Abstract
Precise balance between phosphorylation, catalyzed by protein kinases, and dephosphorylation, catalyzed by protein phosphatases, is essential for cellular homeostasis. Deregulation of this balance leads to pathophysiological states that drive diseases such as cancer, heart disease, and diabetes. The recent discovery of the PHLPP (pleckstrin homology domain leucine-rich repeat protein phosphatase) family of Ser/Thr phosphatases adds a new player to the cast of phosphate-controlling enzymes in cell signaling. PHLPP isozymes catalyze the dephosphorylation of a conserved regulatory motif, the hydrophobic motif, on the AGC kinases Akt, PKC, and S6 kinase, as well as an inhibitory site on the kinase Mst1, to inhibit cellular proliferation and induce apoptosis. The frequent deletion of PHLPP in cancer, coupled with the development of prostate tumors in mice lacking PHLPP1, identifies PHLPP as a novel tumor suppressor. This minireview discusses the structure, function, and regulation of PHLPP, with particular focus on its role in disease.
Keywords: Akt/PKB, Phosphatase, Phosphatidylinositol 3-Kinase, Protein Kinase C (PKC), Signal Transduction, PHLPP
Structure
The PHLPP (pleckstrin homology domain leucine-rich repeat protein phosphatase) enzymes are novel members of the protein phosphatase 2C (PP2C)3 grouping in the protein phosphatase metal-dependent (PPM) family of Ser/Thr phosphatases (1). The catalytic core of PPMs has a signature set of conserved Asp residues that coordinate Mg2+ or Mn2+, metals required for catalytic activity (2). Like other PPMs, but in contrast to the more abundant and better characterized PPPs (phosphoprotein phosphatase), PHLPP contains regulatory modules within the same polypeptide as the phosphatase domain.
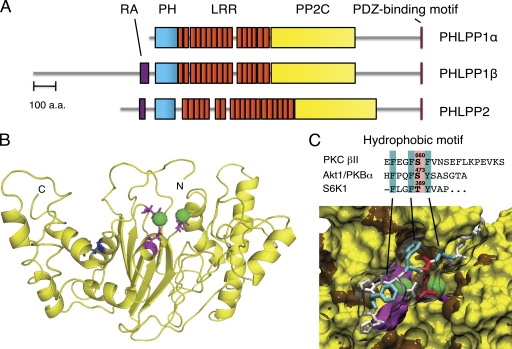
The PHLPP family comprises three isozymes, the alternatively spliced PHLPP1α and PHLPP1β (also referred to as suprachiasmatic nucleus circadian oscillatory protein (SCOP) (3)) and PHLPP2, a separate gene product. All three isozymes share a similar domain structure (Fig. 1A): an N-terminal PH domain, a leucine-rich repeat (LRR) region, a PP2C phosphatase domain, and a C-terminal PDZ-binding motif. In addition, both PHLPP1β and PHLPP2 have a predicted N-terminal Ras association domain that is not present in PHLPP1α. The PH domain contains only the middle Arg of the RXRSF motif required for phosphoinositide binding and, as such, is a weak membrane-binding module (4). The PDZ ligands are type 1 PDZ ligands (TPL for PHLPP1 and TAL for PHLPP2). PHLPP is evolutionarily conserved, with the PH domain and PDZ ligands being later additions. Curiously, the yeast homolog, Cyr1, is fused to the N terminus of adenylate cyclase (5).
FIGURE 1.
Domain structure of PHLPP. A, domain structure of PHLPP family members showing predicted Ras association domain (RA; purple), PH domain (cyan), LRRs (orange), PP2C domain (yellow), and C-terminal PDZ-binding motif (pink). a.a., amino acids. B, model of the PP2C domain of PHLPP2 showing active site acidic residues (Asp-806, Glu-990, and Asp-1024; pink) that coordinate two Mn2+ ions (green spheres) (13). Leu-1016, which is present as Ser in 30% of the population, is highlighted in blue. C, surface rendition of the active site of PHLPP2 docked with a phosphorylated hydrophobic motif peptide from Akt (HFPQFpSYSAS; with phosphorylated Ser (Ser-473) in red and underlined hydrophobic residues in cyan). Active site residues and Mn2+ are colored as described for B. The alignment of hydrophobic phosphorylation motif sequences of PKCβII, Akt1, and S6K1 is indicated.
Function
Based on the reasoning that so many players in the Akt/PKB pathway have a PH domain (notably Akt itself and its upstream kinase, PDK1), a rational search of the NCBI database was performed to identify a gene predicted to encode a PH domain and phosphatase (6). This led to the discover of PHLPP, which was shown to dephosphorylate a key regulatory site on the C terminus of Akt, the hydrophobic motif, and thus inactivate the kinase (6). PHLPP was subsequently shown to serve as the hydrophobic motif phosphatase for three AGC kinase family members, Akt, PKC, and S6K1 (ribosomal protein S6 kinase 1).
The AGC kinase family mediates an array of important cellular functions, and their dysregulation is strongly associated with the pathogenesis of many human diseases, most notably cancer (7). A common property of many AGC kinases is that their activation is induced by phosphorylation at two conserved segments: 1) the activation loop, which is phosphorylated by PDK1 and required to correctly align residues for catalysis, and 2) a C-terminal segment with two phosphorylation sites named the “turn motif” and the “hydrophobic motif.” Phosphorylation at both segments is necessary for maximal activation of these enzymes (8). Signal transduction by the AGC kinases is opposed by cellular phosphatases that dephosphorylate these enzymes, leading to their inactivation.
Initial studies identified PHLPP as a hydrophobic motif phosphatase, unveiling a new player to oppose prosurvival signaling pathways. Both PHLPP1 and PHLPP2 selectively dephosphorylate the hydrophobic motif site of Akt isozymes, PKC isozymes, and S6K (6, 9, 10). In the case of Akt, dephosphorylation of this site (Ser-473 of Akt1) reduces its intrinsic catalytic activity, leading to increased apoptosis and decreased proliferation (6, 11). In the case of PKC, dephosphorylation of this site (Ser-657 of PKCα) destabilizes PKC and shunts it to degradation pathways. In the case of S6K, dephosphorylation of the hydrophobic motif (Thr-389) reduces its activity, with the functional consequence of reducing protein translation (12). This selectivity for the hydrophobic motif may reflect unique interactions of the motif with the substrate-binding cavity of the phosphatase domain; molecular docking revealed that the signature Phe at the P−1 position of the motif is accommodated in a hydrophobic pocket (brown residues in Fig. 1C) (13). Thus, PHLPP terminates signaling by three AGC kinases by selective dephosphorylation of their hydrophobic motif.
Another substrate that PHLPP dephosphorylates to inhibit cell survival is Mst1 (mammalian sterile 20-like kinase 1) (14). The Mst1 signaling pathway is a critical regulator of cell survival, and it is frequently inactivated in tumorigenesis (15). By binding to and dephosphorylating Mst1 at an autoinhibitory site (Thr-387), PHLPP activates the Mst1 signaling pathway, leading to activation of the downstream pro-apoptotic MAPK pathway (14). Therefore, the antiproliferative and apoptosis-inducing effects of PHLPP are mediated not only through the inactivation of the Akt/PKC prosurvival pathways but additionally through the activation of Mst1-controlled pro-apoptotic pathways.
Protein interactions mediated by the regulatory modules on PHLPP likely play a determining role in dictating substrate specificity. The ability of PHLPP to dephosphorylate PKC is dependent upon the PH domain, as deletion of this domain results in increased PKC levels (9). In contrast, the ability of PHLPP to properly regulate Akt depends on an intact PDZ-binding motif, as deletion of the four amino acids delineating this motif greatly impairs the ability of PHLPP to terminate Akt signaling (6). Furthermore, knockdown of PHLPP1 and PHLPP2 revealed isozyme-specific interactions with individual Akt isozymes that regulate their activity toward downstream substrates (11). Specifically, PHLPP1 controls the activity of Akt2 and Akt3 (but not Akt1), and PHLPP2 regulates Akt1 and Akt3 (but not Akt2). As a result, the PHLPP family constitutes a unique signaling axis that regulates both the amplitude and direction of Akt signaling.
A number of protein scaffolds that serve as signaling platforms to direct downstream signaling by PHLPP have been identified recently. PHLPP has been shown to be targeted to Akt through interaction with three distinct scaffolding proteins, FKBP51 (FK506-binding protein of 51 kDa) (16), Scribble (17), and NHERF1 (Na+/H+ exchanger regulatory factor 1) (18), all of which enhance the ability of PHLPP to dephosphorylate Akt by co-localizing the two enzymes. Additionally, overexpression of the membrane proteins Scribble and NHERF1 forces the relocalization of PHLPP1, which is normally localized primarily in the cytosol, to the plasma membrane. Using both purified proteins and immunoprecipitation studies, it was determined that the PDZ-binding motif of PHLPP1 and PHLPP2 is necessary for their interaction with NHERF1. Specifically, PHLPP1 binds to both PDZ domains of NHERF1, whereas PHLPP2 binds primarily to PDZ2 of NHERF1. In contrast, the interaction between PHLPP1 and Scribble is independent of the PDZ-binding motif of PHLPP1 but is lost upon deletion of the entire C-terminal region. These scaffolding proteins are commonly lost in tumorigenesis, resulting in the impaired ability of PHLPP to terminate Akt signaling, thus promoting cell survival and, importantly, resistance to chemotherapy. Thus, poising PHLPP near its substrates through scaffolding interactions is crucial for its cellular function.
In addition to its influence on cell survival, PHLPP is involved in a diverse array of physiological processes, including the circadian rhythm, memory formation, and T cell development. PHLPP1β was identified by Shimizu et al. (3) and originally termed SCOP because mRNA levels were demonstrated to oscillate in a circadian manner, suggestive of a role in controlling the central clock. Subsequent studies by Sassone-Corsi and co-workers (19) determined that in response to a light-induced phase shift, PHLPP1−/− mice display delayed shortening of circadian period length (tau) and impaired capacity to stabilize the circadian rhythm. Additionally, in neuronal cell systems, PHLPP1β was reported to bind the nucleotide-free form of K-Ras, through interaction within the LRR region of PHLPP1, to prevent GTP binding to K-Ras and ultimately diminish signaling through the MEK/ERK pathway (20). Consequently, overexpression of PHLPP1 in the hippocampi of mice reduces ERK1/2 signaling, which has been reported to inhibit cAMP response element-mediated transcription and impair long-term memory formation (21). These findings implicate a role for PHLPP in maintaining neural plasticity and in resetting the circadian clock in response to changing stimuli.
Through their opposition of Akt, PHLPP isozymes also serve a critical role in the development and function of regulatory T cells (Tregs) (22). Inactivation of Akt is known to be a functional requirement of Tregs, and both PHLPP1 and PHLPP2 mRNAs are significantly up-regulated in Tregs compared with conventional T cells. Knockdown or genetic deletion of PHLPP1 results in the activation of Akt, an effect that dramatically impairs the development and function of Tregs. Interestingly, stimulation of conventional T cells with the immunosuppressive cytokine TGF-β enhances Smad3 binding to the PHLPP1 promoter and induces PHLPP1 expression, indicating that up-regulation of PHLPP1 may be a critical aspect of the Treg developmental program.
Although the majority of the existing research on PHLPP has focused on its opposition of Akt and other prosurvival signaling pathways, there is growing evidence that PHLPP is a central player in a broad range of cellular processes. Unlike the majority of protein phosphatases, PHLPP contains multiple regulatory domains, which raises the possibility that it can control various effectors through mechanisms independent of its phosphatase activity. Further study will likely reveal novel interacting proteins and unique functions for the PHLPP isozymes.
Regulation
Maintenance of PHLPP activity and expression is crucial to sustain cellular homeostasis in response to various stimuli and to preserve the balance between cell survival and apoptosis. Thus, understanding how the stability and activity of PHLPP are regulated is an important area of research.
PHLPP regulates hydrophobic motif phosphorylation in both the absence and presence of growth factor stimulation (6, 11), suggesting constitutive phosphatase activity. However, whether the intrinsic catalytic activity of PHLPP is acutely modulated by agonist stimulation is largely unknown. Given the large number of predicted phosphorylation sites, it is likely that future studies will unveil multiple mechanisms for agonist-dependent regulation of activity or subcellular location of PHLPP.
Interestingly, PHLPP2 activity is highly sensitive to a common polymorphism affecting 30% of the human population: a single amino acid change (Leu-1016 to Ser) (Fig. 1B) within the phosphatase domain of PHLPP2 reduces its rate of dephosphorylation of Akt and PKC by ∼5-fold in vitro, thus rendering it ineffective toward Akt and PKC in cells and reducing PHLPP2-dependent apoptosis (23). Whether this polymorphism predisposes to cancer is unclear; however, it is noteworthy that in three of three heterozygous patients with high-grade breast cancers, the Leu variant (active toward Akt and PKC) was shown to be lost in the tumor. This residue is situated in the predicted substrate-binding cleft of PHLPP2 (Fig. 1B), suggesting that the less abundant Ser variant has a lower affinity for substrates such as Akt and PKC.
Recent drug discovery efforts using chemical and virtual screening methods have identified small molecules that specifically inhibit PHLPP at micromolar concentrations (13). These compounds are selective for PHLPP compared with other PP2C family members, including PP2Cα. Biochemical and cellular assays demonstrated that the two most promising compounds, which are structurally diverse, effectively inhibit PHLPP in vitro, increase Akt signaling, and prevent etoposide-induced apoptosis in cells. Furthermore, molecular modeling of the phosphatase domain of PHLPP2, based on the structure of PP2Cα, identified several residues that are likely to be important for its catalytic activity (Fig. 1B). This molecular modeling suggests that two Mn2+ ions bind the active site and are coordinated by Asp-806, Glu-989, and Asp-1024 (13). The identification of PHLPP-specific inhibitors provides a powerful tool to enhance the understanding of PHLPP at the molecular and cellular levels and provides leads for potential therapeutic application.
In recent years, our knowledge of the mechanisms controlling PHLPP expression levels has greatly improved. At the translational level, PHLPP1 and PHLPP2 expression is controlled by way of mammalian target of rapamycin (mTOR)-dependent protein translation (24). Specifically, pharmacological or genetic inhibition of mTOR activity reduces PHLPP expression via S6K and 4E-BP1, as expression of constitutively active, rapamycin-insensitive S6K or knockdown of 4E-BP1 is sufficient to overcome the ability of mTOR inhibition to reduce PHLPP expression. This finding has broad implications because physiological stimuli such as amino acid availability and glucose levels are known to affect the activity of mTOR (25) and, in turn, influence the steady-state levels of PHLPP.
PHLPP expression is also controlled at the protein level through multiple degradation pathways. First, a study examining the role of PHLPP1β (SCOP) in the regulation of MAPK signaling demonstrated that an increase in intracellular Ca2+ causes a rapid reduction in the cellular levels of PHLPP1β (21). It has been established previously that a class of proteases, calpains, is activated in the presence of Ca2+ (26). Purified calpains induce the proteolysis of PHLPP1β in vitro, and pharmacological inhibition of calpains was reported to cause a significant increase in steady-state levels of PHLPP1β. These findings suggest a role for calpain-mediated proteolysis in regulating the steady-state levels of PHLPP1β in neurons. Second, PHLPP1 levels are modulated by proteasomal degradation. Recent work by Gao and co-workers (27) identified PHLPP1 as a proteolytic target of β-transducin repeat-containing protein (β-TrCP), an E3 ligase that serves as the substrate recognition subunit in the SCF (Skp1/cullin 1/F-box) protein complex. Specifically, phosphorylation of PHLPP1 by casein kinase 1 (CK1) and glycogen synthase kinase 3 (GSK-3) at multiple residues within its PP2C domain generates a phosphodegron motif that promotes PHLPP degradation. CK1 constitutively phosphorylates PHLPP1, an event that is essential for subsequent phosphorylation by GSK-3 at Ser-847. Upon phosphorylation by both CK1 and GSK-3, PHLPP1 is recognized and bound by β-TrCP, polyubiquitinated, and degraded by the 26 S proteasome. Importantly, it has been established previously that the activity of GSK-3β is impaired upon phosphorylation by its upstream kinase, Akt. Thus, Akt stabilizes its negative regulator, PHLPP1, resulting in a negative feedback loop through which PHLPP1 serves to dampen Akt signaling. Proteasomal degradation is largely responsible for the basal turnover of PHLPP1, as 1) inhibition of Akt, CK1, or GSK-3, 2) siRNA knockdown of β-TrCP, or 3) mutation of the Ser/Thr phosphodegron sites on PHLPP1 to Ala results in a marked increase in PHLPP1 stability. Furthermore, an inverse correlation between PHLPP1 and β-TrCP levels was observed in colon cancer cell lines, suggesting that high levels of this E3 ligase correlate with the low level of PHLPP1 often associated with tumors.
A screen of the NCI60 panel of tumor cell lines identified a subset of tumors in which the negative feedback loop between Akt and PHLPP1 is lost (28). Specifically, the cellular levels of PHLPP1 are insensitive to manipulation of Akt activity in high-grade glioblastoma (GBM) cell lines. Cellular fractionation revealed that in astrocytoma cell lines and normal brain tissue, β-TrCP1 is predominantly cytoplasmic, whereas in GBM cell lines and patient-derived tumor neurospheres, β-TrCP1 is confined to the nucleus and thus spatially separated from PHLPP1, which is cytoplasmic. As a result, in GBM, although PHLPP1 is properly phosphorylated by upstream kinases, it can no longer interact with β-TrCP1, and PHLPP1 levels are no longer sensitive to Akt activity. Consistent with this, reintroduction of β-TrCP1 to the cytosol of GBM cells is sufficient to restore the ability of Akt to control PHLPP1 levels. This study indicates a novel mechanism for the dysregulation of PHLPP1 levels, as well as other SCFβ-TrCP substrates, in cancer. Interestingly, PHLPP1 mRNA levels were consistently reduced in a majority of the GBM cell lines tested compared with low-grade astrocytomas, suggesting that dysregulation of PHLPP at the transcriptional level may be responsible for promoting Akt signaling in this disease.
Inhibitors of the mTOR and PI3K/Akt signaling pathways have shown promise as a therapeutic strategy in the clinic and are the subject of a great deal of anticancer research. However, as an off-target effect, we note that the use of these inhibitors may decrease PHLPP levels and, as a result, promote survival signaling through Akt, PKC, and Mst1 (Fig. 2). Thus, inhibitors targeting the PI3K signaling pathway may display greater efficacy in tumors that have lost the feedback loop between Akt and PHLPP1 such as GBM because PHLPP levels will remain constant despite the inhibition of Akt activity. Therefore, it is important to determine whether the negative effect on PHLPP expression caused by inhibitors of PI3K, mTOR, and Akt negates some of their antitumorigenic effects.
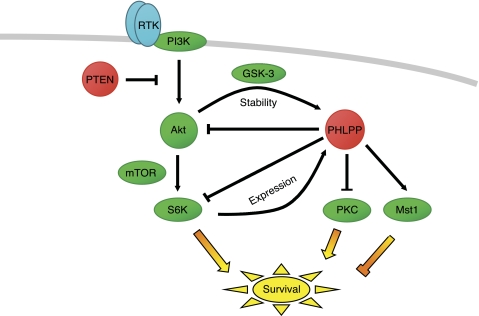
FIGURE 2.
Role of PHLPP in terminating oncogenic signaling pathways. PHLPP (red) directly dephosphorylates and inactivates Akt, PKC, and S6K, thus opposing cell survival and proliferation signals, and directly dephosphorylates and activates Mst1, thus promoting pro-apoptotic signals. Indicated are two negative feedback loops: high Akt activity up-regulates PHLPP levels by suppressing GSK-3-dependent degradation, and high S6K activity up-regulates PHLPP expression. Note that targeting the PI3K pathway within these feedback loops will have the consequence of decreasing PHLPP levels, thus promoting prosurvival pathways mediated by PKC and inhibiting pro-apoptotic pathways mediated by Mst1. Protein kinases are shown in green, and tumor suppressors are shown in red (the lipid phosphatase PTEN, which opposes the lipid kinase PI3K, and the protein phosphatase PHLPP, which controls the indicated protein kinases). RTK, receptor tyrosine kinase.
Role in Disease
Increasing evidence supports a crucial role for PHLPP in the onset and progression of various disease states. The best documented role of PHLPP is as a tumor suppressor protein in cancer (29). Furthermore, several reports suggest that PHLPP not only blocks tumorigenesis by inactivation of oncogenic pathways but also sensitizes cancer cells to chemotherapy (16, 30, 31).
Loss of PHLPP1 is sufficient to cause prostate tumors in mice, and its genetic deletion or mRNA repression is prevalent in prostate cancer patients (29). Indeed, PHLPP is deleted as often as the phosphatase and tensin homolog (PTEN; ∼40%) in metastatic prostate cancers (32). Of potential clinical relevance, low levels of both PHLPP1 and PTEN transcripts were shown to correlate strongly with relapse in patients after prostate surgery, suggesting that levels of these tumor suppressors could serve as a predictor for disease recurrence after prostectomy (29). Reductions in PHLPP1 or PHLPP2 mRNA and/or protein levels are associated with a broad spectrum of additional cancers, including chronic lymphocytic leukemia (33), breast carcinomas (34, 35), GBM (36), and melanoma (37) among others. Aberrations in PHLPP may be particularly prevalent in GBM, where mutations have been found in human tumors and where higher levels of PHLPP correlate with higher survival rates (38). PHLPP has also been characterized as a tumor suppressor in colon cancer: immunohistochemical staining of colon tumors revealed 78 and 86% decreases in PHLPP1 and PHLPP2 expression, respectively, compared with normal tissue. Furthermore, stable overexpression of PHLPP in colon cancer cells decreases proliferation and sensitizes cells to growth inhibition induced by PI3K inhibition. Reduced Akt signaling is largely responsible for the inhibition of cell growth observed upon PHLPP overexpression, as a constitutively active and PHLPP-resistant Akt construct (S473E) negates these effects (39). Importantly, reconstitution of either PHLPP1 or PHLPP2 into colon cancer cells strongly inhibits tumor growth in vivo (39). Thus, PHLPP is a major tumor suppressor, and mechanisms to enhance its activity provide a potentially promising novel chemotherapeutic approach.
In contrast to the detrimental effect of enhanced Akt activity in promoting tumorigenesis, enhancing Akt activity can be advantageous in disease states such as ischemic heart disease and metabolic disorders. In this regard, PHLPP1 has been shown to be an important regulator of Akt signaling in the heart (40): knockdown or genetic deletion of PHLPP1 enhances Akt activity in cardiac myocytes and, in turn, provides protection against ischemic injury. Thus, acute inhibition of PHLPP1 after cardiac injury may be of therapeutic benefit. Similarly, Akt is a key modulator of insulin signaling. Impaired activation of Akt isozymes, specifically Akt2, reduces glucose transport and ultimately leads to insulin resistance, which is commonly associated with obesity and type 2 diabetes (41, 42). Indeed, PHLPP1 protein levels are significantly higher in skeletal muscle and adipose tissue from obese human subjects compared with non-obese subjects, correlating with lower Akt (Ser-473) phosphorylation (43). Consistent with this, PHLPP1 mRNA is enhanced in type 2 diabetic individuals, correlating with decreased hydrophobic motif phosphorylation of Akt2 (43). Overexpression of PHLPP1 in insulin-responsive cell lines results in a decrease in insulin-induced Akt (Ser-473) phosphorylation, diminished glycogen synthesis, and reduced glucose transport (44). These reports suggest that increased PHLPP1 expression promotes insulin resistance associated with diabetes and obesity.
In summary, mechanisms that control the amount of PHLPP in the cell are often aberrant in disease. Notably, loss of PHLPP drives proliferative/survival pathways and is frequently associated with cancer, whereas gain of PHLPP drives signal termination and is associated with insulin resistance and obesity. Thus, maintenance of PHLPP levels is essential for cellular homeostasis.
Conclusion
Identified originally as the hydrophobic motif phosphatase for Akt, PHLPP has emerged as a central player in the onset and progression of major diseases because of its control of the balance between cell survival and apoptosis. Noting that there are close to 60,000 phosphorylation sites (PhosphoSitePlus) and fewer than 50 Ser/Thr phosphatases (2), PHLPP is likely to catalyze the dephosphorylation of a plethora of other substrates that await identification. As a result, efforts to identify novel substrates and gain a more complete understanding of the mechanisms governing the activity, localization, and stability of PHLPP are burgeoning research areas likely to provide novel insight into the biological function of this new player in cell signaling. The development of both pharmacological inhibitors and activators of PHLPP or its regulators, particularly if designed to target PHLPP activity at specific protein scaffolds, holds much promise for intervention in the diverse pathophysiologies resulting from aberrant PHLPP signaling.
Acknowledgments
We thank Bill Sinko for Fig. 1C and members of the Newton laboratory for helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM067946 (to A. C. N.). This work was also supported by Department of Defense Breast Cancer Research Program Award BC093021 from the United States Army Medical Research Acquisition Activity (to N. A. W.).
- PP2C
- protein phosphatase 2C
- PPP
- phosphoprotein phosphatase
- PH
- pleckstrin homology
- PHLPP
- pleckstrin homology domain leucine-rich repeat protein phosphatase
- PPM
- protein phosphatase metal-dependent
- SCOP
- suprachiasmatic nucleus circadian oscillatory protein
- S6K
- ribosomal protein S6 kinase
- Mst1
- mammalian sterile 20-like kinase 1
- FKBP51
- FK506-binding protein of 51 kDa
- NHERF1
- Na+/H+ exchanger regulatory factor
- SCF
- Skp1/cullin 1/F-box
- LRR
- leucine-rich repeat
- Treg
- regulatory T cell
- mTOR
- mammalian target of rapamycin
- β-TrCP
- β-transducin repeat-containing protein
- CK
- casein kinase
- GSK
- glycogen synthase kinase
- GBM
- glioblastoma
- PTEN
- phosphatase and tensin homolog.
REFERENCES
- 1. Brognard J., Newton A. C. (2008) PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol. Metab. 19, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 3. Shimizu K., Okada M., Takano A., Nagai K. (1999) SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 458, 363–369 [DOI] [PubMed] [Google Scholar]
- 4. Park W. S., Heo W. D., Whalen J. H., O'Rourke N. A., Bryan H. M., Meyer T., Teruel M. N. (2008) Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell 30, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsumoto K., Uno I., Oshima Y., Ishikawa T. (1982) Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 79, 2355–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao T., Furnari F., Newton A. C. (2005) PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell 18, 13–24 [DOI] [PubMed] [Google Scholar]
- 7. Pearce L. R., Komander D., Alessi D. R. (2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 8. Alessi D. R., Cohen P. (1998) Mechanism of activation and function of protein kinase B. Curr. Opin. Genet. Dev. 8, 55–62 [DOI] [PubMed] [Google Scholar]
- 9. Gao T., Brognard J., Newton A. C. (2008) The phosphatase PHLPP controls the cellular levels of protein kinase C. J. Biol. Chem. 283, 6300–6311 [DOI] [PubMed] [Google Scholar]
- 10. Liu J., Stevens P. D., Li X., Schmidt M. D., Gao T. (2011) PHLPP-mediated dephosphorylation of S6K1 inhibits protein translation and cell growth. Mol. Cell. Biol. 31, 4917–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brognard J., Sierecki E., Gao T., Newton A. C. (2007) PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell 25, 917–931 [DOI] [PubMed] [Google Scholar]
- 12. Chung J., Kuo C. J., Crabtree G. R., Blenis J. (1992) Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70-kDa S6 protein kinases. Cell 69, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 13. Sierecki E., Sinko W., McCammon J. A., Newton A. C. (2010) Discovery of small molecule inhibitors of the PH domain leucine-rich repeat protein phosphatase (PHLPP) by chemical and virtual screening. J. Med. Chem. 53, 6899–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiao M., Wang Y., Xu X., Lu J., Dong Y., Tao W., Stein J., Stein G. S., Iglehart J. D., Shi Q., Pardee A. B. (2010) Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol. Cell 38, 512–523 [DOI] [PubMed] [Google Scholar]
- 15. de Souza P. M., Lindsay M. A. (2004) Mammalian sterile 20-like kinase 1 and the regulation of apoptosis. Biochem. Soc. Trans. 32, 485–488 [DOI] [PubMed] [Google Scholar]
- 16. Pei H., Li L., Fridley B. L., Jenkins G. D., Kalari K. R., Lingle W., Petersen G., Lou Z., Wang L. (2009) FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16, 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X., Yang H., Liu J., Schmidt M. D., Gao T. (2011) Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. EMBO Rep. 12, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Molina J. R., Agarwal N. K., Morales F. C., Hayashi Y., Aldape K. D., Cote G., Georgescu M. M. (August 1, 2011) PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 10.1038/onc.2011.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masubuchi S., Gao T., O'Neill A., Eckel-Mahan K., Newton A. C., Sassone-Corsi P. (2010) Protein phosphatase PHLPP1 controls the light-induced resetting of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 107, 1642–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shimizu K., Okada M., Nagai K., Fukada Y. (2003) Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. J. Biol. Chem. 278, 14920–14925 [DOI] [PubMed] [Google Scholar]
- 21. Shimizu K., Phan T., Mansuy I. M., Storm D. R. (2007) Proteolytic degradation of SCOP in the hippocampus contributes to activation of MAP kinase and memory. Cell 128, 1219–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patterson S. J., Han J. M., Garcia R., Assi K., Gao T., O'Neill A., Newton A. C., Levings M. K. (2011) Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regulatory T cells. J. Immunol. 186, 5533–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brognard J., Niederst M., Reyes G., Warfel N., Newton A. C. (2009) Common polymorphism in the phosphatase PHLPP2 results in reduced regulation of Akt and protein kinase C. J. Biol. Chem. 284, 15215–15223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J., Stevens P. D., Gao T. (2011) mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J. Biol. Chem. 286, 6510–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sengupta S., Peterson T. R., Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan S. L., Mattson M. P. (1999) Caspase and calpain substrates: roles in synaptic plasticity and cell death. J. Neurosci. Res. 58, 167–190 [PubMed] [Google Scholar]
- 27. Li X., Liu J., Gao T. (2009) β-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol. Cell. Biol. 29, 6192–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warfel N. A., Niederst M., Stevens M. W., Brennan P. M., Frame M. C., Newton A. C. (2011) Mislocalization of the E3 ligase, β-transducin repeat-containing protein 1 (β-TrCP1), in glioblastoma uncouples negative feedback between the pleckstrin homology domain leucine-rich repeat protein phosphatase 1 (PHLPP1) and Akt. J. Biol. Chem. 286, 19777–19788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen M., Pratt C. P., Zeeman M. E., Schultz N., Taylor B. S., O'Neill A., Castillo-Martin M., Nowak D. G., Naguib A., Grace D. M., Murn J., Navin N., Atwal G. S., Sander C., Gerald W. L., Cordon-Cardo C., Newton A. C., Carver B. S., Trotman L. C. (2011) Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell 20, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carver B. S., Chapinski C., Wongvipat J., Hieronymus H., Chen Y., Chandarlapaty S., Arora V. K., Le C., Koutcher J., Scher H., Scardino P. T., Rosen N., Sawyers C. L. (2011) Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mulholland D. J., Tran L. M., Li Y., Cai H., Morim A., Wang S., Plaisier S., Garraway I. P., Huang J., Graeber T. G., Wu H. (2011) Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19, 792–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., Wilson M., Socci N. D., Lash A. E., Heguy A., Eastham J. A., Scher H. I., Reuter V. E., Scardino P. T., Sander C., Sawyers C. L., Gerald W. L. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390 [DOI] [PubMed] [Google Scholar]
- 34. Karnoub A. E., Dash A. B., Vo A. P., Sullivan A., Brooks M. W., Bell G. W., Richardson A. L., Polyak K., Tubo R., Weinberg R. A. (2007) Mesenchymal stem cells within tumor stroma promote breast cancer metastasis. Nature 449, 557–563 [DOI] [PubMed] [Google Scholar]
- 35. Richardson A. L., Wang Z. C., De Nicolo A., Lu X., Brown M., Miron A., Liao X., Iglehart J. D., Livingston D. M., Ganesan S. (2006) X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9, 121–132 [DOI] [PubMed] [Google Scholar]
- 36. Bredel M., Bredel C., Juric D., Harsh G. R., Vogel H., Recht L. D., Sikic B. I. (2005) High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer Res. 65, 4088–4096 [DOI] [PubMed] [Google Scholar]
- 37. Talantov D., Mazumder A., Yu J. X., Briggs T., Jiang Y., Backus J., Atkins D., Wang Y. (2005) Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. Cancer Res. 11, 7234–7242 [DOI] [PubMed] [Google Scholar]
- 38. Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., Weiss H. L., Rychahou P., Jackson L. N., Evers B. M., Gao T. (2009) Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene 28, 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miyamoto S., Purcell N. H., Smith J. M., Gao T., Whittaker R., Huang K., Castillo R., Glembotski C. C., Sussman M. A., Newton A. C., Brown J. H. (2010) PHLPP1 negatively regulates Akt activity and survival in the heart. Circ. Res. 107, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 42. Kohn A. D., Summers S. A., Birnbaum M. J., Roth R. A. (1996) Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J. Biol. Chem. 271, 31372–31378 [DOI] [PubMed] [Google Scholar]
- 43. Andreozzi F., Procopio C., Greco A., Mannino G. C., Miele C., Raciti G. A., Iadicicco C., Beguinot F., Pontiroli A. E., Hribal M. L., Folli F., Sesti G. (2011) Increased levels of the Akt-specific phosphatase PH domain leucine-rich repeat protein phosphatase (PHLPP)-1 in obese participants are associated with insulin resistance. Diabetologia 54, 1879–1887 [DOI] [PubMed] [Google Scholar]
- 44. Cozzone D., Fröjdö S., Disse E., Debard C., Laville M., Pirola L., Vidal H. (2008) Isoform-specific defects of insulin stimulation of Akt/protein kinase B (PKB) in skeletal muscle cells from type 2 diabetic patients. Diabetologia 51, 512–521 [DOI] [PubMed] [Google Scholar]