Background: The central carbon metabolism is important for bacterial survival and fitness.
Results: Isotopologue experiments identified amino acid biosynthesis pathways of pneumococci.
Conclusion: The analysis of the carbon metabolism in pneumococci identified an unconventional pathway to synthesize serine.
Significance: Understanding the metabolism of pathogens is critical to gain insights into the adaptation strategies and to identify new drug targets.
Keywords: Amino Acid, Bacterial Metabolism, Carbohydrate Metabolism, Metabolism, Serine, Amino Acid Synthesis, Isotopologue Profiling, Pneumococci, Pyruvate Formate-lyase
Abstract
The metabolism of Streptococcus pneumoniae was studied by isotopologue profiling after bacterial cultivation in chemically defined medium supplemented with [U-13C6]- or [1,2-13C2]glucose. GC/MS analysis of protein-derived amino acids showed lack of 13C label in amino acids that were also essential for pneumococcal growth. Ala, Ser, Asp, and Thr displayed high 13C enrichments, whereas Phe, Tyr, and Gly were only slightly labeled. The analysis of the labeling patterns showed formation of triose phosphate and pyruvate via the Embden-Meyerhof-Parnas pathway. The labeling patterns of Asp and Thr suggested formation of oxaloacetate exclusively via the phosphoenolpyruvate carboxylase reaction. Apparently, α-ketoglutarate was generated from unlabeled glutamate via the aspartate transaminase reaction. A fraction of Phe and Tyr obtained label via the chorismate route from erythrose 4-phosphate, generated via the pentose phosphate pathway, and phosphoenolpyruvate. Strikingly, the data revealed no significant flux from phosphoglycerate to Ser and Gly but showed formation of Ser via the reverse reaction, namely by hydroxymethylation of Gly. The essential Gly was acquired from the medium, and the biosynthesis pathway was confirmed in experiments using [U-13C2]glycine as a tracer. The hydroxymethyl group in Ser originated from formate, which was generated by the pyruvate formate-lyase. Highly similar isotopologue profiles were observed in corresponding experiments with pneumococcal mutants deficient in PavA, CodY, and glucose-6-phosphate dehydrogenase pointing to the robustness of the core metabolic network used by these facultative pathogenic bacteria. In conclusion, this study demonstrates the dual utilization of carbohydrates and amino acids under in vitro conditions and identifies the unconventional de novo biosynthesis of serine by pneumococci.
Introduction
The Gram-positive pathogen Streptococcus pneumoniae (pneumococcus) resides asymptomatically as a harmless commensal in the upper respiratory tract of humans (1). However, pneumococci also cause severe and life-threatening diseases, including otitis media, pneumonia, septicemia, and meningitis (2–4). Similar to other pathogenic bacteria, pneumococci have evolved sophisticated strategies to adhere to host cells, escape the host complement system, and resist the immune responses (3, 5–7). It has been reported that the regulation of virulence determinants such as capsular polysaccharides or toxins, for example, is negatively regulated in nutrient-rich environments, whereas adhesins are positively regulated (8). Compared with the aspects of virulence, only little attention has been paid to the physiology and metabolism of pathogenic bacteria, although the metabolism under in vivo conditions was shown to be profoundly different from that under in vitro and defined culture conditions (9). In addition, the liaison between bacterial metabolism and virulence has been indicated for several pathogenic bacteria, including e.g. Listeria, Legionella, Mycobacteria, Staphylococci, and pneumococci (8, 10–16). Bacterial fitness is closely linked to the ability of the bacteria to take up and catabolize nutrients provided by the respective host niche or a de novo synthesis of amino acids as they need energy and carbon for growth and replication. Pneumococci encounter different host niches during pathogenesis, which differ significantly in temperature, oxygen, pH values, and disposability of nutrients. Consequently, the pneumococcal physiology has to adapt to these various physiological conditions to ensure fitness and virulence factor expression.
A possibility to provide further insights into the conditions required for optimal growth of pneumococci in the host and their physiology is the elucidation of metabolic pathways and metabolic fluxes during catabolism of utilized carbon sources. Although the genome data can be used to postulate breakdown of nutrients by enzymatic reactions, uptake systems of pneumococci, and hypothetical metabolic pathways, the method of isotopologue profiling, offers a possibility to observe directly metabolic processes such as the biosynthesis of amino acids (11). Pneumococci are endowed with a large number of enzymes and transporter systems involved in the uptake of carbohydrates, amino acids, and other compounds. Thirty percent of the pneumococcal transporter systems, including ATP-binding cassette (ABC)3 transporters and classical phosphotransferase systems, are theoretically involved in the uptake of approximately 12 different carbohydrates, which can be metabolized via the Embden-Meyerhof-Parnas (EMP) pathway (glycolysis) or the pentose phosphate pathway (PPP) (17–19). Pneumococcal growth depends on carbohydrates, and compared with other respiratory pathogens such as Haemophilus influenzae and Neisseria meningitidis, S. pneumoniae can metabolize a wider range of carbohydrates (19). The importance of these transport systems for pneumococcal fitness and virulence has been demonstrated in numerous studies (15, 20–24).
Pneumococci are members of the order of Lactobacillales, and consequently the major catabolite of carbohydrate metabolism under microaerophilic conditions is lactate. The lactate dehydrogenase converts pyruvate to lactate and regenerates NADH to NAD+. Further enzymes are available (phosphotransacetylase, acetokinase, and NADH oxidase) to catabolize pyruvate into acetate by an ATP-gaining reaction (17). The analyses of pneumococcal genomes revealed the lack of genes encoding enzymes of the Entner-Doudoroff pathway (ED), the Krebs cycle (tricarboxylic acid cycle (TCA)), or the electron transport chain for aerobe and anaerobe respiration. Moreover, pneumococci do not have the capacity to synthesize de novo all of 20 amino acids. Instead, the genome analysis revealed incomplete pathways for the biosynthesis of Cys, Gly, Gln, Glu, and His and probably Lys and Pro (17, 19). To overcome this deficiency, pneumococci produce cell wall-located peptidases and proteases to digest proteins and transporters for uptake of amino acids and oligopeptides (15, 25, 26).
The metabolism of pneumococci is regulated by global nutritional regulators such as CodY and catabolite control protein A (CcpA). Their role in pneumococcal fitness and virulence has been reported (27, 28). Targets of CodY include operons involved in branched-chain amino acid (BCAA) metabolism and general amino acid metabolism such as the ilv operon, which encodes enzymes that condense threonine and pyruvate or two pyruvates into branched-chain keto acids, precursors of BCAAs. Recently, the involvement of CodY in controlling the rate of synthesis of its own co-effectors, BCAA, has been reported, demonstrating that the endogenous amount of BCAA serves as a signal for CodY-mediated gene regulation (29).
In this study isotopologue profiling was applied for the first time to decipher the metabolic pathways in pneumococci cultured in chemically defined medium with glucose as a carbon source. The 13C-labeled isotopologue patterns in amino acids from [U-13C6]- or [1,2-13C2]glucose as precursors identified the EMP (glycolysis) as the predominant metabolic pathway to catabolize glucose and revealed the pathways to the de novo synthesized amino acids in S. pneumoniae.
EXPERIMENTAL PROCEDURES
Strains, Culture Conditions, Transformations, and Antibiotics
S. pneumoniae D39 (serotype 2; NCTC7466) and isogenic mutants D39Δcps, D39ΔcpsΔpavA, D39ΔcpsΔcodY, and D39ΔcpsΔzwf were cultured to mid-log phase in Todd-Hewitt broth (Oxoid, Basingstoke, UK) supplemented with 0.5% yeast extract (THY, Roth, Karlsruhe, Germany) in chemically defined medium (CDM) (for details see supplemental Table S1) (30, 31) or grown on Columbia blood agar plates at 37 °C, 5% CO2 (Oxoid). Escherichia coli strains used in cloning procedures were cultured on Luria-Bertani (LB) agar or in LB broth. Transformation of E. coli and pneumococci was conducted by standard protocols described recently (32). Pneumococcal mutants were cultured in the presence of the appropriate antibiotics as follows: chloramphenicol (8 μg ml−1), erythromycin (5 μg ml−1), kanamycin (150 μg ml−1), and/or spectinomycin (50 μg ml−1).
Primers, Plasmids, and Molecular DNA Techniques
Primers that were used in this study are provided in supplemental Table S2. Chromosomal DNA used as template in PCRs was isolated and purified using the Qiagen Genomic Tip 100/G kit (Qiagen, Hilden, Germany). The DNA was amplified by PCR using the AmpliTaq Gold® DNA polymerase (PerkinElmer Life Sciences), and PCRs were subjected to denaturation at 94 °C, 30 cycles of 94 °C, primer annealing for 1 min, and elongation at 72 °C. PCR products were purified using the PCR DNA purification kit (Qiagen), and plasmids were extracted according to the QIAprep Spin Midi/Maxiprep kit protocol. Pneumococcal RNA isolation and purification for reverse transcription (RT)-PCR were conducted using the RNeasy mini kit, including a DNase digestion step using the RNase-free DNase set (Qiagen) as described (15). For RT-PCRs, 5–10 μg of RNA (calculated using the NanoDrop® ND-1000) were incubated with 10 nmol of dNTP in 20 μl of RNase-free water for 5 min at 65 °C, kept on ice for 1 min, and then 4 μl of First strand Buffer (five times), 1 μl of random primers (pd(N)6; GE Healthcare), 1 μl of DTT (0.1 m), 1 μl of RNasin (Promega), and 1 μl of SuperScript® III reverse transcriptase (Invitrogen) were added to amplify cDNA. The cDNA was employed as template in the PCRs, and control PCRs were conducted with genomic DNA and total RNA as template.
Construction of Pneumococcal Mutants
Pneumococcal mutants used and constructed in this study are all in the D39Δcps (nonencapsulated serotype 2) genetic background (33). The double mutant D39ΔcpsΔpavA, lacking the capsule and the virulence factor PavA, has been described (34, 35). The mutant deficient in the global regulator CodY and glucose-6-phosphate-dehydrogenase, encoded by zwf, were constructed by applying the insertion deletion mutagenesis strategy (15, 16). Briefly, the 1820-bp DNA fragment comprising the codY ORF (spd_1412), including 5′ and 3′ regions, was amplified by PCR using primers codYfw/codYrev and D39 chromosomal DNA as template. The PCR product was cloned directly into the pGEM-T easy vector (Invitrogen), resulting in plasmid pGcodY (pG619). By inverse PCR with primers codY_revinv/codY_fwdinv incorporating a BamHI and HindIII restriction site, a 674-bp DNA fragment of the codY ORF was deleted. The erythromycin resistance gene cassette ermB, amplified by PCR using primers Eryfor-ClaI/Eryrev-ClaI and plasmid pJDC9 (36) as template, was digested with BamHI/HindIII and cloned into similarly digested pGcodY, resulting in plasmid pGcodY::ermB (pG620). To generate the D39ΔcpsΔzwf (kanR; specR (aad9)) mutant in our used D39 strain, mutant D39ΔcpsΔzwf (kindly provided by W. T. Hendriksen and P. W. M. Hermans, Rotterdam/Nijmegen, The Netherlands) (27) was used as the DNA-template for a PCR with primers zwffw/zwfrev. S. pneumoniae D39Δcps was transformed with pGcodY::ermB or the PCR product zwf::aad9 as described previously (37). The integrity of the antibiotic resistance cassettes in pneumococcal mutants ΔcodY and Δzwf was verified by PCR (data not shown).
13C-Labeled Isotopologue Labeling Experiments
In all experiments, pneumococci were thawed on blood agar and incubated overnight at 37 °C and 5% CO2. To ensure fitness of pneumococci before using the bacteria to inoculate a 50-ml CDM preculture, an additional 10-h culture step was conducted on new blood agar plates. The starting A600 in the preculture was 0.07, and the labeling experiment was started by inoculation of 500 ml of CDM supplemented with 55.5 mm [U-13C6]glucose, 27.8 mm [1,2-13C2]glucose, or 1.33 mm [U-13C2]glycine with pneumococci cultured to A600 0.5 in the preculture. All experiments with [13C]glucose were performed in the absence of [12C]glucose. The absorbance was measured at regular intervals, and pneumococcal growth was stopped at mid-log phase, meaning an A600 of 0.5. To eliminate the possibility of contamination, a defined aliquot of bacteria was incubated on blood agar plates and monitored for contamination after 14 h at 37 °C and 5% CO2. Pneumococci were killed with 10 mm sodium azide and sedimented at 5250 × g at 5 °C for 15 min. The bacteria were washed with 40 mm Tris/HCl, pH 7.4, and the supernatant was discarded, and bacteria were autoclaved at 120 °C for 20 min.
Protein Hydrolysis and Amino Acid Derivatization
The bacterial cells were resuspended in 6 m hydrochloric acid and heated at 105 °C for 24 h under an inert atmosphere. The hydrolysate was placed on a cation exchange column of Dowex 50-X8 column(H+ form, 200–400 mesh, 5 × 10 mm) that was washed with 70% methanol and water and developed with 4 m ammonium hydroxide. An aliquot of the eluate was dried under a stream of nitrogen, and the residue was dissolved in 50 μl of water-free acetonitrile. A mixture of 50 μl of N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide containing 1% tert-butyldimethylsilyl chloride (Sigma) was added. The mixture was kept at 60 °C for 30 min. The resulting N-(tert-butyldimethylsilyl)-amino acids were then analyzed by GC/MS.
Amino Acid Quantification
Pneumococci of a 200-ml CDM culture were resuspended in 1 ml of 6 m HCl, and protein hydrolysis was carried out for 24 h at 100 °C. The samples were neutralized with 6 m NaOH, and prior to quantitative 1H NMR analysis, the suspension was sterilized using a 0.22-μm Rotilabo® syringe filter (Roth, Karlsruhe, Germany). All NMR spectra were obtained at 600.27 MHz at a nominal temperature of 310 K on a Bruker AVANCE-II 600 NMR spectrometer operated by TOPSPIN 2.1 software (both Bruker Biospin, Rheinstetten, Germany) as described previously (38).
Mass Spectrometry
GC/MS analysis was performed on a GC-QP2010 Plus (Shimadzu, Duisburg, Germany) equipped with a fused silica capillary column (Equity TM-5; 30 m × 0.25 mm, 0.25-μm film thickness; SUPELCO, Bellefonte, PA). The mass detector worked with electron impact ionization at 70 eV. An aliquot (1 μl) of a solution containing N-(tert-butyldimethylsilyl)-amino acids was injected in a 1:10 split mode at an interface temperature of 260 °C and a helium inlet pressure of 70 kPa. The column was developed at 150 °C for 3 min and then with a temperature gradient of 10 °C/min to a final temperature of 280 °C that was held for 3 min. Data were collected using the LabSolutions software (Shimadzu, Duisburg, Germany). Selected ion monitoring data were acquired using a 0.3-s sampling rate. Samples were analyzed at least three times. The overall 13C excess and the relative contribution of isotopomers were computed according to Ref. 39.
RESULTS
Influence of Amino Acid Deprivation on Pneumococcal Growth under Standardized Conditions
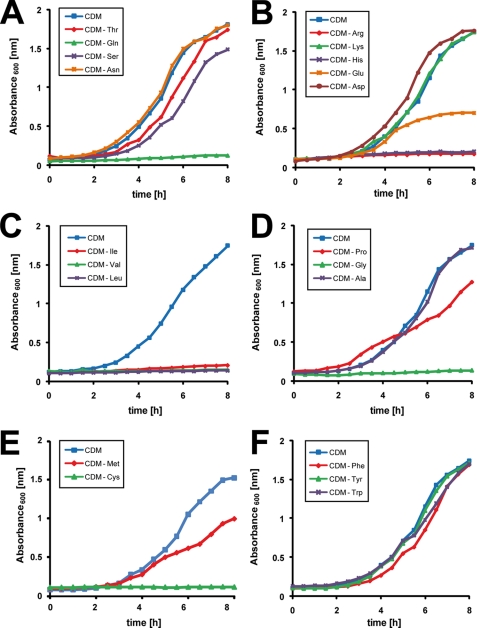
Pneumococci can be cultured in CDM, a medium that can be employed to investigate usage of carbon sources or the necessity for individual amino acids. By omitting separately one of the 20 amino acids, we showed that pneumococci are most likely auxotrophic for Arg, Cys, His, Gly, Gln, and the BCAA Ile, Leu, and Val. In contrast, lack of Thr, Ser, Asn, Asp, Ala, aromatic amino acids, or Lys did not affect growth under in vitro conditions (Fig. 1). As the growth characteristic in CDM depleted of Thr, Ser, and Asn was unchanged compared with complete CDM, these uncharged but polar amino acids are nonessential for the pneumococcal growth in the presence of glucose as carbon source, although Gln is essential (Fig. 1A). The charged amino acids, Asp and Lys, are nonessential amino acids for pneumococci, although Arg and His are absolutely required for growth, and the absence of Glu reduces pneumococcal fitness in CDM (Fig. 1B). Pneumococci produce ABC uptake systems for BCAA referred to as LivJGHMF (40). However, the bioinformatic analysis of the genomes (D39 and TIGR4) suggests that pneumococci are also able to synthesize branched amino acids out of pyruvate and Thr with the key enzymes acetolactate synthase (EC 2.2.1.6) and branched-chain-amino-acid transaminase (EC 2.6.1.42) encoded by spd_0404 (ilvB) and spd_0749 (ilvE). However, under the defined conditions used for isotopologue profiling pneumococci use preferentially BCAA uptake systems as indicated by the necessity of Ile, Val, and Leu as supplements in CDM (Fig. 1C). Out of the nonpolar amino acids Gly, Ala, and heterocyclic Pro, only Gly is essential, although lack of Pro has a growth limiting effect, and Ala is a nonessential amino acid (Fig. 1D). Within the sulfur-containing amino acids, pneumococcal growth is decelerated in the absence of Met, although Cys is essential under these in vitro conditions (Fig. 1E). Finally, the aromatic amino acids are not required for pneumococcal growth (Fig. 1F).
FIGURE 1.
S. pneumoniae D39 cultured in CDM with glucose as carbon source is auxotrophic for amino acids Cys, Arg, Gln, Gly, His, Ile, Leu, and Val. Pneumococcal growth was monitored in chemically defined medium (CDM) with glucose as the sole carbohydrate source, and cultures were individually depleted for uncharged (Asn, Gln, Ser, or Thr) (A), charged (Glu, Asp, His, Lys, or Arg) (B), branched chained (Ile, Leu, or Val) (C), aliphatic (Pro, Gly, or Ala) (D), sulfur containing (Met or Cys) (E), or aromatic amino acids (Phe, Tyr, or Trp) (F). Growth of S. pneumoniae D39 in complete CDM is included in all panels, and the experiments were performed at least three times. Results are shown for representative growth curves monitored in parallel.
Identification of Amino Acids Synthesized de Novo by S. pneumoniae
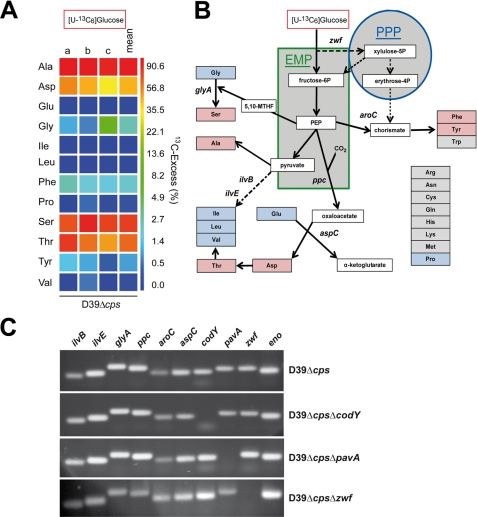
To investigate the de novo synthesis of amino acids by S. pneumoniae D39Δcps cultured in CDM with glucose as the carbon source, the carbon metabolism was analyzed by 13C-labeled isotopologue profiling. Pneumococci were grown in CDM containing [U-13C6]glucose (55.5 mm), and the 13C incorporation into protein-derived amino acids was measured by GC/MS after reaching an A600 of 0.5. The 13C excess was displayed in a logarithmic color map with values between 0 mol % (dark blue) and 90 mol % (red) (Fig. 2A; for numerical values see supplemental Table S3). It has to be mentioned that 13C labeling of amino acids Arg, Asn, Gln, Met, and Trp is not detectable due to their degradation during the acidic treatment of protein hydrolysis. The data revealed that none of the amino acids essential for pneumococcal growth in CDM had any 13C labeling (Fig. 2, A and B). The apparent lack of 13C labeling in Glu, Pro, and Gly suggested that pneumococci are auxotrophic for these amino acids. In accordance with the requirement for Val, Leu, and Ile for pneumococcal growth in CDM, none of the BCAAs was 13C-labeled. However, the following amino acids were labeled in the order Ala (87.2% overall 13C excess) > Ser (71.3%) > Thr (56.8%) > Asp (39.5%) > Phe (2.2%) > Tyr (1.0%) (Fig. 2A). From these data and the underlying isotopologue compositions, a schematic metabolic model of amino acid transport and de novo synthesis pathways in S. pneumoniae is derived (Fig. 2B). The measured 13C-labeling patterns demonstrated that glucose is catabolized via the EMP and PPP. The isotopologue profile revealed further that Asp is not synthesized via the general pathway, including pyruvate and acetyl-CoA. Instead, oxaloacetate is synthesized from phosphoenolpyruvate (PEP) and CO2 by the PEP carboxylase, demonstrating one aspect of CO2 consumption for pneumococcal growth. Oxaloacetate is then converted into Asp, although simultaneously the essential glutamate is converted into α-ketoglutarate. These reactions are catalyzed by the aspartate transaminase (Figs. 2B and 3A). It is noteworthy that pneumococci lack all enzymes of the TCA cycle (Krebs cycle) and the Entner-Doudoroff (ED) pathway. Completely labeled aromatic amino acids such as Phe and Tyr are synthesized from erythrose 4-phosphate and two molecules of PEP via the intermediate chorismate (Figs. 2B and 3B).
FIGURE 2.
Profiling of the metabolism in S. pneumoniae on the basis of 13C incorporation into amino acids from [U-13C6]glucose. A, overall 13C excess (%) of labeled isotopologues in amino acids derived from proteins after cultivation of S. pneumoniae D39 with 55.5 mm [U-13C6]glucose in CDM. Multiple 13C-labeled isotopologues were determined by GC/MS spectroscopy. The color map indicates 13C excess in a logarithmic form. The results of three independent biological experiments each measured in triplicate (a–c) and the means are shown. B, schematic overview of a metabolic model for the de novo synthesis of amino acids in S. pneumoniae D39 as concluded from the incorporation of 13C incorporation experiments and from the genome data. The EMP and PPP as well as the pyruvate formate-lyase reaction serve as the core metabolic pathways in pneumococci. Important intermediates and products of these pathways are shown as white boxes. Amino acids in red boxes indicate incorporation of 13C label from [U-13C6]glucose; amino acids in blue boxes displayed no significant 13C label, and amino acids in gray boxes could not be detected with the GC/MS method. C, RT-PCR of genes encoding central metabolic enzymes of the EMP, PPP, and amino acids synthesis (ppc, aspC, aroC, zwf, glyA, ilvB, or ilvE), regulator codY, virulence factor pavA, or zwf mutant using chromosomal DNA of S. pneumoniae D39Δcps and isogenic mutants D39ΔcpsΔcodY, D39ΔcpsΔpavA, or D39ΔcpsΔzwf. The abbreviations used are as follows: aspC, aspartate aminotransferase (SPD_1373 (EC 2.6.1.1)); aroC, chorismate synthase (SPD_1208 (EC 4.2.3.5)); glyA, SHMT (SPD_0910 (EC 2.1.2.1)); ppc, phosphoenolpyruvate carboxylase (SPD_0953 (EC 4.1.1.31)); zwf, glucose-6-phosphate dehydrogenase (SPD_1100 (EC 1.1.1.49)); ilvB, acetolactate synthase (SPD_0404 (EC 2.2.1.6)); ilvE, BCAA aminotransferase (SPD_0749 (EC 2.6.1.42)); eno, enolase (SPD_1012 (EC 4.2.1.11)); codY (SPD_1412), and pavA (SPD_0854).
FIGURE 3.
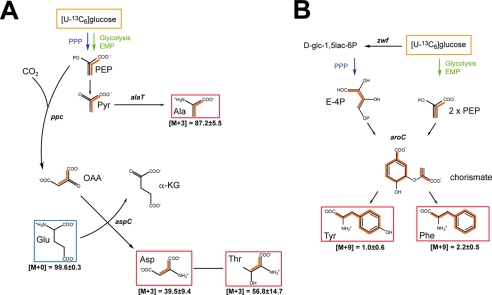
Schematic models of de novo syntheses of amino acids derived from [U-13C6]glucose. A, model of the de novo synthesis of alanine, aspartate, and threonine. B, model of the de novo synthesis of aromatic acids by the shikimate pathway. Multiple 13C labeling is indicated by orange bars connecting 13C-labeled atoms in a given molecule. Amino acids shown in red boxes indicate 13C-enriched amino acids, and the blue box indicates uptake of the amino acid from the medium. The relative contributions in % of the isotopomer groups comprising different numbers of 13C-labeled atoms, as determined by mass spectrometry (M+0, M+3, and M+9), are given. Tryptophan is not detectable by the applied GC/MS method. The abbreviations used are as follows: E-4P, erythrose 4-phosphate; Pyr, pyruvate; OAA, oxaloacetate; α-KG, α-ketoglutarate; aspC, aspartate aminotransferase (SPD_1373 (EC 2.6.1.1)); aroC, chorismate synthase (SPD_1208 (EC 4.2.3.5)); ppc, phosphoenolpyruvate carboxylase (SPD_0953 (EC 4.1.1.31)); zwf, glucose-6-phosphate dehydrogenase (SPD_1100 (EC 1.1.1.49)).
The expression of genes encoding important metabolic enzymes involved in processes depicted in the schematic model in Fig. 2B was assessed by reverse transcription-PCR to confirm the genetic basis of the de novo synthesis pathways. The results confirmed the expression of ilvB, ilvE, glyA, ppc, aroC, aspC, zwf, and eno in the nonencapsulated D39Δcps and its isogenic mutants deficient for the virulence factor PavA (SPD_0854), glucose-6-phosphate dehydrogenase (Zwf; SPD_1100), or metabolic regulator CodY (SPD_1412) (Fig. 2C).
Reconstruction of the de Novo Synthesis of Alanine, Aspartate, and Threonine by 13C- Isotopologue Profiling
Reconstruction of the anabolic amino acid synthesis pathways revealed that pneumococci synthesize alanine through conversion of pyruvate into alanine by transamination of pyruvate. The labeling patterns of PEP, pyruvate, and Ala were dominated by the universally 13C-labeled isotopologue when pneumococci were cultured in CDM supplemented with [U-13C6]glucose. Specifically, 87.2 ± 5.5 mol % of Ala was 13C3-labeled demonstrating its formation from the supplied [U-13C6]glucose via [U-13C3]PEP and [U-13C3]pyruvate (Fig. 2). Approximately 10% of Ala was not labeled, which points to some additional uptake of unlabeled Ala from the medium via a transport system. It is plausible that the sodium:alanine symporter (SPD_0372) of pneumococci mediates Ala uptake. The [U-13C3]PEP is converted into [U-13C3]pyruvate by dephosphorylation of PEP via the pyruvate kinase (pyk, SPD_0790 (EC 2.7.1.40), and the final formation of [U-13C3]Ala is catalyzed by the alanine transaminase (alaT, SPD_1791 (EC 2.6.1.2)) which transfers the amino group of Glu to pyruvate (Fig. 3A). This results in universally 13C3-labeled Ala and unlabeled α-ketoglutarate as glutamate was taken up from the medium.
The proportion of 13C3-labeled Asp, which is synthesized in a two-step reaction from PEP, is 39.5 ± 9.4 mol % (Fig. 2). In a first reaction, the PEP carboxylase (ppc, SPD_0953 (EC 4.1.1.31)) carboxylates [U-13C3]PEP to [1,2,3-13C3]oxaloacetate. Hence, oxaloacetate contains three 13C-labeled atoms from PEP and one unlabeled carbon because of fixation of CO2, which is derived from the environment (Fig. 3A). This pathway is similar to the PEP carboxylation in Enterococcus faecalis (41–43). In a second transamination reaction, the aspartate transaminase (aspC, SPD_1373 (EC 2.6.1.1)) converts [1,2,3-13C3]oxaloacetate into [1,2,3-13C3]Asp and thereby unlabeled glutamate into α-ketoglutarate, which is therefore unlabeled. In the AspC-catalyzed reactions, the l-amino group of glutamate is transferred to oxaloacetate, and this forms aspartate and α-ketoglutarate (Fig. 3A). Glutamate is the dominant amino acid formed from glutamine, which is one of the essential amino acids (Fig. 1), and is taken up by specific ABC transporter systems (15, 26). Threonine showed a corresponding labeling pattern with 56.8 mol % of a 1,2,3-13C3-labeled isotopologue. This is in line with Thr biosynthesis from aspartate via several catalytic reactions and homoserine as an intermediate. The genes encoding the essential enzymes aspartate kinase (SPD_0377), aspartate semialdehyde dehydrogenase (asd, SPD_0900), homoserine dehydrogenase (hom, SPD_1195), homoserine kinase (thrB, SPD_1194), and threonine synthase (thrC, SPD_1877) are annotated in the pneumococcal genomes (17).
De Novo Synthesis of Aromatic Amino Acids from [U-13C6]Glucose
In general, aromatic amino acids of microorganisms are synthesized via the shikimate pathway (44). Classically, PEP and erythrose 4-phosphate are the starting reactants and assembled to generate in seven metabolic steps the precursor chorismate. All enzymes of the shikimate pathway are encoded by pneumococci (data not shown), which is in accordance with the finding that pneumococci can be cultured in the absence of Phe, Trp, and Tyr (Fig. 1). [U-13C4]Erythrose 4-phosphate is generated in the experiment with [U-13C6]glucose in the PPP with glucose-6-phosphate dehydrogenase (zwf, (EC 1.1.1.49)) as the central enzyme (Figs. 2C and 3B). As mentioned above, PEP, as an intermediate of the EMP, is also triple labeled in the same experiment. The ultimate enzyme required to generate chorismate in three enzymatic reactions from shikimate is the chorismate synthase (aroC, SPD_1208 (EC 4.2.3.5)). Using completely 13C-labeled precursors [U-13C10]chorismate is formed (Fig. 3B). Chorismate is converted by chorismate mutase (SPD_1151 (EC 5.4.99.5)) to prephenate, and after two additional reactions, Phe and Tyr are produced (Fig. 3B). The fraction of universally 13C-labeled Phe or Tyr is relatively low with 2.2 and 1.0 mol %, respectively. These results demonstrate that uptake of aromatic amino acids is preferred by S. pneumoniae (45).
Hypothetical Synthesis Pathway of the Branched Amino Acids
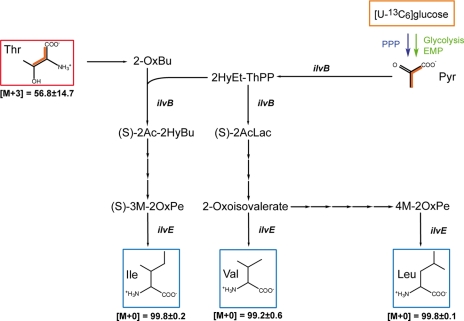
Pneumococci cultured in CDM are auxotrophic for branched-chain amino acids (Fig. 1C), and the isotopologue experiments showed no 13C-labeling of BCAA, confirming that Ile, Leu, and Val are indeed taken up by the ABC transporter LivJHMGF under these in vitro conditions (40). However, analysis of pneumococcal genomes revealed the presence of genes encoding all enzymes required to synthesize BCAA with Thr and pyruvate, respectively, as precursors as shown in Fig. 4. Without going into the details of BCAA synthesis, the genes encoding threonine dehydratase (ilvA, SPD_0409 (EC 4.3.1.19)), acetolactate synthase (ilvB, SPD_0404 (EC 2.2.1.6)), ketol-acid reductoisomerase (ilvC, SPD_0406 (EC 1.1.1.86)), dihydroxy-acid dehydratase (ilvD, SPD_1956 (EC 4.2.1.9)), and BCAA transaminase (ilvE, SPD_0749 (EC 2.6.1.42)) were identified in the pneumococcal genomes, and RT-PCR analysis indicated the expression of ilvB and ilvE (Fig. 2C). The functional activity of these enzymes and the conditions under which pneumococci conduct a de novo synthesis are yet unknown and require in-depth analysis.
FIGURE 4.
Hypothetical synthetic pathways leading to branched amino acids in S. pneumoniae cultured in CDM supplemented with [U-13C6]glucose. Multiple 13C labeling in potential precursors is indicated by orange bars connecting 13C-labeled atoms in a given molecule. 13C enrichment in Thr is detected and indicated by the red box. The 13C enrichment in pyruvate is predicted from the corresponding labeling of Ala. Blue boxes indicate the absence of 13C enrichment in Ile, Val, and Leu. For more details, see legend to Fig. 3B. The most essential enzymes are the acetolactate synthase (ilvB), and BCAA aminotransferase (ilvE). The abbreviations used are as follows: 2-OxBu, 2-oxobutanoate; 2HyEt-ThPP, 2-hydroxyethylthiamine pyrophosphate; (S)-2Ac-2HyBu, (S)-2-aceto-2-hydroxybutanoate; (S)-2AcLac, (S)-2-acetolactate; (S)-3M-2OxPe, (S)-3-Methyl-2-oxopentanoate; 4M-2OxPe, 4-methyl-2-oxopentanoate.
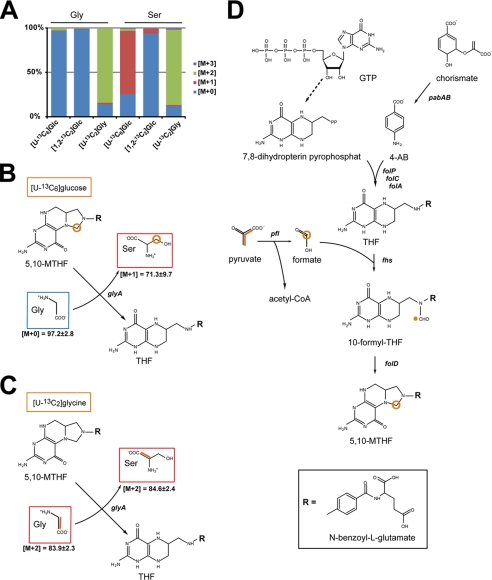
Serine Biosynthesis by Hydroxymethylation of Glycine Is Linked with the Pyruvate Formate-lyase Reaction
Nonessential serine is typically synthesized in bacteria from 3-phosphoglycerate in three steps via NAD+ and glutamate, which is required for transamination of 3-phosphohydroxypyruvate. The product 3-phosphoserine is then hydrolyzed resulting in serine. In a catabolic reaction, serine is deaminated to pyruvate or decomposed to glycine and a hydroxymethyl group to replenish the one carbon pool by generating N5,N10-methylene tetrahydrofolate (5,10-MTHF) (46). On this basis, one would expect the formation of [U-13C3]Ser and [U-13C2]Gly in the experiment with [U-13C6]glucose. Strikingly, unlabeled Gly and [13C1]Ser (71.3% 13C), but no significant amounts of [U-13C3]Ser or [U-13C2]Gly, were detected. These data suggested that Ser, which is not essential for pneumococcal growth (Fig. 1A), is very efficiently synthesized from (unlabeled) Gly by hydroxymethylation using a one-carbon precursor that acquired 13C label from [U-13C6]glucose at high rates (Fig. 5, A and B). Glycine, an essential amino acid for S. pneumoniae (Fig. 1D) and most likely assimilated by a glycine transporter system (proVWX; SPD_1642/1643), showed an almost 100% unlabeled state in experiments with [U-13C6]glucose and [1,2-13C2]glucose, respectively. The hydroxymethylation of assimilated Gly was confirmed in experiments using 1.33 mm [U-13C2]glycine as a tracer (Fig. 5, A and C; for numerical values see supplemental Table S4). The enzyme required for this reaction is the serine hydroxymethyltransferase (SHMT (EC 2.1.2.1)), and the expression of the SHMT-encoding gene glyA (SPD_0910) in pneumococci was shown by RT-PCR (Fig. 2C). The SHMT catalyzes the conversion of 5,10-MTHF to tetrahydrofolate (THF), and the generated hydroxymethyl group is transferred to glycine, which results in serine. In the presence of [U-13C6]glucose, the hydroxymethyl group derived from 5,10-MTHF has to be 13C-labeled as glycine is unlabeled and the product serine is 3-13C1-labeled. The labeling of the hydroxymethyl donor in 5,10-MTHF can be explained by linking glucose catabolism with the synthesis and regeneration of the one carbon pool. An essential reaction in the synthesis of 13C-labeled 5,10-MTHF in the experiment with [U-13C6]glucose is the conversion of [U-13C3]pyruvate to [13C]formate and [U-13C2]acetyl-CoA via the pyruvate formate-lyase (pfl, SPD_0420 (EC 2.3.1.54)). The key role of PFL in the formation of formate has previously been reported (47). [13C]Formate is added to THF leading via 5-formyl-THF to 5,10-MTHF containing 13C at the methylene position (Fig. 5D). The enzymes formate-tetrahydrofolate ligase (fhs, SPD_1087 (6.3.4.3)) and methylenetetrahydrofolate cyclohydrolase (folD, SPD_0721 (EC 3.5.4.9)) catalyze under ATP consumption the formation of 5,10-MTHF. All the enzymes required to synthesize folate and THF are annotated in the pneumococcal genomes. For example, the enzyme dihydropteroate synthase (folP, SPD_0269 (EC 2.5.1.15)) catalyzes the formation of 7,8-dihydropteroate from condensation of 4-aminobenzoate with 2-amino-4-hydroxy-6-hydroxymethyl-7,8-dihydropteridine diphosphate (7,8-dihydropterin PP). The dihydrofolate synthase and dihydrofolate reductase (folC/folA, SPD_0183/1401) then catalyze the conversion of 7,8-dihydropteroate via 7,8-dihydrofolate to folate and/or THF. The precursor of 4-aminobenzoate is chorismate, whereas 7,8-dihydropterin-PP is generated in seven reactions from guanosine 5′-triphosphate (Fig. 5D).
FIGURE 5.
Pneumococcal synthesis of serine by hydroxymethylation of glycine. A, relative fractions of 13C-labeled isotopologues in serine and glycine in the experiments with [U-13C6]glucose, [1,2-13C2]glucose, or [U-13C2]glycine, respectively. The colored boxes indicate the relative contributions in percent of isotopologues with 0, 1, 2, or 3 13C-atoms (M+0, M+1, M+2, and M+3, respectively) in the overall enrichments of serine and glycine. The values represent the means of three technical replicates. B, reaction model of de novo synthesis of serine in CDM supplemented with [U-13C6]glucose. The molar abundances of isotopomers (M+0, M+1, and M+2) is listed. C, reaction model of de novo synthesis of serine in CDM supplemented with [U-13C2]glycine. Multiple 13C labeling is indicated by orange bars connecting 13C-labeled atoms in a molecule, and labeling of one carbon atom by 13C in a given molecule is depicted by a red circle. D, biosynthesis and regeneration of 5,10-MTHF in pneumococci. 13C labeling is shown for pyruvate and carbon atoms derived thereof in formate and 5,10-MTHF. The abbreviations used are as follows: glyA, SHMT (SPD_0910 (EC 2.1.2.1)); THF, tetrahydrofolate; pabAB, para-aminobenzoate synthase; folA, dihydrofolate reductase; folC, folylpolyglutamate synthase; folD, 5,10-methylenetetrahydrofolate cyclohydrolase; folP, dihydropteroate synthase; pfl, pyruvate formate-lyase; fhs, formate tetrahydrofolate ligase; R, N-benzoyl-l-glutamate.
In the experiments with [1,2-13C2]glucose, Ser acquired the 13C label only at low rates (5.7% 13C1) (Fig. 5A; for numerical values see supplemental Table S5). This is in accordance with a reaction trajectory starting from [1,2-13C2]glucose to [2,3-13C2]pyruvate via the EMP pathway that gives rise to an unlabeled formate in the PFL reaction. Taken together, Ser formation is linked with the PFL reaction in pneumococci where its substrate pyruvate is derived from glucose via glycolysis.
Embden-Meyerhof-Parnas Pathway Is the Major Pathway for Glucose Catabolism
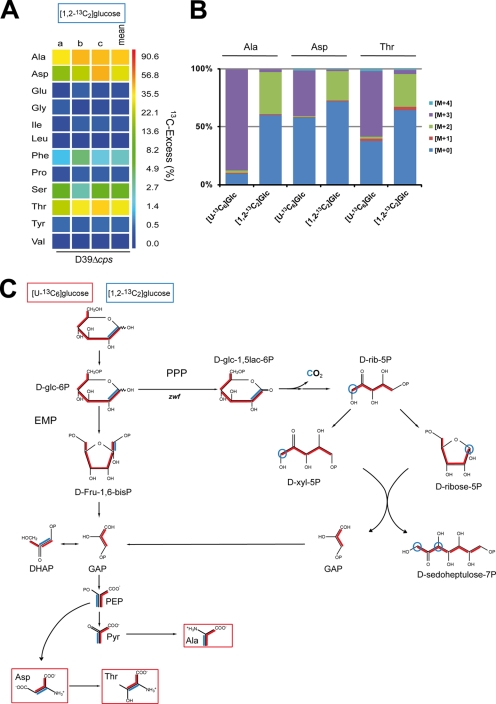
To dissect the metabolic pathways used to catabolize glucose, the relative amounts of Ala, Asp, and Thr isotopologues were compared after growth of pneumococci in CDM with [U-13C6]glucose or [1,2-13C2]glucose, respectively. Pneumococci take up glucose by the PTS (glucose-specific IIA component) and phosphorylate it to glucose 6-phosphate. Pneumococci can catabolize glucose only via the EMP and PPP as they lack the ED pathway (17). As mentioned, Ala is generated through conversion of pyruvate, which represents a major intermediate of the glycolysis. Isotopologue experiments with [U-13C6]glucose revealed that 87.2% of Ala is universally 13C3-labeled (Figs. 3A and 6). The precursors of Asp and Thr are PEP and oxaloacetate, both containing three 13C-labeled isotopologues when using [U-13C6]glucose. The GC/MS analysis indeed showed 39.5 mol % [13C3]Asp and 56.8 mol % [13C3]Thr (Figs. 3A and 6).
FIGURE 6.
Metabolic profiling of fluxes via the EMP and the PPP on the basis of 13C-labeling experiments. A, overall 13C excess (%) in amino acids derived from proteins after cultivation of S. pneumoniae D39 in CDM supplemented with 27.8 mm [1,2-13C2]glucose. The color map indicates 13C excess in a logarithmic form. The results of three independent experiments measured in triplicate (a–c) and the means are shown. B, relative fractions of isotopologues in certain amino acids relevant to discriminate fluxes via the EMP and the PPP in experiments with [U-13C6]glucose (cf. also Fig. 2A) and [1,2-13C2]glucose. The colored boxes indicate the relative contributions in percent of carbon isotopologues (M+0, M+1, M+2, M+3, and M+4 are in different colors) in the overall enrichments. C, metabolic model for glucose utilization of S. pneumoniae D39 in CDM supplemented with [U-13C6]glucose (13C label shown by red lines) or [1,2-13C2]glucose (13C label shown by blue lines). Labeling patterns were detected in protein-derived amino acids (shown in boxes). Multiple 13C-labeled isotopologues determined by GC/MS are depicted as colored bars connecting 13C atoms in a molecule. Red bars indicate labeling in EMP and PPP with [U-13C6]glucose as carbon source and blue bars with [1,2-13C2]glucose as C-source in CDM. The abbreviations used are as follows: α-d-glc-6P, α-d-glucose-6P; d-glc-1,5lac-6P, d-glucono-1,5-lactone-6P; DHAP, dihydroxyacetone phosphate; d-Fru-1,6-bisP, d-fructose-1,6-bisP; d-rib-5P, d-Ribulose-5P; d-xyl-5P, d-xylulose-5P; OAA, oxaloacetate; α-KG, α-ketoglutarate.
The relative contributions of EMP and PPP in glucose utilization were estimated on the basis of the labeling patterns of Ala, Asp, and Thr from the experiment with [1,2-13C2]glucose (Fig. 6B). The GC/MS analysis showed that both 13C atoms of the tracer were transferred to Ala, Asp, and Thr at similar rates of 36.2, 25.2, and 28.1%, respectively (Fig. 6; for all numerical values see supplemental Table S5). In comparison, the incorporation rates into these amino acids were approximately two times higher in the experiment with of [U-13C6]glucose (Fig. 2A and supplemental Table S3). The lower 13C enrichment from [1,2-13C2]glucose can be easily explained by the fact that both halves of the supplied glucose, i.e. C1–C3 and C4–C6, respectively, contribute to the formation of the early precursors for Ala, Asp, and Thr. As a consequence, only the transfer rates will be 50% in the experiment with [1,2-13C2]glucose in comparison with [U-13C6]glucose. In accordance to the metabolic model shown in Fig. 6, it can be concluded that the major fraction of glyceraldehyde 3-phosphate (GAP) molecules, representing the precursor of PEP/Asp/Thr and pyruvate/Ala, are generated via the EMP and not via the PPP involving the glucose-6-phosphate dehydrogenase reaction.
During glycolysis (EMP) the glucose 6-phosphate is metabolized to fructose 1,6-bisphosphate, still containing C1 and C2 of the carbon source [1,2-13C2]glucose. Then fructose 1,6-bisphosphate is cleaved by the fructose-bisphosphate aldolase (fba, SPD_0526 (EC 4.1.2.13)) into GAP and dihydroxyacetone phosphate, which contains both 13C atoms. The rapid triose-phosphate isomerase (SPD_1404 (EC 5.3.1.1)) catalyzes the interconversion between the isomers dihydroxyacetone phosphate and GAP, so that both 13C atoms of the carbon source are found in 50% of GAP. In contrast, GAP molecules generated by the oxidative PPP will remain unlabeled in experiments with [1,2-13C2]glucose as the precursors of GAP, the xylulose 5-phosphate and d-ribulose 5-phosphate, contain at position 1 only one 13C atom arising from the C-2 of [1,2-13C2]glucose. In the subsequent transketolase reaction, the 13C label of xylulose 5-phosphate is transferred to d-ribose 5-phosphate yielding 13C2-labeled sedulose 7-phosphate and unlabeled GAP (Fig. 6C). Notably, during the oxidative PPP, the 13C atom at position C-1 of [1,2-13C2]glucose is lost during decarboxylation of the 6-phosphogluconate, which is catalyzed by the 6-P-gluconate dehydrogenase (SPD_0343 (EC 1.1.1.44)). The products are 13CO2 and d-[1-13C]ribulose-5-P (Fig. 6C). Considering a potential efficient carbon flux via this route of the PPP, the formed 13CO2 should be re-incorporated at least in part into those products that are formed by carboxylation, e.g. oxaloacetate/Asp/Thr. However, our experimental data do not support this hypothesis. In conclusion, the results strongly suggest that under the conditions used in the isotopologue experiments glucose is mainly catalyzed via the EMP.
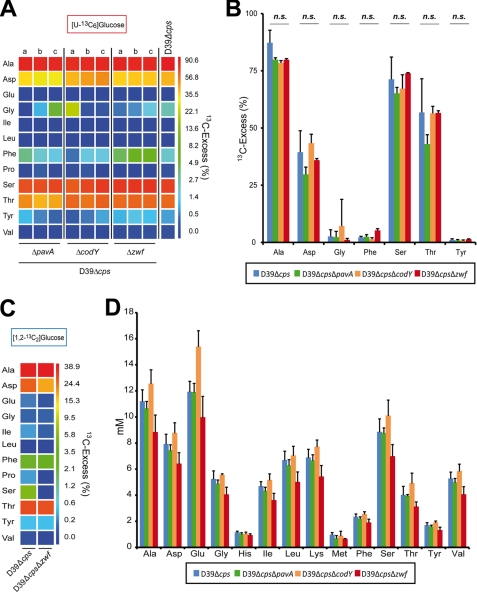
Isotopologue Profiling of Virulence Attenuated ΔpavA, ΔcodY, and Δzwf Pneumococcal Mutants
To assess the impact of the metabolic regulator CodY, the enzyme glucose-6-phosphate dehydrogenase (encoded by zwf), which is a central enzyme in the PPP and converts glucose 6-phosphate into d-glucono-1,5-lactone 6-phosphate, and the virulence factor PavA on the central amino acid biosynthesis, isotopologue experiments were conducted with universally labeled [U-13C6]glucose. All three pneumococcal mutants D39ΔpavA, D39ΔcodY, and D39Δzwf showed no growth defects in the complex medium THY; however, in CDM the codY mutant showed a significantly reduced growth rate (data not shown). Previous studies demonstrated that the mutants were impaired in adherence and/or attenuated in mouse infection models (16, 27, 35, 48, 49). It is noteworthy here that the codY mutant generated in this study contains, similar to the mutant constructed by Hendriksen et al. (27), a suppressor mutation in amiC (data not shown), which is part of the amiACDEF operon encoding an ABC transporter (50). This confirms a previous study, which suggested that CodY is essential for pneumococcal viability due to its involvement in amino acid transport systems, regulation of oxidative stress, and iron consumption (51). The comparison of the isotopologue profiles between the wild-type and isogenic mutants demonstrated similar levels of amino acid labeling for all four strains (Fig. 7; for numerical values see supplemental Tables S3 and S6–S8). Importantly, the total amino acid content was also similar for all strains (Fig. 7D). These results suggest that the carbon metabolism and glucose utilization were not affected due to loss of function of CodY, PavA, or glucose-6-phosphate dehydrogenase. Furthermore, isotopologue experiments conducted with [1,2-13C2]glucose and the zwf mutant confirmed that glucose is predominantly catabolized via the EMP as the percental allocation of isotopologues in Ala, Asp, and Thr, respectively, showed no significant differences between the wild-type and the zwf mutant (Fig. 7C).
FIGURE 7.
Impact of PavA, CodY, or Zwf deficiency on pneumococcal fitness and 13C-labeled isotopologue profiles in amino acids after growth in CDM supplemented with [U-13C6]- or [1,2-13C2]glucose. A and B, overall 13C excess (%) of 13C-labeled amino acids of pneumococcal mutants ΔcodY, ΔpavA, and Δzwf, respectively, compared with the isogenic wild-type D39Δcps after cultivation of bacteria in CDM supplemented with 55.5 mm [U-13C6]glucose. The color map (A) indicates 13C excess in a logarithmic form. Each experiment was performed in triplicate (a–c), and the wild-type column shows the mean value of three independent experiments. The columns in B show the same values and the standard deviations from the three technical replicates. n.s., not significant. C, overall 13C excess (mol %) of 13C-labeled amino acids of the mutant D39ΔcpsΔzwf compared with its isogenic D39Δcps after cultivation of bacteria in CDM supplemented with 27.8 mm [1,2-13C2]glucose. D, amino acid concentrations (millimolar) in pneumococci after growth in CDM. The total amino acid content was determined by 1H NMR.
DISCUSSION
In this study the central carbon and amino acid metabolism of pneumococci were analyzed by comprehensive isotopologue profiling under defined culture conditions with amino acids and glucose as potential carbon sources. Pneumococci conduct a fermentative metabolism regardless of oxygen, which is consistence with the lack of genes encoding enzymes of the TCA. The central carbon metabolism generates predominantly pyruvate or lactate in homolactic fermentation of glucose, NADH, and ATP. However, under sugar limitation or in the presence of carbon sources such as galactose, a mixed acid fermentation is preferred, which results in the formation of ethanol, acetate, and formate (47). Given the fact that the host niches encountered by pneumococci represent, with the exception of the blood, nutrient-limited niches but are rich in glycoproteins with O- and N-linked glycans containing e.g. galactose, pneumococci have to adapt to these different sets of nutrients and carbon sources (17, 19, 52). In fact, pneumococci are equipped with a broad range of transporters whose impact on bacterial fitness and virulence has been reported to some extent. Moreover, pneumococci are able to utilize mucin as a carbon source (15, 40, 47, 53–58). This also raises the hypothesis that pneumococci prefer uptake, breakdown, and catabolism of nutrients, peptides, and amino acids instead of their de novo synthesis.
The analysis of pneumococci cultured in chemically defined medium, whose complete mixture contains glucose and all 20 amino acids, allowed us to study the effect of amino acid deprivation on cell proliferation. The data revealed that under these conditions pneumococci are auxotrophic for Arg, Cys, Gln, Gly, His, and the branched-chain amino acids Leu, Ile, and Val. The remaining amino acids may be converted from available amino acids or are synthesized from intermediates of the carbohydrate catabolism. The results of the amino acid deprivation experiments (Fig. 1) and the proposed de novo synthesis of amino acids by pneumococci were confirmed by the 13C incorporation rates measured for individual amino acids after culturing pneumococci in the presence of [U-13C6]glucose. The isotopologue profiling showed the formation of 13C3-labeled isotopologues in Ala, Asp, and Thr at high rates, universally labeled Tyr and Phe at low rates, and surprisingly, a 13C1-labeled Ser again at a high rate. In contrast, Gly, Glu, His, Pro, and the BCAAs Ile, Leu, and Val remained unlabeled. The results corroborate previous studies showing that a deficiency of ABC transporters for Gln or BCAA (LivJHMGF) impairs pneumococcal growth and virulence (15, 26, 40). Pneumococci seem to prefer uptake of BCAAs under in vitro conditions, although genes encoding enzymes for a de novo synthesis of BCAAs were annotated in the genomes. The expression of these genes and their functional role warrants therefore further analysis. As a general rule, one can predict that alterations in amino acid metabolism influence the virulence potential of pathogenic microorganisms and most likely also the physiology and fitness of nonpathogenic microorganisms. For example, deficiencies in amino acid synthesis in Salmonella enterica serovar Typhimurium impaired survival in murine infection models (59–62). Furthermore, microorganisms utilize amino acids as carbon sources such as the lung pathogen Legionella pneumophila (13, 63, 64).
The isotopologue profiles and analysis of the genome data indicate that pneumococci synthesize Ala conventionally from pyruvate. The alanine transaminase transfers the amino group from glutamate to pyruvate, which forms alanine and α-ketoglutarate. The precursor of Asp and Thr, oxaloacetate, is synthesized by carboxylation of PEP by the PEP carboxylase in S. pneumoniae, a reaction that requires endogenously produced or externally gained CO2. Oxaloacetate is then converted into Asp, catalyzed by the aspartate transaminase, whereas Glu is simultaneously converted into α-ketoglutarate. Threonine is then synthesized from Asp via several intermediates such as e.g. homoserine.
In part, the metabolic network is similar to Listeria monocytogenes, where oxaloacetate is also made by carboxylation. However, in L. monocytogenes oxaloacetate is obtained by carboxylation of pyruvate and not PEP. In contrast to pneumococci, the PEP carboxylase is not produced by L. monocytogenes (12).
In pneumococci, Gln is an essential amino acid, and Glu deprivation results in growth defects. According to these results, the isotopologue profiling showed no 13C excess for Glu suggesting that also α-ketoglutarate remained unlabeled, which is synthesized from glutamate via the aspartate transaminase. Pneumococci produce Gln uptake systems, which are regulated by GlnR and the glutamine synthetase GlnA (15, 26). These data suggest that pneumococci are unable to synthesize these amino acids de novo, and as both amino acids are major intermediates of biosynthetic pathways such as the de novo synthesis of aspartate or nucleotides, insufficient Gln levels decelerate pneumococcal proliferation. In contrast, Glu is nonessential in L. monocytogenes and synthesized from α-ketoglutarate, which is formed via condensation of oxaloacetate and acetyl-CoA followed by decarboxylation (14). Similarly, α-ketoglutarate and oxaloacetate serve as precursors for Glu and Asp, respectively, in L. pneumophila. In these microorganisms, oxaloacetate is formed by the complete TCA or by carboxylation of pyruvate (13).
Despite the low 13C excess of nonessential aromatic amino acids Phe and Tyr in isotopologue experiments with [U-13C6]glucose, the data and genome analysis indicated their de novo synthesis in S. pneumoniae via chorismate. This pathway is also used by L. monocytogenes, whose isotopologue composition of Tyr and Phe further reflected the isotopologue composition of PEP and erythrose 4-phosphate, the precursors of chorismate and hence Phe and Tyr (14).
A remarkable and untypical 13C labeling was measured for Ser and Gly. Pneumococci cultured in CDM supplemented with [U-13C6]glucose showed unlabeled Gly, which is in accordance with their auxotrophy for Gly, and only a 13C1-labeled isotopologue in Ser. The analysis by isotopologue profiling with [U-13C2]glycine confirmed that serine is not synthesized from 3-phosphoglycerate but by hydroxymethylation of Gly as reflected by 13C2-labeled isotopologues in Gly and Ser. The reaction used by pneumococci to fill up their Ser pool is rather unconventional among bacterial species. Most bacterial species prefer the reverse reaction, where the SHMT catalyzes the conversion of Ser to Gly. The de novo synthesis of Ser in bacteria typically involves dehydrogenation of 3-phosphoglycerate, which is followed by reductive transamination and dephosphorylation. This pathway has been reported for Streptococcus thermophilus (65, 66) and also L. monocytogenes (12, 14). In L. pneumophila, Ser supports the growth and serves as a major carbon substrate (13). In pneumococci, the conversion of Gly to Ser is catalyzed by the SHMT encoded by glyA (SPD_0910), and gene expression was confirmed by RT-PCR. As Gly is essential under the used in vitro conditions, one may conclude that loss of function of SHMT is lethal for pneumococci and that this enzyme may represent a novel drug target. Moreover, enzymes linked to the serine biosynthetic pathway are enzymes involved in the regeneration of the one carbon pool by folate and the PFL. The latter generates formate from pyruvate, and formate is then added to THF to produce formyl-THF, which is then converted to 5,10-MTHF. The importance of PFL for pneumococcal fermentative metabolism and virulence has recently been reported (47). As the genes for the pyruvate dehydrogenase complex, converting pyruvate to CO2 and acetyl-CoA, are absent in pneumococci, the PFL is also essential for the mixed acid fermentation of sugars others than glucose (17, 19, 47, 67). The PFL deficiency in Staphylococcus aureus had pleiotropic effects, and the pathogen was unable to produce formate. Apparently, the S. aureus PFL is essential to refill the formyl-THF pool (68). In this study, we further show now a link between the function of PFL and Ser synthesis. As formate is essential in pneumococci due to its role in Ser synthesis, this is probably one of the reasons for the importance of PFL for pneumococcal metabolism and virulence. In turn, PFL may also represent a promising drug target.
The genome sequences of pneumococci indicated the presence of the EMP and the PPP but not of the ED and TCA. Pneumococci are incapable of respiratory metabolism, and the only nutrients from which they can get sufficient energy are carbohydrates (17, 69). Pneumococci have a wide substrate utilization range for sugars, and the genome data suggest that they can use the glycolysis (EMP) for catabolism of over 12 different sugars, including glucose, sucrose, or cellobiose and the PPP to catabolize C5 sugar alcohols (pentitols). Pneumococci use PEP-dependent phosphotransferase systems and energetically less efficient ABC transporters to import carbohydrates (17). The isotopologue profiling using [U-13C6]- and [1,2-13C2]glucose suggested the EMP as the main route of glucose catabolism. A major role of the PPP involving the oxidative branch can be excluded on the basis of the labeling patterns in amino acids from the experiment with [1,2-13C2]glucose. However, genes encoding enzymes of the nonoxidative part of the PPP are also existent in pneumococci such as the transketolase (tk, SPD_1839 (EC 2.2.1.1)) and transaldolase (talC, SPD_0236 (2.2.1.2)). This implies that pneumococci have the capability to synthesize fructose 6-phosphate from GAP. Assuming carbon flux in the PPP using [1-13C]ribulose phosphate and [2,3-13C2]GAP, a mixture of [5,6-13C2]- and [1,3-13C2]fructose 6-phosphate is generated. Taking [1,3-13C2]fructose 6-phosphate as substrate in the EMP pathway, GAP may also occur as a 1,3-13C2-labeled isotopologue. Following the reactions of the metabolic model shown in Fig. 6, PEP, pyruvate, and oxaloacetate are also labeled at C-1 and C-3 positions. Direct experimental evidence for a minor flux via this complex metabolic route is provided by the detection of a 13C1 species at low concentrations in Ser from the experiment with [1,2-13C2]glucose (Fig. 5A). In the absence of flux via the PPP, Ser would be unlabeled as formate is derived from the C-1 atom of pyruvate, which is unlabeled when generated only via the EMP in presence of [1,2-13C2]glucose (Fig. 6C). In accordance with carbon metabolism predominantly via the EMP, the 13C excess in amino acids of the zwf mutant, encoding the first enzyme of the PPP, namely glucose-6-phosphate dehydrogenase, was similar to that measured for the wild type. The pavA and codY mutant cultured with [U-13C6]glucose also showed no differences in the labeling pattern of amino acids. This strengthens the hypothesis that PavA modulates only the functional activity of surface-exposed virulence factors (35) and that loss of function of the global regulator CodY is lethal in pneumococci. CodY has an essential role in regulating the amino acid transport, oxidative stress, and iron consumption as well, and a defect in CodY was can only be compensated by a suppressor mutation (51).
In conclusion, this study indicates that pneumococci use an unconventional pathway to synthesize Ser and that glucose and most likely also other carbon sources are metabolized predominantly via the EMP. The high number of uptake systems and the simplified biosynthesis pathways may point to the robustness and adaptation of this extracellular pathogen to its different host niches. In pneumococci, the dual utilization of carbohydrates and amino acids seems to be a prerequisite for the success of the pathogen to encounter the various host niches and cause severe infectious diseases. However, this study also provides evidence that the elucidation of bacterial metabolism and metabolic flux offers the possibility to find novel drug targets such as the SHMT or PFL in pneumococci.
Supplementary Material
Acknowledgment
We are grateful to M. Hecker (Greifswald) for critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants DFG HA 3125/3-1 and EI-384/5-1 within the SPP1316 and EU FP7 CAREPNEUMO Grant EU-CP223111 from the European Union.

This article contains supplemental Tables S1–S8.
- ABC
- ATP-binding cassette
- BCAA
- branched chain amino acids
- CDM
- chemical defined medium
- EMP
- Embden-Meyerhof-Parnas
- 5,10-MTHF
- N5,N10-methylene tetrahydrofolate
- PEP
- phosphoenolpyruvate
- PFL
- pyruvate formate-lyase
- PPP
- pentose phosphate pathway
- RT
- reverse transcriptase
- SHMT
- serine hydroxymethyltransferase
- ED
- Entner-Doudoroff pathway
- GAP
- glyceraldehyde 3-phosphate.
REFERENCES
- 1. Bogaert D., De Groot R., Hermans P. W. (2004) Streptococcus pneumoniae colonization. The key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154 [DOI] [PubMed] [Google Scholar]
- 2. Cartwright K. (2002) Pneumococcal disease in western Europe. Burden of disease, antibiotic resistance, and management. Eur. J. Pediatr. 161, 188–195 [DOI] [PubMed] [Google Scholar]
- 3. Hammerschmidt S. (2006) Adherence molecules of pathogenic pneumococci. Curr. Opin. Microbiol. 9, 12–20 [DOI] [PubMed] [Google Scholar]
- 4. Musher D. M. (1992) Infections caused by Streptococcus pneumoniae. Clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14, 801–807 [DOI] [PubMed] [Google Scholar]
- 5. Bergmann S., Hammerschmidt S. (2006) Versatility of pneumococcal surface proteins. Microbiology 152, 295–303 [DOI] [PubMed] [Google Scholar]
- 6. Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W. (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301 [DOI] [PubMed] [Google Scholar]
- 7. Nobbs A. H., Lamont R. J., Jenkinson H. F. (2009) Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Somerville G. A., Proctor R. A. (2009) At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muñoz-Elías E. J., McKinney J. D. (2006) Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8, 10–22 [DOI] [PubMed] [Google Scholar]
- 10. Beste D. J., Bonde B., Hawkins N., Ward J. L., Beale M. H., Noack S., Nöh K., Kruger N. J., Ratcliffe R. G., McFadden J. (2011) 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria that requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 7, e1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eisenreich W., Dandekar T., Heesemann J., Goebel W. (2010) Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412 [DOI] [PubMed] [Google Scholar]
- 12. Eisenreich W., Slaghuis J., Laupitz R., Bussemer J., Stritzker J., Schwarz C., Schwarz R., Dandekar T., Goebel W., Bacher A. (2006) 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U.S.A. 103, 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eylert E., Herrmann V., Jules M., Gillmaier N., Lautner M., Buchrieser C., Eisenreich W., Heuner K. (2010) Isotopologue profiling of Legionella pneumophila. Role of serine and glucose as carbon substrates. J. Biol. Chem. 285, 22232–22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eylert E., Schär J., Mertins S., Stoll R., Bacher A., Goebel W., Eisenreich W. (2008) Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69, 1008–1017 [DOI] [PubMed] [Google Scholar]
- 15. Härtel T., Klein M., Koedel U., Rohde M., Petruschka L., Hammerschmidt S. (2011) Impact of glutamine transporters on pneumococcal fitness under infection-related conditions. Infect. Immun. 79, 44–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hendriksen W. T., Kloosterman T. G., Bootsma H. J., Estevão S., de Groot R., Kuipers O. P., Hermans P. W. (2008) Site-specific contributions of glutamine-dependent regulator GlnR and GlnR-regulated genes to virulence of Streptococcus pneumoniae. Infect. Immun. 76, 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoskins J., Alborn W. E., Jr., Arnold J., Blaszczak L. C., Burgett S., DeHoff B. S., Estrem S. T., Fritz L., Fu D. J., Fuller W., Geringer C., Gilmour R., Glass J. S., Khoja H., Kraft A. R., Lagace R. E., LeBlanc D. J., Lee L. N., Lefkowitz E. J., Lu J., Matsushima P., McAhren S. M., McHenney M., McLeaster K., Mundy C. W., Nicas T. I., Norris F. H., O'Gara M., Peery R. B., Robertson G. T., Rockey P., Sun P. M., Winkler M. E., Yang Y., Young-Bellido M., Zhao G., Zook C. A., Baltz R. H., Jaskunas S. R., Rosteck P. R., Jr., Skatrud P. L., Glass J. I. (2001) Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183, 5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paulsen I. T., Nguyen L., Sliwinski M. K., Rabus R., Saier M. H., Jr. (2000) Microbial genome analyses. Comparative transport capabilities in 18 prokaryotes. J. Mol. Biol. 301, 75–100 [DOI] [PubMed] [Google Scholar]
- 19. Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 [DOI] [PubMed] [Google Scholar]
- 20. Hava D. L., Camilli A. (2002) Large scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 21. Lau G. W., Haataja S., Lonetto M., Kensit S. E., Marra A., Bryant A. P., McDevitt D., Morrison D. A., Holden D. W. (2001) A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40, 555–571 [DOI] [PubMed] [Google Scholar]
- 22. Marra A., Asundi J., Bartilson M., Lawson S., Fang F., Christine J., Wiesner C., Brigham D., Schneider W. P., Hromockyj A. E. (2002) Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. Infect. Immun. 70, 1422–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Obert C., Sublett J., Kaushal D., Hinojosa E., Barton T., Tuomanen E. I., Orihuela C. J. (2006) Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74, 4766–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polissi A., Pontiggia A., Feger G., Altieri M., Mottl H., Ferrari L., Simon D. (1998) Large scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerr A. R., Adrian P. V., Estevão S., de Groot R., Alloing G., Claverys J. P., Mitchell T. J., Hermans P. W. (2004) The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 72, 3902–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kloosterman T. G., Hendriksen W. T., Bijlsma J. J., Bootsma H. J., van Hijum S. A., Kok J., Hermans P. W., Kuipers O. P. (2006) Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281, 25097–25109 [DOI] [PubMed] [Google Scholar]
- 27. Hendriksen W. T., Bootsma H. J., Estevão S., Hoogenboezem T., de Jong A., de Groot R., Kuipers O. P., Hermans P. W. (2008) CodY of Streptococcus pneumoniae. Link between nutritional gene regulation and colonization. J. Bacteriol. 190, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iyer R., Baliga N. S., Camilli A. (2005) Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187, 8340–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brinsmade S. R., Kleijn R. J., Sauer U., Sonenshein A. L. (2010) Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J. Bacteriol. 192, 6357–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leonard C. G., Ranhand J. M., Cole R. M. (1970) Competence factor production in chemically defined media by noncompetent cells of group H Streptococcus strain Challis. J. Bacteriol. 104, 674–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mickelson M. N. (1964) Chemically defined medium for growth of Streptococcus pyogenes. J. Bacteriol. 88, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jensch I., Gámez G., Rothe M., Ebert S., Fulde M., Somplatzki D., Bergmann S., Petruschka L., Rohde M., Nau R., Hammerschmidt S. (2010) PavB is a surface-exposed adhesin of Streptococcus pneumoniae contributing to nasopharyngeal colonization and airways infections. Mol. Microbiol. 77, 22–43 [DOI] [PubMed] [Google Scholar]
- 33. Rennemeier C., Hammerschmidt S., Niemann S., Inamura S., Zähringer U., Kehrel B. E. (2007) Thrombospondin-1 promotes cellular adherence of Gram-positive pathogens via recognition of peptidoglycan. FASEB J. 21, 3118–3132 [DOI] [PubMed] [Google Scholar]
- 34. Noske N., Kämmerer U., Rohde M., Hammerschmidt S. (2009) Pneumococcal interaction with human dendritic cells. Phagocytosis, survival, and induced adaptive immune response are manipulated by PavA. J. Immunol. 183, 1952–1963 [DOI] [PubMed] [Google Scholar]
- 35. Pracht D., Elm C., Gerber J., Bergmann S., Rohde M., Seiler M., Kim K. S., Jenkinson H. F., Nau R., Hammerschmidt S. (2005) PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation. Infect. Immun. 73, 2680–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen J. D., Morrison D. A. (1988) Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64, 155–164 [DOI] [PubMed] [Google Scholar]
- 37. Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C., Zipfel P. F. (2007) The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J. Immunol. 178, 5848–5858 [DOI] [PubMed] [Google Scholar]
- 38. Liebeke M., Dörries K., Zühlke D., Bernhardt J., Fuchs S., Pané-Farré J., Engelmann S., Völker U., Bode R., Dandekar T., Lindequist U., Hecker M., Lalk M. (2011) A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol. Biosyst. 7, 1241–1253 [DOI] [PubMed] [Google Scholar]
- 39. Lee W. N., Sorou S., Bergner E. A. (1991) Glucose isotope, carbon recycling, and gluconeogenesis using [U-13C]glucose and mass isotopomer analysis. Biochem. Med. Metab. Biol. 45, 298–309 [DOI] [PubMed] [Google Scholar]
- 40. Basavanna S., Khandavilli S., Yuste J., Cohen J. M., Hosie A. H., Webb A. J., Thomas G. H., Brown J. S. (2009) Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect. Immun. 77, 3412–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goff R. C., Hartman R. E. (1970) Carbon dioxide fixation by cells of Streptococcus faecalis var. liquefaciens. J. Bacteriol. 104, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hartman R. E. (1970) Carbon dioxide fixation by extracts of Streptococcus faecalis var. liquefaciens. J. Bacteriol. 102, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lachica V. F., Hartman P. A. (1969) Inhibition of CO2 fixation in group D streptococci. Can. J. Microbiol. 15, 57–60 [DOI] [PubMed] [Google Scholar]
- 44. Herrmann K. M., Weaver L. M. (1999) The shikimate pathway. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 50, 473–503 [DOI] [PubMed] [Google Scholar]
- 45. Panina E. M., Vitreschak A. G., Mironov A. A., Gelfand M. S. (2003) Regulation of biosynthesis and transport of aromatic amino acids in low-GC Gram-positive bacteria. FEMS Microbiol. Lett. 222, 211–220 [DOI] [PubMed] [Google Scholar]
- 46. Pizer L. I. (1965) Glycine synthesis and metabolism in Escherichia coli. J. Bacteriol. 89, 1145–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yesilkaya H., Spissu F., Carvalho S. M., Terra V. S., Homer K. A., Benisty R., Porat N., Neves A. R., Andrew P. W. (2009) Pyruvate formate-lyase is required for pneumococcal fermentative metabolism and virulence. Infect. Immun. 77, 5418–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holmes A. R., McNab R., Millsap K. W., Rohde M., Hammerschmidt S., Mawdsley J. L., Jenkinson H. F. (2001) The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41, 1395–1408 [DOI] [PubMed] [Google Scholar]
- 49. Kadioglu A., Brewin H., Härtel T., Brittan J. L., Klein M., Hammerschmidt S., Jenkinson H. F. (2010) Pneumococcal protein PavA is important for nasopharyngeal carriage and development of sepsis. Mol. Oral Microbiol. 25, 50–60 [DOI] [PubMed] [Google Scholar]
- 50. Claverys J. P., Grossiord B., Alloing G. (2000) Is the Ami-AliA/B oligopeptide permease of Streptococcus pneumoniae involved in sensing environmental conditions? Res. Microbiol. 151, 457–463 [DOI] [PubMed] [Google Scholar]
- 51. Caymaris S., Bootsma H. J., Martin B., Hermans P. W., Prudhomme M., Claverys J. P. (2010) The global nutritional regulator CodY is an essential protein in the human pathogen Streptococcus pneumoniae. Mol. Microbiol. 78, 344–360 [DOI] [PubMed] [Google Scholar]
- 52. Philips B. J., Meguer J. X., Redman J., Baker E. H. (2003) Factors determining the appearance of glucose in upper and lower respiratory tract secretions. Intensive Care Med. 29, 2204–2210 [DOI] [PubMed] [Google Scholar]
- 53. Brown J. S., Gilliland S. M., Holden D. W. (2001) A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40, 572–585 [DOI] [PubMed] [Google Scholar]
- 54. Marra A., Lawson S., Asundi J. S., Brigham D., Hromockyj A. E. (2002) In vivo characterization of the psa genes from Streptococcus pneumoniae in multiple models of infection. Microbiology 148, 1483–1491 [DOI] [PubMed] [Google Scholar]
- 55. Tseng H. J., McEwan A. G., Paton J. C., Jennings M. P. (2002) Virulence of Streptococcus pneumoniae. PsaA mutants are hypersensitive to oxidative stress. Infect. Immun. 70, 1635–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brown J. S., Gilliland S. M., Ruiz-Albert J., Holden D. W. (2002) Characterization of pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect. Immun. 70, 4389–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sampson J. S., O'Connor S. P., Stinson A. R., Tharpe J. A., Russell H. (1994) Cloning and nucleotide sequence analysis of psaA, the Streptococcus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect. Immun. 62, 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shafeeq S., Yesilkaya H., Kloosterman T. G., Narayanan G., Wandel M., Andrew P. W., Kuipers O. P., Morrissey J. A. (2011) The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270 [DOI] [PubMed] [Google Scholar]
- 59. Fields P. I., Swanson R. V., Haidaris C. G., Heffron F. (1986) Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoiseth S. K., Stocker B. A. (1981) Aromatic-dependent Salmonella typhimurium is nonvirulent and effective as live vaccines. Nature 291, 238–239 [DOI] [PubMed] [Google Scholar]
- 61. McFarland W. C., Stocker B. A. (1987) Effect of different purine auxotrophic mutations on mouse virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3, 129–141 [DOI] [PubMed] [Google Scholar]
- 62. O'Callaghan D., Maskell D., Liew F. Y., Easmon C. S., Dougan G. (1988) Characterization of aromatic and purine-dependent Salmonella typhimurium. Attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect. Immun. 56, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chien M., Morozova I., Shi S., Sheng H., Chen J., Gomez S. M., Asamani G., Hill K., Nuara J., Feder M., Rineer J., Greenberg J. J., Steshenko V., Park S. H., Zhao B., Teplitskaya E., Edwards J. R., Pampou S., Georghiou A., Chou I. C., Iannuccilli W., Ulz M. E., Kim D. H., Geringer-Sameth A., Goldsberry C., Morozov P., Fischer S. G., Segal G., Qu X., Rzhetsky A., Zhang P., Cayanis E., De Jong P. J., Ju J., Kalachikov S., Shuman H. A., Russo J. J. (2004) The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968 [DOI] [PubMed] [Google Scholar]
- 64. Pine L., George J. R., Reeves M. W., Harrell W. K. (1979) Development of a chemically defined liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 9, 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gutierrez M. L., Garrabou X., Agosta E., Servi S., Parella T., Joglar J., Clapés P. (2008) Serine hydroxymethyltransferase from Streptococcus thermophilus and l-threonine aldolase from Escherichia coli as stereocomplementary biocatalysts for the synthesis of β-hydroxy-α,ω-diamino acid derivatives. Chemistry 14, 4647–4656 [DOI] [PubMed] [Google Scholar]
- 66. Vidal L., Calveras J., Clapés P., Ferrer P., Caminal G. (2005) Recombinant production of serine hydroxymethyltransferase from Streptococcus thermophilus and its preliminary evaluation as a biocatalyst. Appl. Microbiol. Biotechnol. 68, 489–497 [DOI] [PubMed] [Google Scholar]
- 67. Smith A. W., Roche H., Trombe M. C., Briles D. E., Håkansson A. (2002) Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Mol. Microbiol. 44, 431–448 [DOI] [PubMed] [Google Scholar]
- 68. Leibig M., Liebeke M., Mader D., Lalk M., Peschel A., Götz F. (2011) Pyruvate formate-lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J. Bacteriol. 193, 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Poolman B. (1993) Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12, 125–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.