wide variety of cellular events, including vesicular trafficking and endocytosis.
Results: Inhibition of ARF6 by dominant mutants and siRNA impaired CpG ODN/TLR9-mediated response.
Conclusion: A novel class III PI3K-ARF6 axis pathway mediates TLR9 signaling by regulating the cellular uptake of CpG ODN.
Significance: Determining the physiological roles of ARF6 in CpG ODN/TLR9-mediated responses.
Keywords: Cell Biology, Cell Signaling, Cytokines Induction, Dendritic Cells, Macrophages, Toll-like Receptors (TLR), ADP-ribosylation Factor 6, CpG Oligodeoxynucleotides Cellular Uptake
Abstract
CpG oligodeoxynucleotide (CpG ODN) cellular uptake into endosomes, the rate-limiting step of Toll-like receptor 9 (TLR9) signaling, is critical in eliciting innate immune responses. ADP-ribosylation factor 6 (ARF6) is a member of the Ras superfamily, which is critical to a wide variety of cellular events including endocytosis. Here, we found that inhibition of ARF6 by dominant mutants and siRNA impaired CpG ODN-mediated responses, whereas cells expressing the constitutively active ARF6 mutant enhanced CpG ODN-induced cytokine production. Inhibition of ARF6 impaired TLR9 trafficking into endolysosomes, thereby inhibiting proceed functional cleavage of TLR9. Additional studies showed that CpG ODN uptake was increased in ARF6-activated cells but impaired in ARF6-defective cells. Furthermore, cells pretreated with CpG ODN but not GpC ODN had increased CpG ODN uptake due to CpG ODN-induced ARF6 activity. Further studies with ARF6-defective and ARF6-activated cells demonstrated that class III phosphatidylinositol 3-kinases (PI3K) was required for downstream ARF6 regulation of CpG ODN uptake. Together, our findings demonstrate that a novel class III PI3K-ARF6 axis pathway mediates TLR9 signaling by regulating the cellular uptake of CpG ODN.
Introduction
Bacterial DNA, known as immunostimulatory pathogen-associated molecular patterns, can directly activate B cells to proliferate and secrete immunoglobulins in a T cell-independent manner (1–3). It also induces B cells and monocytes to activate transcription factor NF-κB and secrete cytokines, including interleukin 12 (IL-12), tumor necrosis factor α (TNF-α), and interferon α/β (IFN-α/β) (4–7). The immunostimulatory activity of bacterial DNA has been assigned to unmethylated CpG motifs (GACGTT for murine, GTCGTT for humans) (8). Much evidence shows that synthetic oligodeoxynucleotides containing a CpG motif (CpG ODN),2 like bacterial DNA with the CpG moiety (CpG DNA), induce potent T helper 1-like immune responses that are protective against several infectious agents and immune disorders in animal models (9–11). Biologically active CpG ODN, such as bacteria DNA, activates macrophages and immature dendritic cells (DCs) to increase the expression of major histocompatibility complex class II and co-stimulatory molecules, thereby transcribing cytokine mRNAs and producing pro-inflammatory cytokines, including TNF-α, IL-1, IL-6, and IL-12 (11, 12). Unmethylated CpG ODN can, therefore, serve as an adjuvant and immunomodulator in vaccines against a wide variety of targets, including infectious agents, cancer antigens, and allergens (13, 14).
Toll-like receptor 9 (TLR9) was first cloned and identified as a receptor for bacterial DNA and unmethylated CpG DNA in 2000 (15). Analysis of TLR9-deficient mice revealed that TLR9 is essential for proinflammatory cytokine production and other inflammatory responses, and it also plays a role in the induction of T helper 1 adaptive immune responses and the proliferation of B cells. The binding of bacterial CpG DNA to TLR9 (16–18) and the subsequent endosomal maturation are thought to be essential for bacterial CpG DNA-driven immunostimulatory activity (19). After CpG DNA binding, TLR9 signaling is initiated by recruitment of the adaptor molecule myeloid differentiation factor 88 (MyD88). Recruitment of MyD88 is followed by engagement of IL-1 receptor-associated kinases (e.g. IRAK-1 and IRAK-4) and the adaptor protein TNF receptor-associated factor 6 (TRAF6). Oligomerization of TRAF6 can activate the inhibitor of κB (IκB) kinase complex (20–23) and subsequently activate the NF-κB-dependent genes, such as TNF-α, IL-1, and IL-6, thus leading to increased production of these cytokines (11, 24).
Although NF-κB is one of the key factors that affects cytokine production, CpG DNA has been shown to activate NF-κB and other transcription factors that are important regulators controlling the expression of many proinflammatory cytokines. These transcription factors include ATF2, CREB (cAMP-response element-binding protein), and C/EBP (24). In addition, CpG DNA activates stress kinases such as p38 mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K). Stress kinase activation is essential for CpG DNA-induced cytokine release of TNF-α and IL-12 (19). Our studies have indicated that CpG ODN induces the expression of heat shock proteins 70 (Hsp70) and 90β via a PI3K-dependent pathway. Furthermore, the up-regulation of heat shock proteins 70 and 90β plays a critical role in CpG ODN-mediated responses (25, 26). Although the molecular mechanism leading to the activation of TLR9 signaling is not fully understood, the cellular uptake of unmethylated CpG DNA/ODN into endosomes is thought to be the rate-limiting step for CpG DNA/TLR9-mediated signaling (27, 28). The uptake mechanism is one of the least well understood steps in CpG DNA/TLR9-mediated signaling. Accumulated evidence has shown that class III PI3K is specifically involved in TLR9 signaling by regulating the uptake of CpG ODN (29), but the precise mechanisms of CpG ODN uptake require further investigation.
ADP-ribosylation factors (ARFs) are members of the Ras superfamily of 20-kDa guanine nucleotide-binding proteins. There are six related gene products, ARF1 to ARF6, that have been divided into 3 classes on the basis of sequence homology (30): class I, ARF1 and ARF3; class II, ARF4 and ARF5; class III, ARF6. The function of ARF proteins depends on binding and hydrolyzing GTP with the protein forms, therefore cycling between GTP-bound (ARF-GTP) and GDP-bound (ARF-GDP). Class I and II ARFs localize primarily to intracellular organelles and have been implicated in many types of intracellular membrane vesicle trafficking events, such as vesicular transport between the endoplasmic reticulum (ER) and the Golgi and receptor recycling from endosomes to the plasma membrane (30–32). In contrast, class III ARF6 localizes on the plasma membrane and has been found to affect endocytosis, phagocytosis, receptor recycling, and the formation of actin-rich protrusions and ruffles (30–32). Although ARFs have important functions in cellular processes, studies demonstrating the precise role of each ARF in cellular biological responses have been limited because of a lack of specific inhibitors to individual ARFs. Recent reports have shown that the ARF-inhibitor brefeldin A impaired CpG ODN-induced NF-κB activation by blocking TLR9 trafficking through Golgi but not by inhibiting cellular CpG ODN uptake (33), suggesting that brefeldin A-sensitive ARFs plays a critical role in CpG ODN-mediated responses. Although the roles of individual brefeldin A-sensitive ARFs in TLR9-mediated signaling remain elusive, another important issue regarding the functions of brefeldin A-resistant ARF, ARF6, in CpG ODN/TLR9-mediated signaling is also unresolved. For example, CpG ODN uptake and TLR9 trafficking from the ER to endosomes are required for activation of CpG ODN/TLR9 signaling (27, 28, 34). Therefore, investigating the involvement of ARF6 in the process of both CpG ODN uptake and TLR9 trafficking is of interest.
TLR9 plays a critical role in unmethylated CpG DNA/ODN-induced innate immunity and is linked with a role in adaptive immunity by inducing the activation of various immune cell types. Thus, understanding the TLR9 signaling pathway will shed light on how the immune response is activated and will be of importance for developing specific therapies that can efficiently fight against infectious diseases. In this study we used dominant mutants and small interfering RNA (siRNA) to determine the physiological roles of ARF6 in CpG ODN/TLR9-mediated responses as well as the molecular mechanisms by which ARF6 regulates CpG ODN/TLR9-mediated responses. Our findings indicate that ARF6 is involved in CpG ODN/TLR9-mediated responses by regulating cellular CpG ODN uptake as well as the proteolytic processing of TLR9.
EXPERIMENTAL PROCEDURES
Reagents
Phosphorothioate-modified CpG ODN1668, CpG ODN1668-FITC, and GpC ODN were purchased from InvivoGen. IL-1β and TNF-α were from ProSpec. Wortmannin was from Sigma, and anti-Akt, -phospho-Akt (Ser473), -p38, -phospho-p38, -NF-κB, and -phospho-NF-κB antibodies were from Cell Signaling Technology. Anti-ARF6 and -MyD88 antibodies were from Santa Cruz Biotechnology. Anti-TLR9 and anti-TLR9 (C-term) was from InvivoGen or Zymed Laboratories Inc. Laboratories Inc., respectively. Anti-actin and -HA antibodies were from Millipore. The following antibodies were used for immunofluorescence staining: anti-TLR9-FITC (Imgenex), -LAMP-1 (BD Pharmingen), Dylight 488-conjugated goat anti-mouse IgG, and Dylight 549-conjugated donkey anti-rat IgG (Jackson ImmunoResearch Laboratories).
Cell Culture and Treatment
Mouse RAW264.7 macrophages and human embryonic kidney 293T cells (HEK293T) were from the American Type Culture Collection. RAW264.7 and HEK293T cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, 200 mmol/liter l-glutamine, and 50 μm β-mercaptoethanol in a humidified atmosphere of 5% CO2 at 37 °C. The medium was changed every 2 days for all experiments.
Cells were typically preincubated with or without inhibitors for 30 min before CpG ODN1668 treatment unless specified otherwise. The duration of CpG ODN treatment varied depending on the experiment.
Plasmacytoid Dendritic Cells (pDCs) and Monocyte/Macrophage Preparation
Normal 8-week-old male BALB/c mice from the National Laboratory Animal Center (Taipei, Taiwan) were sacrificed for the experiments by instant neck dislocation. Monocytes/macrophages and pDCs were then prepared from splenocytes. In brief, spleen cells were depleted of erythrocytes in 0.25× Hanks' balanced salt solution (HBSS) for 15 s followed by the addition of 2× HBSS and centrifugation. The cell pellet was suspended in RPMI1640 medium, and then pDCs and monocytes/macrophages were isolated with use of a pDC isolation kit and CD11b microBeads, respectively (Miltenyi Biotec), according to the manufacturer's instructions.
Stable Transfectants
HEK293T and mouse RAW264.7 cells were transfected with the plasmids pcDNA3-mTLR9, pcDNA3-ARF1T31N-HA, pcDNA3-ARF6-HA, pcDNA3-ARF6T27N-HA, pcDNA3-ARF6Q67L-HA, or pcDNA3.1-hvps34 by use of FuGENE 6 or FuGENE HD (Roche Applied Science) according to the manufacturer's instructions. Two days after transfection, G418 antibiotic (1.0 mg/ml) was used for clonal selection.
RNA Interference
Double-stranded RNAs were synthesized by Invitrogen. The following siRNA sequences were used for mouse ARF6: msiRNA-1 sense (5′-UAG AAC AAG CAG CGC AGC CUG UAA A-3′) and msiRNA-2 sense (5′-AAG UUC AAC GUG UGG GAU GUG-3′). The negative control sequence was 5′-UUC UCC GAA CGU GUC ACG UTT-3′. The siRNA sequence for human ARF6 was 5′-CGG CAU UAC ACU GGG AUU-3′. Cells were transfected with siRNA duplexes using Lipofectamine 2000 (Invitrogen). The transfection medium was removed 6 h later. The cells were then washed twice with phosphate-buffered saline (PBS) and maintained in culture medium for 48 to 72 h.
RAW264.7 cells were transiently transfected with a control plasmid (psiRNA-lucCL3) or psiRNA-tlr9 plasmid (InvivoGen) using FuGENE HD (Roche Applied Science) for 24 h. Cells were treated with GpC ODN or CpG ODN for 30 min.
Luciferase Reporter Assay
Cells in 24-well plates were co-transfected with 0.05 μg of p5xNF-κB-luc (Stratagene) and 0.01 μg of pcDNA3.1-β-galactosidase plus pcDNA3.1, pcDNA3-ARF1T31N-HA, or pcDNA3- ARF6T27N-HA using FuGENE 6 (Roche Applied Science) overnight. The cells were incubated with or without 1.0 μm CpG ODN for 8 h and then lysed. NF-κB luciferase activity assays were performed according to the procedures recommended by the manufacturer (Promega). β-Galactosidase activity was used to normalize the data.
Cytokine-specific ELISA
Microtiter plates (96-well) were coated with anti-mouse IL-6, TNF-α, and IFN-β (BIOSOURCE) in PBS at 4 °C overnight. After the plates were blocked and washed, supernatants from stimulated cells (1 × 106 cell/ml) were added, and the plates were incubated for 1 h at room temperature. The plates were then washed and treated with biotinylated anti-cytokine followed by streptavidin-HPR (BIOSOURCE). A standard curve generated with recombinant IL-6 and TNF-α was used to determine cytokine concentration.
Protein Extraction and Western Blot Analysis
The cells (2 × 106/well) were treated on ice for 15 min with 300 μl of lysis buffer (Pierce) supplemented with protease inhibitor mixture (Sigma). The lysates were subjected to centrifugation at 12,000 × g for 15 min at 4 °C, and protein concentrations in the supernatants were determined by use of the Bio-Rad Protein Assay. The supernatants (25–50 μg of protein/lane) were resolved by 5–20% SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare) according to the manufacturer's instructions.
Subcellular Fractionation
Cells were harvested in 1× PBS, centrifuged at 1500 rpm for 7 min, resuspended in 1× subcellular fractionation buffer (250 mm sucrose, 20 mm HEPES, pH 7.4, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, and 1 mm EGTA), and passed through a 27-gauge needle 6 times. The lysate was left on ice for 20 min. Cell lysate was then centrifuged at 1500 rpm for 10 min to pellet cell debris and unbroken cells and 7300 rpm for 10 min to pellet endolysosomal fractions. Differential centrifugation was performed in a Beckman TL100 ultracentrifuge. Collected supernatants were centrifuged at 80,000 rpm for 1 h in a Beckman TLA 100.4 rotor to pellet the endoplasmic reticulum fractions. All procedures were maintained at 4 °C or on ice.
Confocal Microscopy
Cells were fixed by incubation in PBS containing 3.5% paraformaldehyde at room temperature for 10 min. Cells were washed extensively and blocked with Image-iT FX signal enhancer (Invitrogen) for 30 min at room temperature. Cells were subsequently permeabilized with 0.1% Triton X-100 for 1.5 min at room temperature, then cells were washed with PBS and stained with anti-TLR9-FITC and anti-LAMP-1 at room temperature. After 1 h of staining, cells were washed and counterstained with Dylight 488-conjugated goat anti-mouse IgG and Dylight 549-conjugated donkey anti-rat IgG for 1 h. All cells were visualized under a Leica TCS SP5 laser-scanning confocal microscope.
CpG ODN Uptake
In brief, ODN1668-FITC-stimulated cells were washed with PBS, and surface-bound FITC was quenched by use of 0.04% trypan blue. The fluorescence intensity of ODN1668-FITC cellular uptake was measured at 530 nm using flow cytometry (BD Biosciences) at an excitation wavelength of 488 nm and was expressed as the mean value of the fluorescence intensity of 5000 cells.
ODN1668-FITC-stimulated cells were washed with PBS. The ODN1668-FITC cellular uptake (1.2 × 105 cells) was imaged by confocal microscopy.
Statistical Analysis
All values were given as the means ± S.D. The statistical significance of difference between treatment groups was calculated with a t test. p values of less than 0.05 were considered statistically significant.
RESULTS
ARF6 Is Associated with CpG ODN/TLR9 Signaling
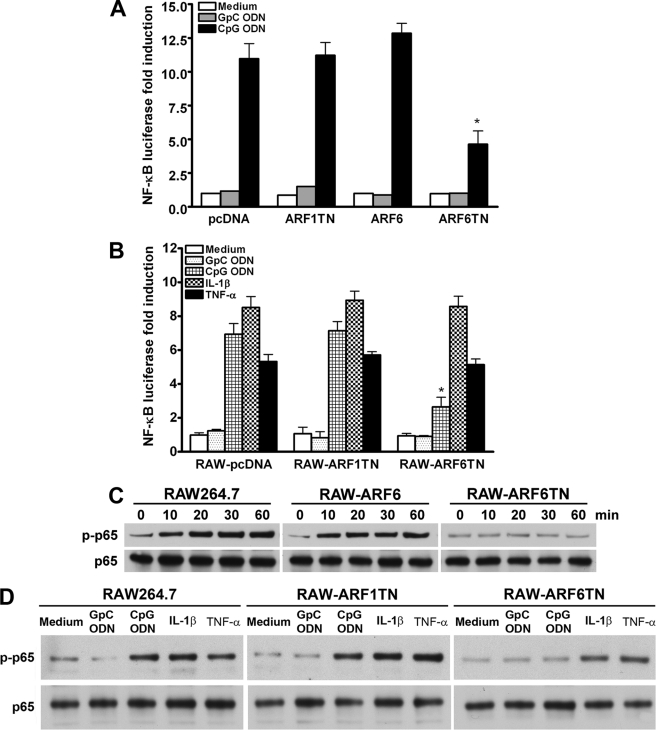
To evaluate the influence of ARF6 on CpG ODN/TLR9-mediated responses, we first examined whether ARF6 is involved in CpG ODN-stimulated NF-κB activation. As shown in Fig. 1, A and B, NF-κB luciferase activity induced by CpG ODN but not by GpC ODN (control ligand) in TLR9-overexpressing HEK293T or mouse macrophage RAW264.7 cells. The CpG ODN-induced NF-κB luciferase activity was reduced by transient expression of the dominant-negative form of ARF6 (ARF6T27N) but not expression of wild type ARF6 or the dominant-negative form of ARF1 (ARF1T31N) in TLR9-overexpressing HEK293T (Fig. 1A). Similarly, CpG ODN-induced NF-κB luciferase activity was impaired in RAW264.7 cells stably expressing ARF6T27N (RAW-ARF6T27N) but not in RAW-ARF1T31N cells (Fig. 1B). To further examine the role of ARF6 in TLR9-mediated NF-κB activation, siRNA for human ARF1 and ARF6 was used to knock down ARF1 and ARF6 expression in HEK293T cells (supplemental Fig. 1). NF-κB activation induced by CpG ODN was impaired in cells transfected with specific ARF6 siRNA but not ARF1 siRNA (supplemental Fig. 1). We also examined the role of ARF6 in NF-κB activation induced by IL-1R or TNFR. NF-κB luciferase activity induced by IL-1β or TNF-α was not affected by ARF6T27N (Fig. 1B). In addition to NF-κB luciferase results, the phosphorylation level of NF-κB p65 (Ser-536) induced by CpG ODN was decreased in RAW-ARF6T27N but not in RAW-ARF6 and RAW-ARF1T31N cells (Fig. 1, C and D). Interestingly, IL-1β- or TNF-α-induced phosphorylation of NF-κB p65 (Ser-536) was not substantially altered in RAW-ARF6T27N cells (Fig. 1D), suggesting that the effect of ARF6T27N is specific in CpG ODN/TLR9-mediated signaling.
FIGURE 1.
ARF6 is involved in CpG ODN-induced NF-κB activation and phosphorylation. A, HEK293T transfectants stably transfected with mouse TLR9 (293T-mTLR9) were transiently transfected with empty vector (pcDNA3.1), ARF1T31N, ARF6, or ARF6T27N plasmids plus p5xNF-κB-luciferase overnight, then stimulated with 1.0 μm GpC ODN (negative control) or CpG ODN for 8 h. NF-κB luciferase activities were then measured. Data represent the mean ± S.D. from three experiments. *, p < 0.05 for ARF6T27N versus pcDNA3.1. B, RAW-pcDNA3.1, RAW-ARF1T31N, and RAW-ARF6T27N cells were transiently transfected with p5xNF-κB-luciferase overnight and then stimulated with GpC ODN, CpG ODN, IL-1β (20 ng/ml), or TNF-α (10 ng/ml) for 8 h. NF-κB luciferase activities were then measured. Data represent the mean ± S.D. from two experiments. *, p < 0.01 for RAW-ARF6T27N versus RAW-pcDNA3.1. C, RAW264.7, RAW-ARF6, and RAW-ARF6T27N cells were treated with 1 μm CpG ODN for the indicated times. D, RAW264.7, RAW-ARF1T31N, and RAW-ARF6T27N cells were treated with GpC ODN, CpG ODN, IL-1β, or TNF-α for 30 min. Cell lysates were immunoblotted with antibodies specific for phospho-NF-κB p65 (Ser-536) or NF-κB p65 as indicated. The experiment was repeated three times with similar results.
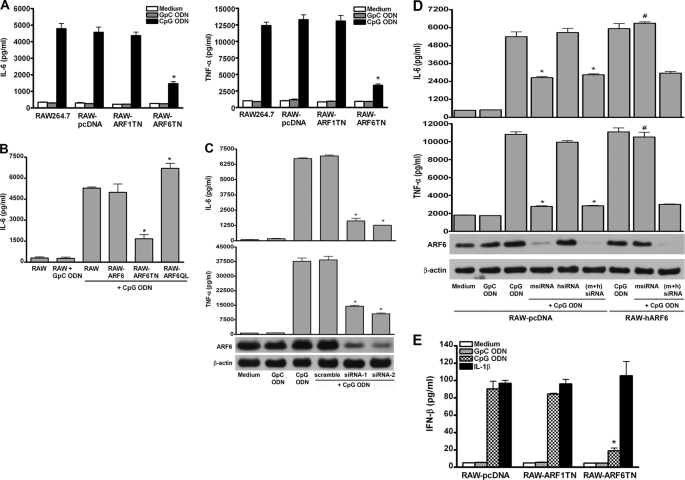
Because NF-κB is one of the key factors affecting cytokine production and CpG DNA has been shown to induce cytokine production, we next examined the role of ARF6 in CpG ODN-induced cytokine production. Cytokine production (e.g. IL-6 and TNF-α) induced by CpG ODN was impaired in RAW-ARF6T27N as compared with control RAW264.7, RAW-pcDNA, RAW-ARF6, and RAW-ARF1T31N cells (Figs. 2, A and B), whereas IL-6 production induced by IL-1β or TNF-α was not affected in RAW-ARF6T27N (supplemental Fig. 2). In addition, RAW264.7 cells expressing constitutively active human ARF6 (RAW-ARF6Q67L) enhanced the production of IL-6 induced by CpG ODN (Fig. 2B). These results support the proposal that the inhibitory effect of ARF6T27N on CpG ODN-mediated cytokine production is not due to nonspecific expression of ARF6 but to the specific blocking of ARF6 activation.
FIGURE 2.
ARF6 is involved in CpG ODN-induced IL-6 and TNF-α production. A, RAW264.7, RAW-pcDNA, RAW-ARF1T31N, and RAW-ARF6T27N cells were stimulated with medium alone, GpC ODN, or CpG ODN for 20 h as indicated. *, p < 0.001 for ARF6T27N versus RAW264.7. B, RAW264.7, RAW-ARF6, RAW-ARF6T27N, or RAW-ARF6Q67L cells were stimulated with medium alone, GpC ODN, or CpG ODN for 20 h as indicated. *, p < 0.001 for ARF6T27N or RAW-ARF6Q67L versus RAW264.7. C, pDCs were transfected with scramble siRNA, mouse ARF6 siRNA-1 (msiRNA-1) or ARF6 siRNA-2 (msiRNA-2) and incubated in medium or medium supplemented with GpC ODN or CpG ODN for 20 h as indicated. *, p < 0.001 for msiRNA-1 or -2 versus scramble. D, RAW264.7 or RAW-hARF6 cells were transiently transfected with mouse ARF6 siRNA (msiRNA), human ARF6 siRNA (hsiRNA) or mouse ARF6 siRNA plus human ARF6 siRNA ((m+h)-siRNA) for 48 h, then medium alone, GpC ODN, or CpG ODN stimulation for 20 h. IL-6 or TNF-α levels in the culture supernatants were measured by ELISA. Data represent the mean ± S.D. of three experiments. E, RAW-pcDNA, RAW-ARF1T31N, and RAW-ARF6T27N cells were stimulated with medium alone, GpC ODN, CpG ODN, or IL-1β for 20 h. IFN-β levels in the culture supernatants were measured by ELISA. Data represent the mean ± S.D. of 3 experiments. * p < 0.001 for ARF6T27N versus RAW-pcDNA.
Given the critical role of ARF6, determining the role of ARF6 in primary cells such as mouse spleen pDCs or macrophages is important. Production of IL-6 and TNF-α induced by CpG ODN was impaired in mouse spleen pDCs, macrophages, and RAW264.7 cells transfected with ARF6 siRNA directed to the mouse ARF6 gene as compared with those of cells transfected with scramble siRNA (Figs. 2, C and D, and supplemental Fig. 3).
To gain further insight into the relationship between ARF6 and CpG ODN-mediated cytokine production, we next examined whether stable expression of human ARF6 in RAW264.7 cells could restore mouse ARF6 siRNA-mediated inhibition of CpG ODN-induced cytokine production. Fig. 2D showed that RAW-ARF6 resistant to mouse ARF6 siRNA mediated the suppression of ARF6 protein level as well as CpG ODN-induced IL-6 and TNF-α production as compared with RAW264.7 cells. However, the restored effect was impaired in RAW-ARF6 cells cotransfected with mouse ARF6 siRNA and human ARF6 siRNA. In addition to cytokine production, we also found that IFN-β production induced by CpG ODN was impaired in RAW-ARF6T27N (Fig. 2E). In contrast, the IL-1β-induced IFN-β was not altered in RAW-ARF6T27N cells (Fig. 2E). Taken together these results suggest that ARF6 is required for TLR9-mediated immune activation, including NF-κB activation and production of cytokine and IFN-β.
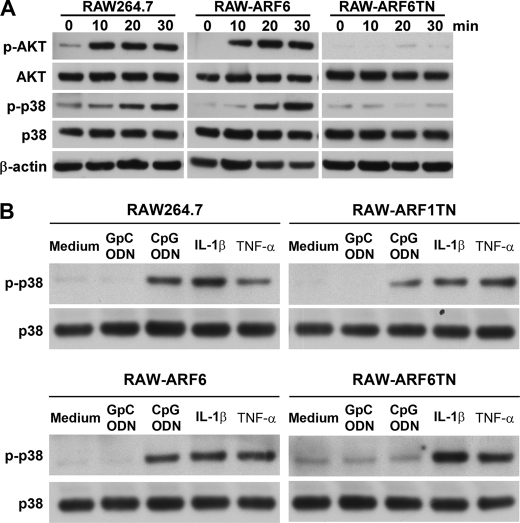
The activation of immune cells by bacterial CpG DNA or CpG ODN is through the TLR9 signaling cascade that regulates immune responses via the activation of many kinases, most notably, p38 MAPK, and PI3K. To evaluate the influence of ARF6 on CpG ODN-mediated activation of kinases, we measured the phosphorylation level of kinases (e.g. p38 MAPK and AKT) in RAW264.7, RAW-ARF1T31N, RAW-ARF6, and ARF6T27N cells. CpG ODN strongly induced the phosphorylation of p38 MAPK and AKT in RAW264.7, RAW-ARF1T31N, and RAW-ARF6 cells, whereas the effect of CpG ODN on inducing kinase activation was impaired in RAW-ARF6T27N (Fig. 3). In contrast, IL-1β- or TNF-α-induced phosphorylation of p38 MAPK was not substantially altered in RAW-ARF6T27N cells (Fig. 3B).
FIGURE 3.
The influence of ARF6 in CpG ODN-mediated activation of kinases. RAW264.7, RAW-ARF6, and RAW-ARF6T27N cells were treated with 1.0 μm CpG ODN for the indicated times. B, RAW-pcDNA3.1, RAW-ARF1T31N, RAW-ARF6, and RAW-ARF6T27N cells were stimulated with GpC ODN, CpG ODN, IL-1β (20 ng/ml), or TNF-α (10 ng/ml) for 30 min. Cell lysates were immunoblotted with antibodies specific for Akt, phospho-Akt, p38 MAPK, phospho-p38 MAPK, or β-actin. The experiment was repeated twice with similar results.
CpG ODN Stimulation Promotes the Activation of ARF6
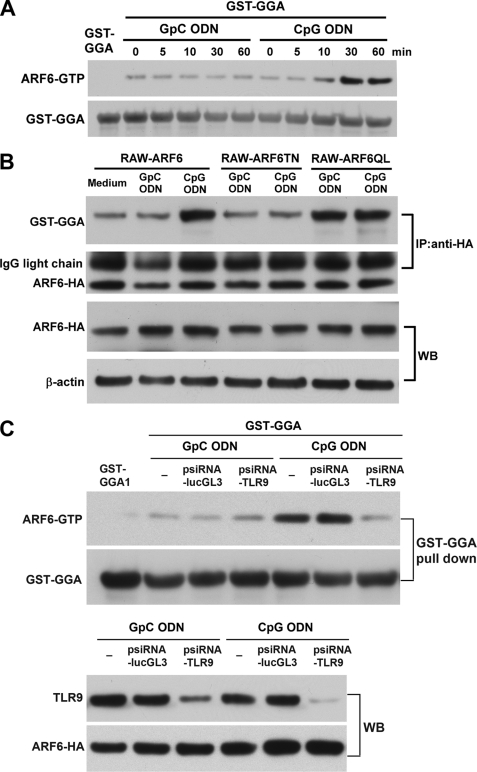
As we demonstrated, ARF6 plays a critical role in CpG ODN-mediated responses. We next examined the activation of ARF6 with CpG ODN treatment. To evaluate the ARF6 activation profile, we monitored the levels of GTP-bound ARF6 upon CpG ODN stimulation by using the ARF binding domain of GGA1 protein fused to GST, which binds the active form of ARF6 (ARF6-GTP). At different times during CpG ODN stimulation, the RAW264.7 cells expressing C-terminal HA-tagged wild type ARF6 were lysed on ice, and the activated ARF6 was pulled down by GST-GGA1 (Fig. 4A). A detectable degree of basal level of active ARF6 (ARF6-HA-GTP) was observed in untreated or GpC ODN-treated cells; furthermore, the ARF6-HA-GTP level was steadily increased in RAW-ARF6 cells treated with CpG ODN as compared with cells treated with GpC ODN (Fig. 4A). No precipitation of ARF6 was detected when GST alone was used in the pulldown assay (data not shown). In addition, the anti-HA immunoprecipitated assay also demonstrated that CpG ODN induced ARF6 activation in RAW-ARF6 cells (Fig. 4B). Our results indicated that GST-GGA1 was more obviously precipitated by anti-HA in RAW-ARF6 cells treated with CpG ODN as compared with cells treated with GpC ODN (Fig. 4B). However, the increase in GST-GGA1 level by CpG ODN was significantly inhibited in RAW-ARF6T27N cells, but the precipitated level of GST-GGA1 by anti-HA was stronger in RAW-ARF6Q67L cells treated with not only CpG ODN but also GpC ODN. To determine whether CpG ODN-induced ARF6 activation is dependent on TLR9, we used psiRNA-TLR9 to silence TLR9 expression (Fig. 4C) and analyzed CpG ODN-induced ARF6 activation. Silencing of TLR9 resulted in a marked reduction of ARF6-GTP induction, whereas a control plasmid psiRNA-lucGL3 did not (Fig. 4C). Taken together, these results suggest that CpG ODN/TLR9 enhances ARF6 activation, thereby advancing TLR9 signaling.
FIGURE 4.
ARF6 activation upon CpG ODN stimulation. A, RAW-ARF6 cells were treated with either GpC ODN or CpG ODN for the indicated times. The cells were then lysed on ice and subjected to ARF6 pulldown assays with the GST-GGA1 fusion protein. The level of ARF6-HA-GTP was assessed by immunoblotting with anti-HA. B, RAW-ARF6 cells were treated with either GpC ODN or CpG ODN for 30 min. Cell lysates were incubated with 10 μg GST-GGA1 protein and then were subjected to an immunoprecipitation (IP )assay with anti-HA. The precipitated level of GST-GGA1 was determined by immunoblotting (WB) with anti-GST. C, RAW-ARF6 cells were transfected with control plasmids (psiRNA-lucGL3) or psiRNA-TLR9 plasmid for 24 h and then stimulated with GpC ODN or CpG ODN for 30 min. Cell lysates were subjected to ARF6 pulldown assays with the GST-GGA1 fusion protein. The level of ARF6-HA-GTP was assessed by immunoblotting with anti-HA. The experiment was repeated twice with similar results.
ARF6 Functions Upstream of MyD88
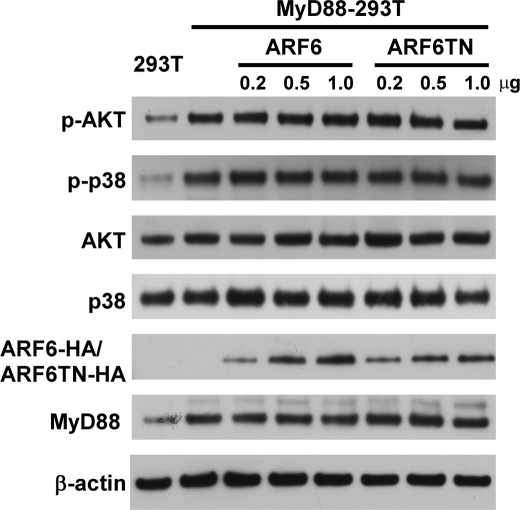
As we demonstrated, ARF6 is involved in CpG ODN-mediated kinase activation. We next attempted to delineate the sequence of action of ARF6 in the TLR9 signaling pathway. MyD88 is known as an important signal adaptor in the TLR9 signaling pathway. Overexpression of MyD88 induces activation of kinases and NF-κB through its death domain (29, 35). To determine whether ARF6 exerts its function upstream or downstream of MyD88, HEK293T cells were transiently co-transfected with a wild type MyD88 expression plasmid and various doses of ARF6-HA or ARF6T27N-HA plasmids. Overexpression of MyD88 induced the phosphorylation level of AKT and p38 (Fig. 5). Interestingly, overexpression of ARF6 or ARF6T27N had no significant effect on MyD88-induced AKT and p38 phosphorylation (Fig. 5). These results suggest that ARF6 regulates CpG ODN-mediated signaling upstream of MyD88.
FIGURE 5.
Inhibiting ARF6 does not affect phosphorylation of p38 and AKT induced by wild type MyD88 overexpression. Wild type MyD88 overexpressing 293T cells were transiently transfected with various concentrations of ARF6-HA or ARF6T27N-HA. After 24 h transfection cell lysates were immunoblotted with antibodies specific for Akt, phospho-Akt, p38 MAPK, phospho-p38 MAPK, or β-actin. The experiment was repeated twice with similar results.
Inhibition of ARF6 Interferes with TLR9 Intracellular Trafficking
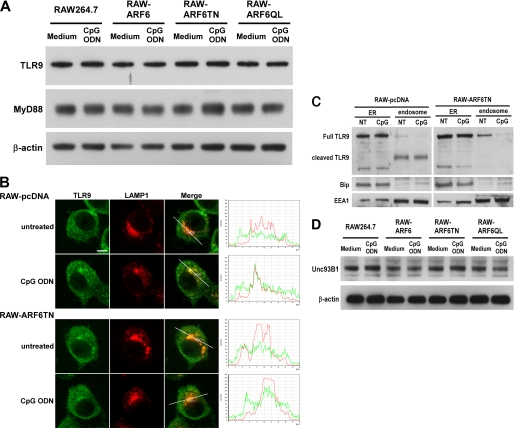
After CpG ODN stimulation, TLR9 recruits MyD88 to initiate CpG ODN-mediated signaling. To further understand the mechanism of action of ARF6 in CpG ODN-mediated signaling, we first assessed the intracellular expression of both TLR9 and MyD88 in RAW264.7, RAW-ARF6, RAW-ARF6T27N, and RAW-ARF6Q67L cells by Western blot analysis. Both TLR9 and MyD88 were constitutively expressed in all cell types, with no substantial difference on TLR9 and MyD88 expression level in CpG ODN-activated cells and resting cells (Fig. 6A), which suggests that the effect of ARF6 is not through altering TLR9 or MyD88 expression in RAW264.7 cells.
FIGURE 6.
Stable expression of ARF6T27N interferes with both trafficking and proteolytic processing of TLR9. A, cell lysates of RAW264.7, RAW-ARF6, RAW-ARF6T27N, or RAW-ARF6Q67L cells incubated in culture medium or medium supplemented with 1.0 μm CpG ODN for 2 h underwent Western blot analysis with antibodies specific for anti-TLR9, MyD88, or β-actin. B, RAW-pcDNA or RAW-ARF6T27N cells were treated with or without CpG ODN (1.0 μm) for 2 h. Cells were then fixed, permeabilized, and labeled with anti-TLR9 (green, FITC) and anti-LAMP1 (red, Dylight 549) antibodies. The histograms depict cross-line scans of the fluorescence intensities of the merged panel. These images are representative of three independent experiments. Scale bar, 5 μm. C, RAW-pcDNA or RAW-ARF6T27N cells were treated with or without (NT) CpG ODN (1.0 μm) for 2 h. Both ER fractions and endolysosomal fractions were immunoblotted with antibodies specific for TLR9, Bip, or EEA1. D, cell lysates of RAW264.7, RAW-ARF6, RAW-ARF6T27N, or RAW-ARF6Q67L cells incubated in culture medium or medium supplemented with 1.0 μm CpG ODN for 2 h underwent Western blot analysis with antibodies specific for anti-Unc93B1 or β-actin. The experiment was repeated three times with similar results.
Stimulation of CpG ODN caused TLR9 translocation from ER to endosomes and binding to its ligand CpG ODN to initiate signaling. As demonstrated above, ARF6 is dispensable for expression of TLR9 and MyD88; the other assumption is that ARF6 may modulate CpG ODN signaling by regulating TLR9 trafficking. To examine this hypothesis, we used fluorescent immunostaining to compare the localization of TLR9 in RAW-pcDNA and RAW-ARF6T27N cells before and after stimulation with CpG ODN. On activation of RAW-pcDNA cells with CpG ODN, TLR9 translocated from the ER to endolysosomes, as inferred from colocalization with the endolysosomal marker protein LAMP1 (Fig. 6B and supplemental Fig. 4). However, the TLR9 trafficking from ER to endolysosomes on CpG ODN stimulation was prevented in RAW-ARF6T27N cells (Fig. 6B and supplemental Fig. 4). Current reports indicate that nascent TLR9 is cleaved to generate a functional receptor by proteolytic processing. Furthermore, the cleaved TLR9, rather than full-length TLR9, recruits MyD88 to activate signal transduction (36, 37). To further investigate whether the effect of ARF6 acts on proteolytic processing of TLR9, we used Western blot to detect the level of cleaved TLR9 in the endolysosomal fraction. As shown in Fig. 6C, full-length TLR9 was mainly expressed in the ER fraction of RAW-pcDNA cells, whereas the cleaved TLR9 was only evident in the endolysosomal fraction of RAW-pcDNA cells. In RAW-ARF6T27N cells, the majority of full-length TLR9 was also observed in the ER fraction of these control cells; however, only a small amount of full-length TLR9 was detected in the endolysosomal fraction. Furthermore, the functional cleaved TLR9 was not expressed in the endolysosomal fraction of RAW-ARF6T27N cells, suggesting that ARF6 may be involved in the proteolytic processing of TLR9. Taken together, these results suggest that ARF6 is associated with the process of full-length TLR9 translocation from the ER to endolysosomes and may affect the proteolytic processing of TLR9.
UNC93B1 is responsible for TLR9 sorting from ER to endolysosomes as well as functional TLR9 production (36, 38); we thus sought to examine the expression level of UNC93B1 in ARF6-defective cells. As shown in Fig. 6D, the expression level of UNC93B1 shows no obvious difference in all cell types treated with or without CpG ODN, suggesting that ARF6 did not alter UNC93B1 expression in RAW264.7 cells. The mechanism by which ARF6 modulates TLR9 trafficking as well as functional TLR9 production is not by decreasing UNC93B1 expression.
ARF6 Is Involved in Cellular Uptake of CpG ODN
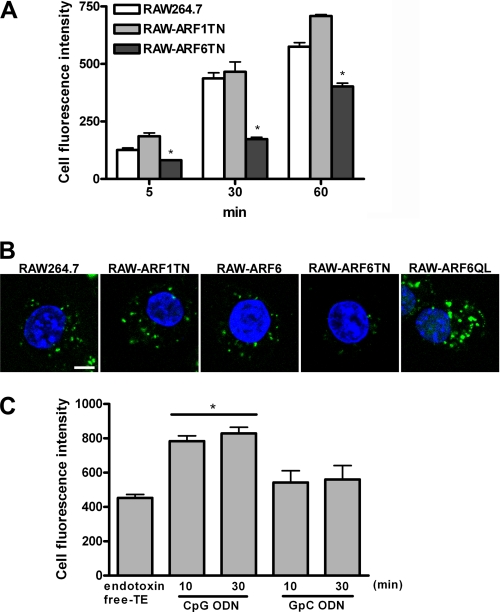
By our demonstration, ARF6 was activated by CpG ODN stimulation but not GpC ODN (Fig. 4); furthermore, CpG ODN-mediated responses were enhanced in RAW-ARF6Q67L cells but impaired in RAW-ARFT27N cells (Fig. 2), suggesting that CpG ODN-mediated responses are correlated with ARF6 activation. Cellular CpG ODN uptake is the limiting step for TLR9 signaling cascades; it is likely that ARF6 modulates CpG ODN/TLR9-mediated responses via regulating CpG ODN uptake. To examine the role of ARF6 in CpG ODN uptake, we used flow cytometry to compare the level of CpG ODN-FITC uptake in RAW264.7, RAW-ARF1T31N, and RAW-ARF6T27N cells. The level of CpG ODN-FITC uptake significantly decreased in RAW-ARF6T27N but not RAW-ARF1T31N cells as compared with RAW264.7 cells (Fig. 7A). Similar results were observed in ARF1, ARF3, or ARF6 siRNA-transfected mouse spleen macrophages, as cells expressing ARF6 siRNA but not scramble ARF1 and ARF3 siRNA inhibited the level of CpG ODN uptake (data not shown). In addition, the uptake level of CpG ODN-FITC was increased in RAW-ARF6Q67L cells but impaired in RAW-ARF6T27N cells as compared with RAW264.7 and RAW-ARF1T31N cells (Fig. 7B). In contrast, the uptake level of CpG ODN-FITC was not substantially altered in RAW-ARF6 cells, which suggests that ARF6 activation may be implicated in cellular CpG ODN uptake.
FIGURE 7.
ARF6 regulates cellular CpG ODN uptake. A, RAW264.7, RAW-ARF1T31N, or RAW-ARF6T27N cells were incubated with 0.5 μm CpG ODN-FITC for the indicated times. Cell fluorescence intensity of CpG ODN-FITC uptake was measured by flow cytometry. *, p < 0.01 for RAW-ARF6T27N versus RAW264.7. Data represent the mean ± S.D. of three experiments. B, RAW264.7, RAW-ARF1T31N, RAW-ARF6, RAW-ARF6T27N, or RAW-ARF6Q67L cells were incubated with 0.5 μm CpG ODN-FITC for 5 min. CpG ODN-FITC was detected by confocal microscopy. The experiment was repeated twice with similar results. Scale bar, 5 μm. C, RAW264.7 cells were pretreated with either CpG ODN or GpC ODN for the indicated times followed by 0.5 μm CpG ODN-FITC for 10 min. Cell fluorescence intensity of CpG ODN-FITC uptake was measured by flow cytometry. *, p < 0.01 for CpG ODN versus endotoxin-free Tris-EDTA buffer or GpC ODN.
To further investigate whether CpG ODN induces ARF6 activation, thereby enhancing cellular CpG ODN uptake, RAW264.7 cells were pretreated with CpG ODN or GpC ODN for 10 or 30 min and then washed twice with PBS; this was followed by the incubation of CpG ODN-FITC for 10 min. Our results showed that the uptake level of CpG ODN-FITC increased in CpG ODN pretreated cells as compared with endotoxin-free Tris-EDTA buffer-treated cells, whereas the CpG ODN-FITC uptake level was not substantially altered in GpC ODN pretreated cells (Fig. 7C). These results suggest that ARF6 activation induced by CpG ODN is implicated in regulating CpG ODN uptake, subsequently modulating CpG ODN/TLR9-mediated responses.
ARF6 Regulates CpG ODN Uptake through Its Upstream Molecule Class III PI3K
Previous reports have explored that class III PI3K specifically regulates TLR9-mediated immune responses by regulating the uptake of CpG ODN (29). We, therefore, used wild type class III PI3K (hvps34) plasmid (39) to evaluate the action sequence of class III PI3K and ARF6 on the mechanism of CpG ODN uptake. Consistent with previous published results (29), the level of CpG ODN-FITC uptake was increased in wild type class III PI3K (hvps34)-overexpressed RAW264.7 cells as compared with RAW264.7 cells transfected with control vector pcDNA3.1; however, the enhancement of CpG ODN-FITC uptake induced by overexpression of hvps34 was inhibited in RAW-ARF6T27N cells (Fig. 8A). With PI3K inhibitor wortmannin treatment, we confirmed that the CpG ODN-FITC uptake was inhibited in RAW-pcDNA cells. Moreover, the inhibitory effect of wortmannin on CpG ODN-FITC uptake was recovered in RAW-ARF6Q67L cells (Fig. 8B). These results suggest that ARF6 regulates CpG ODN uptake through its upstream molecule class III PI3K.
FIGURE 8.
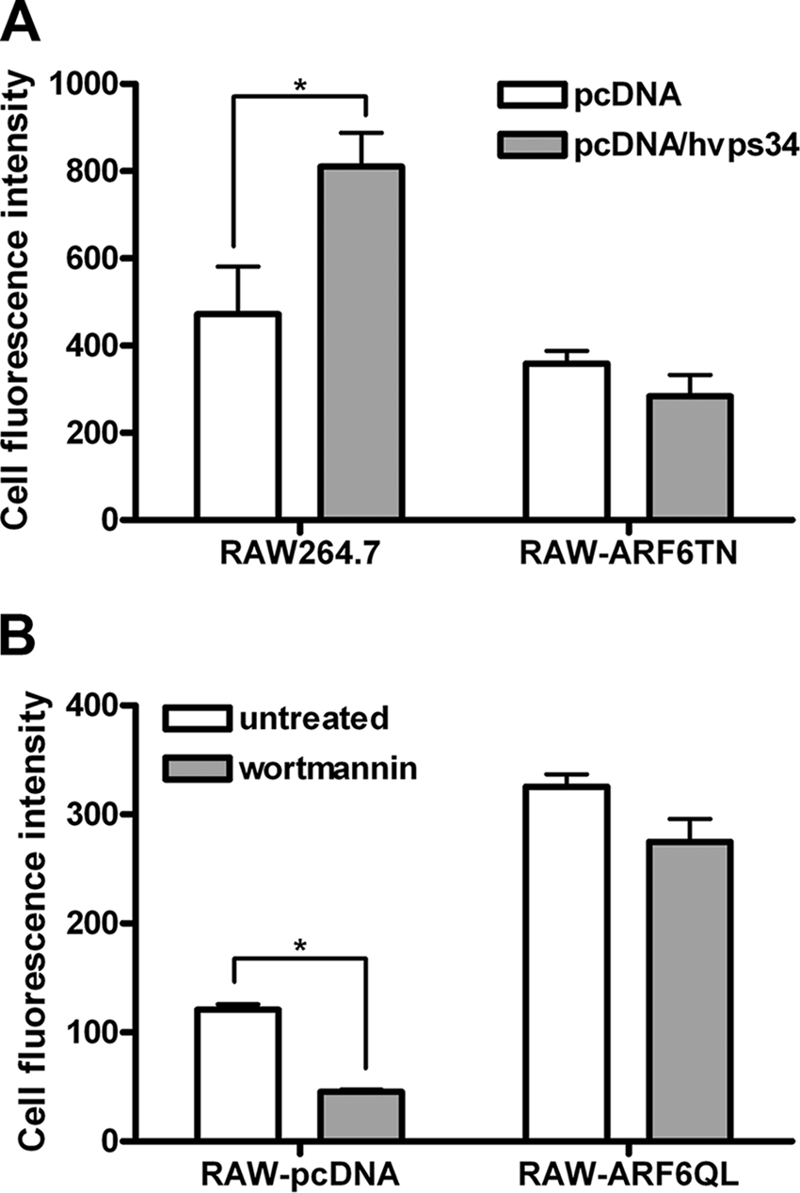
ARF6 regulates CpG ODN uptake through its upstream molecule class III PI3K. A, RAW264.7 and RAW-ARF6T27N cells were transiently transfected with empty vector (pcDNA3.1) or pcDNA3.1-hvps34. After 24 h transfection, cells were treated with 0.5 μm CpG ODN-FITC for 10 min. B, RAW-pcDNA or RAW-ARF6Q67L cells were pretreated with wortmannin for 30 min followed by 0.5 μm CpG ODN-FITC for 10 min. Cell fluorescence intensity of CpG ODN-FITC uptake was measured by flow cytometry. *, p < 0.001. Data represent the mean ± S.D. of three experiments.
DISCUSSION
Much evidence has shown that the ARF family of small GTPases has multiple roles in the regulation of cellular functions, including endocytosis and protein trafficking. The fungal metabolite brefeldin A blocks ARFs, except ARF6, transport to Golgi by preventing their association with the Golgi membrane and the coatomer complex formation. The dependence of the ARF inhibitor brefeldin A on CpG ODN/TLR9 signaling indicated that brefeldin A-sensitive ARFs impaired CpG ODN-induced NF-κB activation by blocking TLR9 trafficking through Golgi (33, 40). The brefeldin A-resistant ARF6 has been found to affect endocytosis as well as phagocytosis; however, the role of ARF6 in CpG ODN/TLR9 signaling remains unclear. In this study we used knockdown and dominate-negative approaches to show that ARF6 is associated with CpG ODN/TLR9-mediated responses, including NF-κB activation and cytokine production (e.g. IL-6 and TNF-α) (Figs. 1 and 2 and supplemental Figs. 1–3). CpG ODN-induced IL-6 increased in RAW264.7 cells expressing the constitutively active ARF6Q67L mutant (Fig. 2B). Furthermore, the rescue studies indicate that stable expression of human ARF6 in RAW264.7 cells reverse the suppressed effect of mouse ARF6 siRNA on CpG ODN-mediated IL-6 and TNF-α production (Fig. 2D). These results suggest that ARF6 activation plays an important role in CpG ODN/TLR9-mediated responses. The enhanced activation of ARF6 on CpG ODN stimulation (Fig. 4) supports this notion as well.
Signaling kinases such as p38 MAPK and PI3K were activated in CpG ODN/TLR9 signaling. The dominant-negative ARF6T27N mutant significantly inhibited CpG ODN-induced kinase phosphorylation (Fig. 3), which suggests that the inhibitory effects of ARF6T27N are due to the specific blocking of ARF6 activation, and the regulation of ARF6 in CpG ODN/TLR9 signaling is upstream of these kinases. TLR9 signaling is initiated by recruitment of MyD88 after CpG ODN binding to TLR9; therefore, it is reasonable to speculate that ARF6T27N may affect the expression of both TLR9 and MyD88. However, Western blot analysis revealed that ARF6T27N significantly inhibited CpG ODN-mediated responses without affecting the expression of TLR9 and MyD88 (Figs. 1–3 and 6A). In addition, experiments with MyD88-overexpressing 293T cells transfected with wild type or dominant-negative mutants of ARF6 demonstrate that ARF6 regulates CpG ODN-mediated signaling upstream of MyD88. It is well known that two critical steps upstream of MyD88 recruitment, CpG ODN uptake as well as TLR9 trafficking and proteolysis, are required for CpG ODN/TLR9-driven responses. Our finding indicates that ARF6T27N interfered with CpG ODN-induced TLR9 trafficking from the ER to endolysosomes (Fig. 6B). Furthermore, the production of functional TLR9 was also impaired in RAW-ARF6T27N cells (Fig. 6C). These results suggest that ARF6 is associated with full-length TLR9 translocation from the ER to endolysosomes and may affect the proteolytic processing of TLR9.
Exposing TLR9-expressing cells to CpG DNA/ODN results in the internalization of CpG DNA/ODN via an endocytic pathway leading to a tubular lysosomal compartment. TLR9, in the meantime, also moves into early endosomes from the ER and binds to CpG DNA/ODN. During the process, CpG DNA/ODN uptake is the rate-limiting step for activation of TLR9 signaling (27, 28, 34); thus, we think ARF6 may be directly involved in regulating TLR9 trafficking, or involvement in TLR9 trafficking may be via regulation of CpG ODN cellular uptake. Inhibition of ARF6 by expression of ARF6T27N or siRNA suppressed cellular CpG ODN uptake, whereas activation of ARF6 in RAW264.7 cells by the expression of ARF6Q67L increased CpG ODN uptake (Figs. 7 and 8). Therefore, we propose that ARF6 is involved in TLR9 trafficking and TLR9 signaling by regulating CpG ODN cellular uptake. Recently, the ARF inhibitor brefeldin A was reported to impair CpG ODN-mediated responses by inhibiting TLR9 trafficking (33). In this study we found that brefeldin A-resistant ARF6 (30) plays a critical role in CpG ODN/TLR9-mediated responses via regulating the uptake of CpG ODN. These observation suggest that brefeldin A-resistant ARF6 and brefeldin A-sensitive ARFs may exhibit different regulatory mechanisms in CpG ODN/TLR9-mediated signaling. However, which classes of brefeldin A-sensitive ARF proteins are involved in TLR9 trafficking and the roles of individual ARFs in TLR9-mediated signaling remain to be determined.
We demonstrated that activation of ARF6 increased, whereas inhibition of ARF6 decreased CpG ODN uptake and CpG ODN-induced responses (Figs. 1–3, 7, and 8). These results indicate that the amounts of CpG ODN uptake are positively correlated with the cellular activity of ARF6. Furthermore, pretreatment of cells with CpG ODN, but not GpC ODN, enhanced CpG ODN-FITC, suggesting that CpG ODN induces ARF6 activation, thereby increasing cellular CpG ODN uptake. We reasoned that there is a basal level of active ARF6 that can initiate CpG ODN uptake upon CpG ODN stimulation. The presence of a basal level of active ARF6 is supported by the factor that we were able to detect a low level of ARF6-GTP in ARF6 activation assay (Fig. 4). Furthermore, treatment with CpG ODN, but not GpC ODN, steadily increased ARF6-GTP level in RAW-ARF6 cells (Fig. 4). In addition, expression of constitutively active ARF6 in RAW264.7 cells exhibited higher levels of ARF6-GTP when compared with that of ARF6 (Fig. 4). Taken together, these finding strongly suggest that the basal level of active ARF6 is responsible for initiation of CpG ODN uptake, thereby inducing TLR9 responses and ARF6 activation. The propagating ARF6 activation by CpG ODN may thus lead to further increase of CpG ODN uptake.
CpG ODNs have shown substantial potency as vaccine adjuvants and as immunotherapeutic molecules for cancer and infectious and allergic diseases. Because cellular CpG ODN uptake efficiency is thought to be one of the challenges that limits its effectiveness, understanding the mechanism of CpG ODN uptake is crucial. Accumulated evidence has shown that class III PI3K is essential for CpG ODN uptake and in TLR9-mediated immune activation (29). In this study ARF6T27N inhibited the increased effect of expression of hvps34 in RAW264.7 cells on CpG ODN uptake (Fig. 8A), whereas ARF6Q67L restored the inhibitory effect of the PI3K inhibitor wortmannin on CpG ODN internalization (Fig. 8B). These results suggest that cellular CpG ODN uptake is regulated by a class III PI3K/ARF6-dependent pathway. Recent studies have reported that several plasma proteins, including KIR3DL2, CXCL16, scavenger receptor AI/II, and α2-macroglobulin, act as CpG ODN-binding proteins and deliver CpG ODN into endosomes (41–44). However, the relevance of a class III PI3K/ARF6-dependent pathway to these CpG ODN-binding proteins in the CpG ODN uptake mechanism requires further investigation, especially whether ARF6 can modulate the expression of these CpG ODN-binding proteins and where they are localized.
In summary, we clearly demonstrate that ARF6 plays a critical role in TLR9 signaling pathways in this study. Our findings suggest that the class III PI3K/ARF6-dependent pathway is involved in regulating cellular CpG ODN uptake, then subsequently modulates CpG ODN/TLR9-signaling cascades such as TLR9 trafficking, kinase activation, and cytokine production. Understanding the mechanism of CpG ODN uptake is valuable for CpG ODN-based therapies; our current results demonstrate that selective activation or inhibition of class III PI3K or ARF6 might be useful in certain immunological or therapeutic applications.
Supplementary Material
Acknowledgments
This research was conducted under the Graduate Program of Biotechnology in Medicine sponsored by National Tsing Hua University and the National Health Research Institutes.
This work was supported by grants from National Health Research Institutes (99A1-CSPP09-014) and the National Science Council (98-2320-B-400-007-MY3) of Taiwan.

This article contains supplemental Figs. 1–4.
- ODN
- oligodeoxynucleotide
- DC
- dendritic cell
- pDC
- plasmacytoid DC
- TLR9
- Toll-like receptor 9
- ARF6
- ADP-ribosylation factor 6
- MyD88
- myeloid differentiation factor 88
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Messina J. P., Gilkeson G. S., Pisetsky D. S. (1991) Stimulation of in vitro murine lymphocyte proliferation by bacterial DNA. J. Immunol. 147, 1759–1764 [PubMed] [Google Scholar]
- 2. Halpern M. D., Kurlander R. J., Pisetsky D. S. (1996) Bacterial DNA induces murine interferon-γ production by stimulation of interleukin-12 and tumor necrosis factor-α. Cell. Immunol. 167, 72–78 [DOI] [PubMed] [Google Scholar]
- 3. Yi A. K., Krieg A. M. (1998) CpG DNA rescue from anti-IgM-induced WEHI-231 B lymphoma apoptosis via modulation of IκBα and IκBβ and sustained activation of nuclear factor-κB/c-Rel. J. Immunol. 160, 1240–1245 [PubMed] [Google Scholar]
- 4. Tokunaga T., Yamamoto H., Shimada S., Abe H., Fukuda T., Fujisawa Y., Furutani Y., Yano O., Kataoka T., Sudo T. (1984) Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J. Natl. Cancer Inst. 72, 955–962 [PubMed] [Google Scholar]
- 5. Yamamoto S., Kuramoto E., Shimada S., Tokunaga T. (1988) In vitro augmentation of natural killer cell activity and production of interferon-α/β and -γ with deoxyribonucleic acid fraction from Mycobacterium bovis BCG. Jpn. J. Cancer Res. 79, 866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stacey K. J., Sweet M. J., Hume D. A. (1996) Macrophages ingest and are activated by bacterial DNA. J. Immunol. 157, 2116–2122 [PubMed] [Google Scholar]
- 7. Yi A. K., Tuetken R., Redford T., Waldschmidt M., Kirsch J., Krieg A. M. (1998) CpG motifs in bacterial DNA activate leukocytes through the pH-dependent generation of reactive oxygen species. J. Immunol. 160, 4755–4761 [PubMed] [Google Scholar]
- 8. Krieg A. M., Yi A. K., Matson S., Waldschmidt T. J., Bishop G. A., Teasdale R., Koretzky G. A., Klinman D. M. (1995) CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374, 546–549 [DOI] [PubMed] [Google Scholar]
- 9. Kline J. N., Waldschmidt T. J., Businga T. R., Lemish J. E., Weinstock J. V., Thorne P. S., Krieg A. M. (1998) Modulation of airway inflammation by CpG oligodeoxynucleotides in a murine model of asthma. J. Immunol. 160, 2555–2559 [PubMed] [Google Scholar]
- 10. Shirota H., Sano K., Hirasawa N., Terui T., Ohuchi K., Hattori T., Shirato K., Tamura G. (2001) Novel roles of CpG oligodeoxynucleotides as a leader for the sampling and presentation of CpG-tagged antigen by dendritic cells. J. Immunol. 167, 66–74 [DOI] [PubMed] [Google Scholar]
- 11. Kumagai Y., Takeuchi O., Akira S. (2008) TLR9 as a key receptor for the recognition of DNA. Adv. Drug Deliv. Rev. 60, 795–804 [DOI] [PubMed] [Google Scholar]
- 12. O'Neill L. A. (2006) How Toll-like receptors signal. What we know and what we don't know. Curr. Opin. Immunol. 18, 3–9 [DOI] [PubMed] [Google Scholar]
- 13. Takeda K., Akira S. (2001) Roles of Toll-like receptors in innate immune responses. Genes Cells 6, 733–742 [DOI] [PubMed] [Google Scholar]
- 14. Krieg A. M. (2007) Development of TLR9 agonists for cancer therapy. J. Clin. Invest. 117, 1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 16. Bauer S., Kirschning C. J., Häcker H., Redecke V., Hausmann S., Akira S., Wagner H., Lipford G. B. (2001) Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. U.S.A. 98, 9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. (2003) Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krug A., French A. R., Barchet W., Fischer J. A., Dzionek A., Pingel J. T., Orihuela M. M., Akira S., Yokoyama W. M., Colonna M. (2004) TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21, 107–119 [DOI] [PubMed] [Google Scholar]
- 19. Häcker H., Mischak H., Miethke T., Liptay S., Schmid R., Sparwasser T., Heeg K., Lipford G. B., Wagner H. (1998) CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 17, 6230–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akira S., Hemmi H. (2003) Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 85, 85–95 [DOI] [PubMed] [Google Scholar]
- 21. Häcker H., Vabulas R. M., Takeuchi O., Hoshino K., Akira S., Wagner H. (2000) Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J. Exp. Med. 192, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi T., Walsh M. C., Choi Y. (2004) The role of TRAF6 in signal transduction and the immune response. Microbes Infect 6, 1333–1338 [DOI] [PubMed] [Google Scholar]
- 23. Vilaysane A., Muruve D. A. (2009) The innate immune response to DNA. Semin. Immunol. 21, 208–214 [DOI] [PubMed] [Google Scholar]
- 24. Krieg A. M. (2002) CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20, 709–760 [DOI] [PubMed] [Google Scholar]
- 25. Kuo C. C., Liang S. M., Liang C. M. (2006) CpG-B oligodeoxynucleotide promotes cell survival via up-regulation of Hsp70 to increase Bcl-xL and to decrease apoptosis-inducing factor translocation. J. Biol. Chem. 281, 38200–38207 [DOI] [PubMed] [Google Scholar]
- 26. Kuo C. C., Liang C. M., Lai C. Y., Liang S. M. (2007) Involvement of heat shock protein (Hsp)90β but not Hsp90α in anti-apoptotic effect of CpG-B oligodeoxynucleotide. J. Immunol. 178, 6100–6108 [DOI] [PubMed] [Google Scholar]
- 27. Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. (2004) TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5, 190–198 [DOI] [PubMed] [Google Scholar]
- 28. Takeshita F., Gursel I., Ishii K. J., Suzuki K., Gursel M., Klinman D. M. (2004) Signal transduction pathways mediated by the interaction of CpG DNA with Toll-like receptor 9. Semin. Immunol. 16, 17–22 [DOI] [PubMed] [Google Scholar]
- 29. Kuo C. C., Lin W. T., Liang C. M., Liang S. M. (2006) Class I and III phosphatidylinositol 3′-kinase play distinct roles in TLR signaling pathway. J. Immunol. 176, 5943–5949 [DOI] [PubMed] [Google Scholar]
- 30. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins. Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 31. Inoue H., Randazzo P. A. (2007) Arf GAPs and their interacting proteins. Traffic 8, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 32. Randazzo P. A., Inoue H., Bharti S. (2007) Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell 99, 583–600 [DOI] [PubMed] [Google Scholar]
- 33. Chockalingam A., Brooks J. C., Cameron J. L., Blum L. K., Leifer C. A. (2009) TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol. Cell Biol. 87, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity. Update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 35. Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 273, 12203–12209 [DOI] [PubMed] [Google Scholar]
- 36. Ewald S. E., Lee B. L., Lau L., Wickliffe K. E., Shi G. P., Chapman H. A., Barton G. M. (2008) The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park B., Brinkmann M. M., Spooner E., Lee C. C., Kim Y. M., Ploegh H. L. (2008) Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim Y. M., Brinkmann M. M., Paquet M. E., Ploegh H. L. (2008) UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452, 234–238 [DOI] [PubMed] [Google Scholar]
- 39. Stein M. P., Feng Y., Cooper K. L., Welford A. M., Wandinger-Ness A. (2003) Human VPS34 and p150 are Rab7 interacting partners. Traffic 4, 754–771 [DOI] [PubMed] [Google Scholar]
- 40. Kuo C. C., Kuo C. W., Liang C. M., Liang S. M. (2005) A transcriptomic and proteomic analysis of the effect of CpG-ODN on human THP-1 monocytic leukemia cells. Proteomics 5, 894–906 [DOI] [PubMed] [Google Scholar]
- 41. Gursel M., Gursel I., Mostowski H. S., Klinman D. M. (2006) CXCL16 influences the nature and specificity of CpG-induced immune activation. J. Immunol. 177, 1575–1580 [DOI] [PubMed] [Google Scholar]
- 42. Butler M., Crooke R. M., Graham M. J., Lemonidis K. M., Lougheed M., Murray S. F., Witchell D., Steinbrecher U., Bennett C. F. (2000) Phosphorothioate oligodeoxynucleotides distribute similarly in class A scavenger receptor knockout and wild type mice. J. Pharmacol. Exp. Ther. 292, 489–496 [PubMed] [Google Scholar]
- 43. Anderson R. B., Cianciolo G. J., Kennedy M. N., Pizzo S. V. (2008) α2-macroglobulin binds CpG oligodeoxynucleotides and enhances their immunostimulatory properties by a receptor-dependent mechanism. J. Leukoc. Biol. 83, 381–392 [DOI] [PubMed] [Google Scholar]
- 44. Sivori S., Falco M., Carlomagno S., Romeo E., Soldani C., Bensussan A., Viola A., Moretta L., Moretta A. (2010) A novel KIR-associated function. Evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood 116, 1637–1647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.