Background: ATG16L1 is associated with the increased susceptibility to Crohn disease.
Results: ATG16L1 suppresses the IL-1β signaling via regulation of p62 stability and mediates ubiquitination of p62.
Conclusion: ATG16L1 suppresses IL-1β signaling by down-regulating p62 levels via both autolysosomal and proteasomal pathways.
Significance: p62 can be a target of intervention for Crohn patients.
Keywords: Autophagy, Inflammatory Bowel Disease, Proteasome, Toll IL-1 Receptor (TIR) Domain, Ubiquitin Ligase, Cullin-3, IL-1β Signaling, Neddylation, Ubiquitination, Autophagy
Abstract
ATG16L1 is an essential component of the autophagasome. The T300A allele of ATG16L1 is associated with the increased susceptibility to Crohn disease. In this study, we identified a novel function of ATG16L1, which suppresses signaling of the pro-inflammatory cytokine IL-1β. Deletion of ATG16L1 in mouse embryonic fibroblasts significantly amplifies IL-1β signal transduction cascades. This amplification is due to elevated p62 levels in ATG16L1-deficient cells. We found that ATG16L1 regulates p62 levels via both autolysosomal and proteasomal pathways. For proteasomal degradation, we found that Cullin-3 (Cul-3) is a E3 ubiquitin ligase of p62 and that ATG16L1 is essential for neddylation of Cul-3, a step required for Cul-3 activation. Taken together our data indicate that loss-of-function of ATG16L1 results in a hyper-responsiveness to the IL-1β signaling because of the increased p62 level.
Introduction
Autophagy is a process that removes damaged proteins and organelles in response to cellular stresses such as starvation. Autophagy also plays a role in cell defense by removing intracellular pathogens (1, 2). The ATG16L1 protein is essential for autophagy and the allele of this gene, T300A, is implicated in the susceptibility to Crohn disease (CD)3 (3), a form of inflammatory bowel disease (IBD). ATG16L1 regulates the granule exocytosis pathway in paneth cells (4), and suppresses induction of IL-1β in response to LPS in macrophages (5). These and other yet unknown functions of ATG16L1 may explain its role in intestinal inflammation.
Ubiquitinated protein aggregates are found in various human diseases, including neurodegenerative, liver, and muscle disorders (6). These protein aggregates contain typically p62 (Sequestosome-1, SQSTM1). p62 acts as a selective autophagy receptor for the ubiquitinated protein aggregates (6). In addition to its role in autophagy, p62 acts as an important scaffold in the IL-1β signaling pathway by promoting oligomerization of ubiquitinated TRAF6 (7) and MyD88 (8), and as an adaptor protein in Nrf2-induced expression of anti-oxidative response genes (9).
Protein ubiquitination is carried out by the sequential action of three enzymes: E1, E2, and E3. Cullin-3 (Cul-3) is a E3 ubiquitin ligase for various substrates and conjugation of Nedd8 to Cul-3 (neddylation) causes conformational changes in Cul-3 (10), which is critical for its dimerization and activation (11, 12). Deletion of Cul-3 causes developmental defects in Drosophila, such as external sensory organ development, pattern formation and cell growth and survival (13). p62 interacts with Keap1, a component of Cullin-3 (Cul-3) ubiquitin ligase for Nrf2 (9).
Here we report a novel function of ATG16L1 in the IL-1β signaling cascade. We discovered that ATG16L1 suppresses IL-1β signaling by promoting degradation of p62 via Cul-3-mediated proteasomal as well as via autolysosomal degradation.
EXPERIMENTAL PROCEDURES
Cells
WT and ATG16L1-deficient MEFs (mouse embryonic fibroblasts) (5) were gifts from Shizou Akira (Osaka University, Osaka, Japan) and ATG5 KO MEF from Noboru Mizushima (Tokyo Medical and Dental University, Japan) (14). MEFs were cultured in DMEM medium supplemented with 10% FBS.
Antibodies and Reagents
MG132, bafilomycin A1 (BA1), 3-methyladenine (3MA), anti-p62, anti-Flag, anti-SMA (smooth muscle actin), anti-V5, and anti-β-actin antibodies were obtained from Sigma. Mouse monoclonal anti-ATG16L1 is from MBL International (Woburn, MA). Anti-pERK, anti-p-JNK, anti-p-p38, anti-LC3, anti-Cul-3, anti-Myc, and anti-IκBα antibodies were from Cell Signaling Technologies (Danvers, MA), anti-vimentin antibody from BD Biosciences (Franklin Lakes, NJ), and anti-ubiquitin antibody and Cullin-3 siRNA from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-nedd8 and anti-GFP antibodies were purchased from Invitrogen (Carlsbad, CA). Anti-Cul-3 antibody for immunoprecipitation was purchased from Epitomics (Burlingame, CA). Anti-Nrf2 antibody was from R&D Systems (Minneapolis, MN). Human recombinant IL-1β was purchased from R&D System. IL-6 and TNF-α ELISA kits were purchased from eBiosciences (San Diego, CA).
Assays
EMSA, immunoblotting, immunoprecipitation, and qPCR were performed as described (15).
Transfection of MEF
Transfection of MEF with siRNA or plasmid DNA was done by electroporation with Nucleofector (Lonza, Basel, Switzerland) using MEF2 solution and protocol T-020.
Stable Transfection of p62 shRNA
Control and p62 shRNA bearing lentivirus particles were purchased from Santa Cruz Biotechnology. Two days post-infection, cells were selected with puromycin (1 μg/ml) for several passages before usage.
Plasmids
Flag-ATG16L1 is described elsewhere (1), and human p62 cDNA was PCR cloned at Bgl2/NotI sites into a modified pCMV (Clontech) containing eGFP. V5-tagged Cul-3 plasmid was obtained from Michael Freeman (Vanderbilt University) (16). Human Myc-Nedd8 expression vector was purchased from OriGene (Rockville, MD). The Flag-ubiquitin construct was obtained from M. Karin (UCSD).
Quantitative PCR (qPCR)
Quantitative PCR was performed as described with SYBR Green and GAPDH as control (15) (Table 1).
TABLE 1.
PCR primers
| Gene names | Forward primer | Reverse primer |
|---|---|---|
| 5′-3′ | 5′-3′ | |
| GAPDH | TCA ACA GCA ACT CCC ACT CTT | ACC CTG TTG GTG TAG CCG TAT |
| IL-6 | CTG CAA GAG ACT TCC ATC CAG TT | AAG TAG GGA AGG CCG TGG TT |
| KC | CTG GGA TTC ACC TCA AGA AC | GAA GCC AGC GTT CAC CAG AC |
| p62 | TGA AAC ATG GAC ACT TTG GCT | ACA TTG GGA TCT TCT GGT GGA |
| IκBα | TGG CCT TCC TCA ACT TCC AGA ACA | TCA GGA TCA CAG CCA GCT TTC AGA |
p62 KO Mice
p62 KO mice were kindly provided by Jose Moscat (7). To address the in vivo IL-1β response, WT or KO mice (n = 3–4) were intraperitoneal injected, sera were collected 90 min post injection, and TNFα or IL-6 was measured by ELISA (eBiosciences).
Densitometry
Densitometry of all blots was performed with Image J software.
Statistical Significance
Statistical significances were calculated with one-way ANOVA or Student's t test. p values less than 0.05 were considered significant.
Immunofluorescence
Immunofluorescence was performed as described (15). Briefly, MEF grown in chamber slides were fixed with 4% paraformaldehyde for 15 min at room temperature, washed with PBS/Triton X-100 (1%, PTX buffer) twice, incubated with the indicated primary antibody for 1 h in PTX, washed twice with PTX, incubated with a secondary antibody conjugated with Alexa Fluor 488 or 546 (Invitrogen) for 1 h, washed twice in PTX, and mounted for confocal imaging (Olympus FV1000).
RESULTS
Autophagy Suppresses IL-1β Signal Transduction
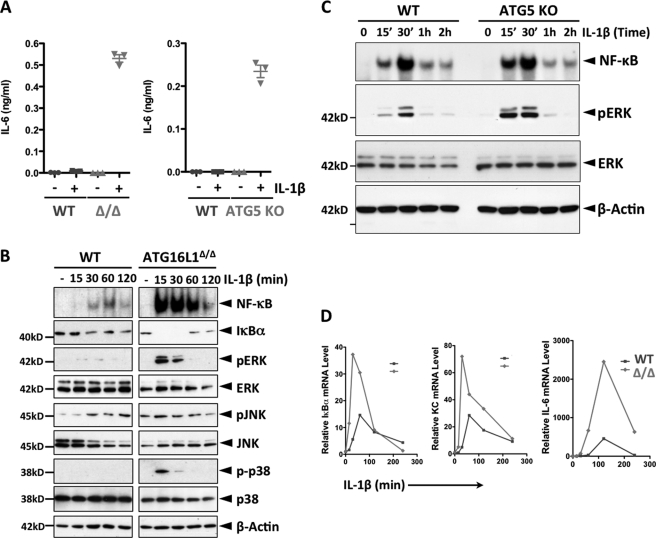
To investigate the role of autophagy in IL-1β signaling, we used WT, ATG16L1-deficient (described in related figures as ATG16L1Δ/Δ or Δ/Δ) MEF (mouse embryonic fibroblasts) (5) and ATG5 KO cells (14). While WT cells produced little IL-6, both ATG16L1-deficient and ATG5-KO cells produced it at a high level in response to IL-1β stimulation (Fig. 1A). Consistently, the levels of IL-1β activated downstream signal transducers were significantly higher in ATG16L1-deficient cells compared with those in WT cells (3-fold in NF-κB activation and 30–50-fold in MAPKs) (Fig. 1B). In subsequent studies, we used activation of ERK as a surrogate for all MAPKs. Similarly, activation of IL-1β-induced NF-κB and ERK in ATG5 KO cells was higher, 2- and 6-fold, respectively, compared with WT cells (Fig. 1C). Additionally, the levels of IL-1β-induced IκBα, IL-6, and KC (keratinocyte-derived chemokine) mRNA were significantly higher in ATG16L1-deficient cells than in WT cells (Fig. 1D). However, the impact of ATG16L1 on the signaling of LPS or TNF-α was minimal (supplemental Fig. S1). Collectively, these data demonstrate that IL-1β signaling is amplified in autophagy-deficient MEF, resulting in transcriptional and translational activation of downstream targets. Because the impact of ATG16L1 or ATG5 deficiency on IL-1β signaling is similar, most of the subsequent studies were performed in ATG16L1-deficient MEF.
FIGURE 1.
Loss of autophagy function enhances IL-1β signaling cascades. A, loss of ATG16L1 or ATG5 function enhances IL-1β-induced induction of IL-6 protein. The indicated MEF cell lines were stimulated with IL-1β (1 ng/ml) for 24 h and the levels of IL-6 were determined (ELISA). B, ATG16L1 deficiency enhances IL-1β-induced activation of NF-κB and MAPKs. WT and ATG16L1-deficient MEF were stimulated with IL-1β (1 ng/ml) as indicated. Activation of NF-κB was measured by EMSA, and degradation of IκBα and phosphorylation of MAPKs by immunoblotting (IB). C, ATG5 deletion enhances IL-1β-induced activation of NF-κB and ERK. WT and ATG5 KO MEF were stimulated with IL-1β (1 ng/ml) as indicated. Activation of NF-κB was measured by EMSA, and phosphorylation of ERK by IB. D, ATG16L1 deficiency enhances IL-1β-induced transcription of downstream target genes. WT and ATG16L1-deficient MEF were stimulated with IL-1β (1 ng/ml) as indicated and the levels of the indicated transcripts were measured by qPCR and plotted. All above data are representative of at least two independent experiments.
ATG16L1 suppresses IL-1β signal transduction via down-regulation of p62. The fact that both NF-κB and MAPK pathways are affected by ATG16L1 deletion indicated that its regulation of IL-1β signaling by ATG16L1 must be at or above the divergence of both pathways, such as at the level of MyD88-IRAK-TRAF6-TAK1 (17). We therefore searched for molecules that are affected by ATG16L1 and influence the MyD88-IRAK-TRAF6-TAK1 activation. p62 accumulates in cells with a defective autophagasome formation (18), and acts as a signaling hub through its ability to recruit and oligomerize signaling molecules (19). For instance, it promotes IL-1β signaling by its association with TRAF6, promoting the oligomerization of ubiquitinated TRAF6 (7, 20).
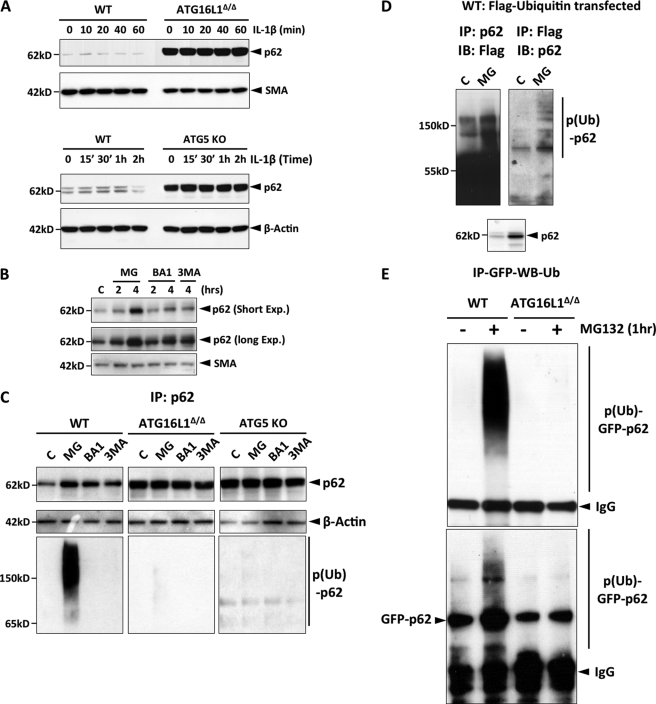
The p62 level in ATG16L1-deficient and ATG5 KO MEF was 20 and 6 fold higher than that observed in WT MEF, respectively (Fig. 2A). However, the p62 transcript levels were not affected by ATG16L1 deficiency (supplemental Fig. S2A). Although it is known that p62 is primarily degraded by the autolysosome, we found that p62 is also degraded by the proteasome: Both proteasome inhibition (5-fold) and autolysosome inhibition (2-fold) induced p62 accumulation (Fig. 2B). Furthermore, ectopically expressed GFP-p62 was also accumulated upon proteasomal inhibition (supplemental Fig. S2B), indicating that the accumulation of p62 by proteasomal inhibition was not due to the transcriptional activation of p62. Both proteasomal and lysosomal p62 degradation was dependent on autophagy (ATG5 and ATG16L1) (Fig. 2C), but these two degradation pathways were distinct because only the proteasomal pathway involved ubiquitination of p62 (Fig. 2C). We further demonstrated the ubiquitination of p62 with overexpression of Flag-tagged ubiquitin by immunoprecipitation and immunoblotting (Fig. 2D). In addition, GFP-p62 underwent ubiquitination in a ATG16L1-dependent manner under the denaturing conditions (Fig. 2D). These data strongly indicate that p62 itself undergoes ubiquitination in an autophagy-dependent manner.
FIGURE 2.
Autophagy mediates both proteasomal and lysosomal degradation of p62. A, p62 protein level is elevated in ATG16L1-deficient and ATG5 KO MEF. The indicated cell lines were stimulated with IL-1β (1 ng/ml), and the levels of the indicated proteins were measured by IB. B, p62 is degraded by both proteasome and autolysosome. WT MEF were treated with MG132 (10 μm), bafiliomycin A1 (BA1, 100 nm), 3-methyladenine (3MA, 1 mm) and the indicated proteins were measured by IB (exp.: exposure). All above data are representative of at least five independent experiments. C, autophagy regulates degradation of p62 by the ubiquitin-proteasome pathway. The cell lines were treated as indicated and p62 and actin levels were measured by IB. To remove the noise from the p62-interacting ubiquitinated proteins, similar amounts of p62 were immunoprecipitated (rather than the same amount of total extracts), subjected to SDS-PAGE, and probed with anti-ubiquitin antibody. D, p62 is ubiquitinated. WT MEF were transfected with Flag-ubiquitin, and either p62 or Flag-ubiquitin was immunoprecipitated and probed for Flag or p62, respectively. All data are representative of at least two independent experiments. E, GFP-p62 is ubiquitinated in a ATG16L1-dependent manner. WT or ATG16L1-deficient MEF were transfected with GFP-p62 and treated with MG132 (10 μm) for 1 h. The lysates were harvested under the denaturing condition (the buffer containing 20 mm HEPES, pH 7.5, 0.15 m NaCl, 1 mm DTT, 1 mm MEM (N-ethylmaleimide), 1% Triton X-100, 1% SDS, protease mixture (Sigma)), and immediately boiled for 10 min. The samples were diluted 3× with the same buffer without SDS (final SDS concentration: 0.33%), p62 was immunoprecipitated with a monoclonal anti-GFP antibody and subjected to IB with anti-ubiquitin and anti-p62 antibodies successively.
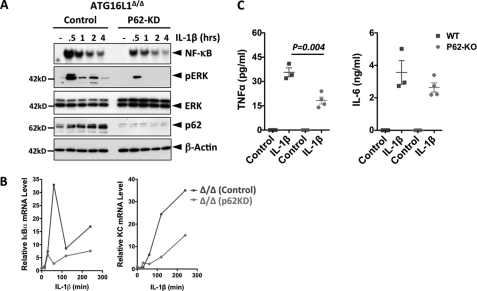
We next tested whether the elevated p62 protein level in ATG16L1-deficient cells is responsible for enhanced IL-1β signaling. Indeed, siRNA-mediated silencing of p62 in ATG16L1-deficient cells inhibited IL-1β-induced activation of NF-κB (2-fold reduction) and ERK (3 fold reduction) (Fig. 3A), and suppressed transcription of IκBα and KC (Fig. 3B). These data indicate that elevated p62 enhances IL-1β signaling in ATG16L1-deficient cells. Furthermore, knockdown or ectopic expression of p62 in WT MEF suppressed or enhanced IL-1β signaling, respectively (supplemental Fig. S3).
FIGURE 3.
p62 enhances the strength of IL-1β signaling. A, p62 knockdown (KD) suppresses IL-1β signaling cascades in ATG16L1-deficient MEF. Cells were transfected with either control or p62 shRNA, and were stimulated with IL-1β (1 ng/ml). Activation of NF-κB was measured by EMSA, and the levels of pERK, p62, and β-actin by IB. B, p62 KD in ATG16L1-deficient MEF suppresses IL-1β-induced transcription of IκBα and KC. The mRNA levels of the indicated genes were measured by qPCR. All data are representative of at least two independent experiments. C, p62 enhances IL-1β signaling in vivo. WT or p62 KO mice (n = 3–4, 10-week-old) were intraperitoneal injected with IL-1β (100 ng/mouse), and the sera were collected 90 min postinjection. The levels of TNFα and IL-6 were measured by ELISA.
We next explored the role of p62 in IL-1β signaling in vivo. We measured serum cytokine levels induced by IL-1β in WT or p62 KO mice and found that TNFα or IL-6 production in p62 KO mice was significantly lower than that in WT mice (Fig. 3C), confirming the amplifying role p62 in IL-1β signaling.
ATG16L1 Induces Ubiquitination of p62
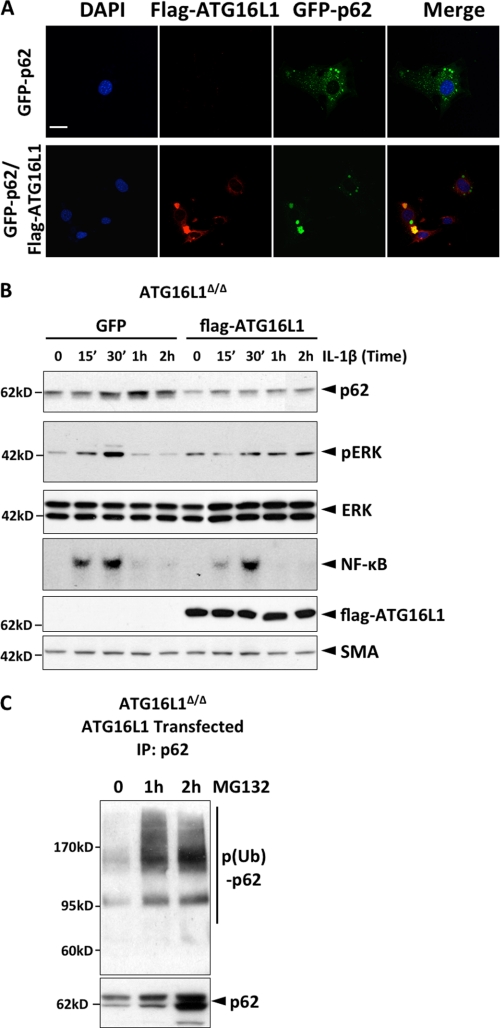
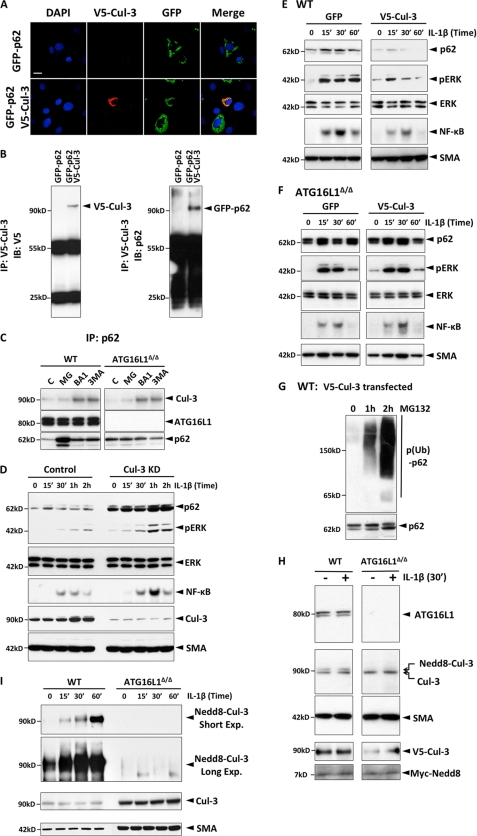
We next investigated how ATG16L1 regulates p62 ubiquitination. First we show that flag-ATG16L1 co-localizes with GFP-p62 in WT MEF (Fig. 4A). In addition, reconstitution of ATG16L1 in KO cells decreased the p62 protein level (2-fold) and suppressed IL-1β-induced activation of NF-κB (1.5-fold) and ERK (3-fold) (Fig. 4B). Blocking proteasomal degradation in the ATG16L1-reconstituted cells increased p62 level (3-fold increase) and induced accumulation of ubiquitinated p62 (Fig. 4C).
FIGURE 4.
ATG16L1 induces ubiquitination of p62. A, ATG16L1 co-localizes with p62. WT MEF were transfected either with GFP-p62 alone or with both flag-ATG16L1 and GFP-p62, and IHC was performed with anti-Flag antibody. (Scale bar represents 50 μm.) B, reconstitution of ATG16L1 suppresses p62 expression and IL-1β signaling cascades in ATG16L1-deficient MEF. Cells were transfected with the vector encoding either GFP or ATG16L1, stimulated with IL-1β (1 ng/ml) and the levels of pERK, p62, ATG16L1 and SMA (smooth muscle actin) were determined by IB. C, reconstitution of ATG16L1 induces ubiquitination of p62 in ATG16L1-deficient MEF. ATG16L1-deficient MEF were transfected with Flag-ATG16L1, treated with MG132, p62 was immunoprecipitated and blotted for ubiquitin. All data are representative of more than two independent experiments.
Cullin-3 Is E3 Ubiquitin Ligase of p62
Because p62 is known to interact with Keap1, a component of Cullin-3 (Cul-3) ubiquitin ligase for Nrf2 (9), we tested whether Cul-3 ubiquitinates p62. Endogenous Cul-3 levels were not significantly affected by ATG16L1 deficiency (supplemental Fig. S4A). Recombinant Cul-3 indeed interacted with p62 (Fig. 5, A and B, and supplemental Fig. S4B) but this interaction did not require ATG16L1 (Fig. 5C). In addition, we found that endogenous ATG16L1, p62, and Cul-3 proteins also form an immunoprecipitable complex (Fig. 5C). Silencing of Cul-3 in WT cells significantly enhanced the p62 level (7-fold) and IL-1β signaling (3-fold in NF-κB and 7-fold in pERK) (Fig. 5D). However, this p62 up-regulation was not due to the increased expression of Nrf2, a transcriptional activator of p62 (21) (supplemental Fig. S5). Furthermore, while overexpression of Cul-3 in WT cells suppressed p62 level (5-fold) and IL-1β signaling (2-fold in NF-κB and 3-fold in pERK) (Fig. 5E), it did not affect p62 level and IL-1β signaling in ATG16L1-deficient cells (Fig. 5F). Blocking proteasome activity in Cul-3-transfected WT cells induced accumulation of ubiquitinated p62 (Fig. 5G). Collectively, these data indicate that Cul-3 ubiquitinates p62 in an ATG16L1-dependent mechanism.
FIGURE 5.
ATG16L1 mediates ubiquitination of p62 by Cul-3. A, p62 co-localizes with Cul-3. GFP-p62 and V5-Cul-3 were expressed in WT MEF and IF was performed with anti-V5 antibody. (Scale bar represents 20 μm.) B, Cul-3 interacts with p62. WT MEF were transfected with either GFP-p62 alone or with GFP-62 plus V5-Cul-3. V5-Cul-3 was immunoprecipitated with anti-V5 antibody, and the precipitates were immunoblotted for p62 and V5-Cul-3 successively. C, endogenous ATG16L1, p62, and Cul-3 form an immunoprecipitable complex. WT or ATG16L1-deficient MEF were treated as indicated, p62 was immunoprecipitated and the levels of ATG16L1 and Cul-3 were measured successively by IB. D, Cul-3 silencing increases the p62 level and amplifies IL-1β signaling. WT MEF were transfected with either control or Cul-3 siRNA. The levels of p62, pERK, ERK, Cul-3, Nrf-2, and SMA were determined by IB and NF-κB by EMSA. E, Cul-3 overexpression in WT MEF decreases the p62 level and suppresses IL-1β signaling. WT MEF were transfected with either GFP or V5-Cul-3. The levels of p62, pERK, ERK, Cul 3, and SMA were measured by IB and NF-κB was measured by EMSA. F, Cul-3 overexpression in ATG16L1-deficient MEF does not affect the p62 level or IL-1β signaling. ATG16L1-deficient MEF were transfected with either GFP or V5-Cul-3. The levels of p62, pERK, Cul-3, and SMA were determined by IB, and NF-κB by EMSA. G, expression of Cul-3 induces ubiquitination of p62. WT MEF were transfected with V5-Cul-3 and treated with MG132 as indicated. p62 was immunoprecipitated and immunoblotted for ubiquitin. All data are representative of more than two independent experiments. H, ATG16L1 is required for Cul-3 neddylation. WT or ATG16L1-deficient MEF were transfected with V5-Cul-3 plus Myc-Nedd8, and the levels of the indicated proteins were measured by IB. I, ATG16L1 is required for Cul-3 neddylation. WT or ATG16L1-deficient MEF were stimulated with IL-1β as indicated, Cul-3 was immunoprecipitated, and probed for Nedd8 or Cul-3 by IB.
ATG16L1 Is Essential for Cullin-3 Neddylation
Because ATG16L1 did not regulate the Cul-3 expression level (supplemental Fig. S4A), we studied whether and how ATG16L1 regulates Cul-3 activity. Conjugation of Nedd8 (neddylation) to Cul-3 is critical for its dimerization and activity (11, 12). To determine the neddylation of Cul-3, we overexpressed V5-Cul-3 and myc-Nedd8, and analyzed the neddylation by immunoblotting. We detected in WT cell lysate a distinct band above Cul-3 (i.e. Nedd8-Cul-3), which was absent in ATG16L1-deficient MEF lysate (Fig. 5H). Using immunoprecipitation, we confirmed that endogenous Cul-3 is neddylated only in WT but not in ATG16L1-deficient cells. Both IL-1β-induced and constitutive neddylation of Cul-3 was completely dependent on ATG16L1 (Fig. 5I). Collectively, these data demonstrate that ATG16L1 is required for Cul-3 neddylation and thus for Cul-3-dependent ubiquitination and degradation of p62.
DISCUSSION
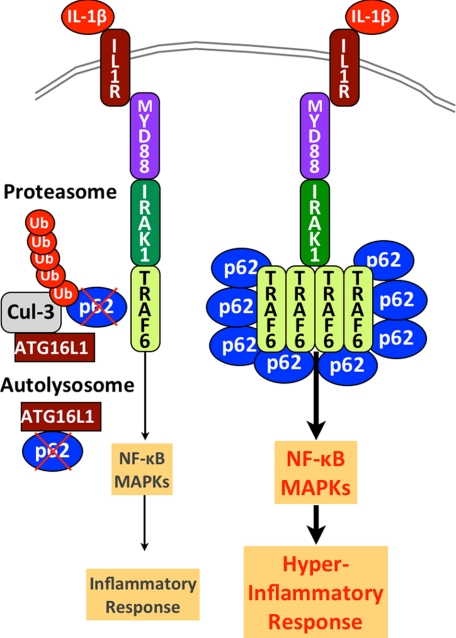
Our data provide the molecular mechanism by which ATG16L1 suppresses a potent pro-inflammatory signal. As summarized in Fig. 6, ATG16L1, most likely in complex with ATG5-ATG12 (22), suppresses IL-1β signaling by down-regulating p62 levels. p62 was proposed to regulate the assembly and delivery of polyubiquitinated, misfolded, or aggregated proteins, or dysfunctional organelles for their clearance through autophagy (23). In addition, it promotes aggregation of ubiquitinated proteins such as TRAF6 for the activation of IL-1β signaling (7). The expression level of p62 is important for these biological functions and it is thought to be regulated solely by autophagy (18). Our data demonstrate that p62 also undergoes proteasomal degradation through Cul-3-mediated ubiquitination and that both ATG16L1 and ATG5 play a key role in both processes. A recent report showed that a significant portion of p62 co-elutes with proteasome when either proteasome or lysosome function is inhibited, and that ATG16L1 associates with proteasome under steady state or upon proteasomal inhibition (24).
FIGURE 6.
ATG16L1 is a negative regulator of IL-1β signaling. Constitutive degradation of p62 by the autolysosome and the proteasome in the presence of WT ATG16L1 restrains IL-1β signaling cascades and the subsequent inflammatory response. In the absence of ATG16L1, p62 levels are increased. This increase in p62 levels promotes oligomerization and activation of TRAF6 (7, 19), resulting in overactivation of NF-κΒ and MAPKs upon IL-1β stimulation that leads to a hyper-inflammatory response.
ATG16L1 deletion in mouse macrophages causes over-production of IL-1β protein upon LPS stimulation (5), which may explain the heightened susceptibility of mice carrying ATG16L1-deficient macrophages in an animal model of ulcerative colitis (5). On the other hand, the reduced expression of ATG16L1 in intestinal epithelial cells impedes the release of antimicrobial peptides from Paneth cells, potentially resulting in altered microbial communities adjacent to epithelial crypts (4). A recent study proposed that viral infection in a host carrying the susceptible ATG16L1 allele can serve as an initial trigger for a CD-like intestinal inflammation of the colon (25).
Our data provide novel insight to underlying molecular mechanisms and demonstrate how ATG16L1 suppresses directly a pro-inflammatory signal. Our model predicts that IL-1β overproduced by macrophages with dysfunctional ATG16L1 (5) also provokes a hyper-inflammatory response in autophagy-deficient cells due to the enhanced TRAF6/p62 oligomerization, which amplifies the downstream signal transduction (Fig. 6). It also suggests that the threshold of IL-1β protein levels to induce inflammatory responses would be much lower in the hosts carrying a CD-susceptible ATG16L1 allele (T300A) than that in those with a CD-resistant allele. Finally, our results suggest that CD patients who carry the mutant form of ATG16L may benefit from the inhibition of IL-1β signaling by neutralizing IL-1β levels or by blocking IL-1R.
Supplementary Material
Acknowledgments
We thank the following investigators for their generous gifts; Shizou Akira (Osaka University, Japan) for WT and ATG16L1-deficient MEFs, Noboru Mizushima (Tokyo Medical and Dental University, Japan) for WT and ATG5 KO MEFs, and Michael Freeman (Vanderbilt University School of Medicine) for V5-Cullin-3 plasmid. We also would like to thank Jennifer Meerloo for assistance in confocal imaging at UCSD Neuroscience Microscopy Shared Facility. We are grateful to Jose Moscat for providing p62 KO mice (7) and Angeles Duran (Sanford-Burnham Medical Research Institute) for technical assistance in IL-1β injection to and in serum collection from p62 KO mice. We thank Scott Herdman for careful editing of the manuscript.
This work was supported by National Institutes of Health Grants A1068685, A1095623, DK35108, and DK080506, and a grant from CCFA.

This article contains supplemental Figs. S1–S5.
- CD
- Crohn disease
- TIR
- Toll IL-1 receptor
- MEF
- mouse embryonic fibroblasts
- SMA
- smooth muscle actin
- qPCR
- quantitative PCR.
REFERENCES
- 1. Kuballa P., Huett A., Rioux J. D., Daly M. J., Xavier R. J. (2008) Impaired autophagy of an intracellular pathogen induced by a Crohn disease-associated ATG16L1 variant. PLoS One 3, e3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lapaquette P., Glasser A. L., Huett A., Xavier R. J., Darfeuille-Michaud A. (2010) Crohn disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 12, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrett J. C., Hansoul S., Nicolae D. L., Cho J. H., Duerr R. H., Rioux J. D., Brant S. R., Silverberg M. S., Taylor K. D., Barmada M. M., Bitton A., Dassopoulos T., Datta L. W., Green T., Griffiths A. M., Kistner E. O., Murtha M. T., Regueiro M. D., Rotter J. I., Schumm L. P., Steinhart A. H., Targan S. R., Xavier R. J., Libioulle C., Sandor C., Lathrop M., Belaiche J., Dewit O., Gut I., Heath S., Laukens D., Mni M., Rutgeerts P., Van Gossum A., Zelenika D., Franchimont D., Hugot J. P., de Vos M., Vermeire S., Louis E., Cardon L. R., Anderson C. A., Drummond H., Nimmo E., Ahmad T., Prescott N. J., Onnie C. M., Fisher S. A., Marchini J., Ghori J., Bumpstead S., Gwilliam R., Tremelling M., Deloukas P., Mansfield J., Jewell D., Satsangi J., Mathew C. G., Parkes M., Georges M., Daly M. J. (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn disease. Nat. Genet. 40, 955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cadwell K., Liu J. Y., Brown S. L., Miyoshi H., Loh J., Lennerz J. K., Kishi C., Kc W., Carrero J. A., Hunt S., Stone C. D., Brunt E. M., Xavier R. J., Sleckman B. P., Li E., Mizushima N., Stappenbeck T. S., Virgin H. W., 4th (2008) A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., Akira S. (2008) Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 [DOI] [PubMed] [Google Scholar]
- 6. Knaevelsrud H., Simonsen A. (2010) Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 584, 2635–2645 [DOI] [PubMed] [Google Scholar]
- 7. Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Into T., Inomata M., Niida S., Murakami Y., Shibata K. (2010) Regulation of MyD88 aggregation and the MyD88-dependent signaling pathway by sequestosome 1 and histone deacetylase 6. J. Biol. Chem. 285, 35759–35769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 10. Liu J., Nussinov R. (2010) Rbx1 flexible linker facilitates cullin-RING ligase function before neddylation and after deneddylation. Biophys. J. 99, 736–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wimuttisuk W., Singer J. D. (2007) The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol. Biol. Cell 18, 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hotton S. K., Callis J. (2008) Regulation of cullin RING ligases. Annu Rev. Plant Biol. 59, 467–489 [DOI] [PubMed] [Google Scholar]
- 13. Mistry H., Wilson B. A., Roberts I. J., O'Kane C. J., Skeath J. B. (2004) Cullin-3 regulates pattern formation, external sensory organ development, and cell survival during Drosophila development. Mech Dev. 121, 1495–1507 [DOI] [PubMed] [Google Scholar]
- 14. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 15. Lee J., Mo J. H., Katakura K., Alkalay I., Rucker A. N., Liu Y. T., Lee H. K., Shen C., Cojocaru G., Shenouda S., Kagnoff M., Eckmann L., Ben-Neriah Y., Raz E. (2006) Maintenance of colonic homeostasis by distinctive apical TLR9 signaling in intestinal epithelial cells. Nat. Cell Biol. 8, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 16. Gao L., Wang J., Sekhar K. R., Yin H., Yared N. F., Schneider S. N., Sasi S., Dalton T. P., Anderson M. E., Chan J. Y., Morrow J. D., Freeman M. L. (2007) Novel n-3 fatty acid oxidation products activate Nrf2 by destabilizing the association between Keap1 and Cullin3. J. Biol. Chem. 282, 2529–2537 [DOI] [PubMed] [Google Scholar]
- 17. Verstrepen L., Bekaert T., Chau T. L., Tavernier J., Chariot A., Beyaert R. (2008) TLR-4, IL-1R, and TNF-R signaling to NF-κB: variations on a common theme. Cell Mol. Life Sci. 65, 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moscat J., Diaz-Meco M. T. (2009) p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seibenhener M. L., Babu J. R., Geetha T., Wong H. C., Krishna N. R., Wooten M. W. (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 24, 8055–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain A., Lamark T., Sjøttem E., Larsen K. B., Awuh J. A., Øvervatn A., McMahon M., Hayes J. D., Johansen T. (2010) p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285, 22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mizushima N., Noda T., Ohsumi Y. (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18, 3888–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 24. Myeku N., Figueiredo-Pereira M. E. (2011) Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: Association with sequestosome 1/p62. J. Biol. Chem., 286, 22426–22440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cadwell K., Patel K. K., Maloney N. S., Liu T. C., Ng A. C., Storer C. E., Head R. D., Xavier R., Stappenbeck T. S., Virgin H. W. (2010) Virus-plus-susceptibility gene interaction determines Crohn disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.