Background: Nei is a DNA glycosylase of the base excision repair pathway.
Results: We present two crystal structures of an Nei bound to thymine glycol or 5-hydroxyuracil.
Conclusion: Mutational analysis of active site residues suggests that lesion recognition happens before the damaged base is everted into the active site.
Significance: These are the first structures of any Nei in complex with a damaged base.
Keywords: Base Excision Repair, Crystal Structure, DNA Damage, DNA Repair, Protein Structure, 5-Hydroxyuracil, DNA Glycosylase, Substrate Specificity, Thymine Glycol
Abstract
Thymine glycol (Tg) and 5-hydroxyuracil (5-OHU) are common oxidized products of pyrimidines, which are recognized and cleaved by two DNA glycosylases of the base excision repair pathway, endonuclease III (Nth) and endonuclease VIII (Nei). Although there are several structures of Nei enzymes unliganded or bound to an abasic (apurinic or apyrimidinic) site, until now there was no structure of an Nei bound to a DNA lesion. Mimivirus Nei1 (MvNei1) is an ortholog of human NEIL1, which was previously crystallized bound to DNA containing an apurinic site (Imamura, K., Wallace, S. S., and Doublié, S. (2009) J. Biol. Chem. 284, 26174–26183). Here, we present two crystal structures of MvNei1 bound to two oxidized pyrimidines, Tg and 5-OHU. Both lesions are flipped out from the DNA helix. Tg is in the anti conformation, whereas 5-OHU adopts both anti and syn conformations in the glycosylase active site. Only two protein side chains (Glu-6 and Tyr-253) are within hydrogen-bonding contact with either damaged base, and mutating these residues did not markedly affect the glycosylase activity. This finding suggests that lesion recognition by Nei occurs before the damaged base flips into the glycosylase active site.
Introduction
Various types of oxidative DNA damage, including abasic sites, base lesions, and strand breaks, are caused by reactive oxygen species generated by endogenous metabolism from a cell and from exposure to ionizing radiation. The resulting DNA damage can interfere with both the efficiency and fidelity of DNA replication and transcription and therefore can lead to mutagenesis, carcinogenesis, and aging. To prevent the toxic and mutagenic effects of oxidative DNA damage, a variety of DNA repair pathways has evolved in all species. One of these pathways is base excision repair, a process initiated by DNA glycosylases, which recognize and remove oxidized DNA damage (1–5). These DNA glycosylases are divided into two families, the HhH superfamily and Fpg3/Nei family, based on the presence of particular structural motifs. The enzymes in both families are also classified based on their substrate specificity. Oxidized purines like 8-oxoguanine (8-oxoG) are substrates for Fpg (Fpg/Nei family) and Ogg (HhH superfamily). Oxidized pyrimidines, however, are recognized and removed by endonuclease VIII (Nei; Fpg/Nei family) and endonuclease III (HhH superfamily).
Members of the Fpg/Nei family share a two-domain architecture. The N-terminal domain consists of a two-layered β-sandwich with two α-helices, and the C-terminal domain contains four α-helices, two of which are involved in a conserved helix-two-turn-helix motif and two β-strands that make up the zinc finger motif (6–10). Escherichia coli formamidopyrimidine DNA glycosylase (Fpg) was the first identified member of this family and was characterized by its ability to recognize and remove methyl-formamidopyrimidines (11). It was later concluded that the principal biological substrate for Fpg is 8-oxoG (12, 13). Recent studies have shown that FapyG is also an important biological substrate (14). Structural studies of Fpg complexed with DNA containing an oxidized base have provided insight into the general features of lesion presentation in the active site of Fpg (15, 16). The everted 8-oxoG is recognized and stabilized by the αF-β9/10 loop, also called the 8-oxoG capping loop (17). Recently, the hypothesis that the recognition of 8-oxoG might occur before it flips out of the DNA helix was put forward (17).
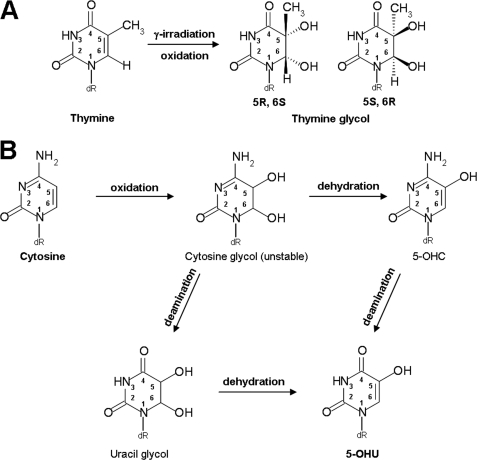
Mimivirus Nei1 (MvNei1) is an ortholog of human NEIL1, and both enzymes belong to the Nei family of DNA glycosylases (18–21). Nei enzymes are bifunctional glycosylases, with glycosylase and lyase activities. Nei glycosylases mainly recognize and cleave oxidized pyrimidines and have also been shown more recently to cleave the further oxidation products of 8-oxoG, spiroiminodihydantoin, and guanidinohydantoin (22–25). Several groups have shown that 8-oxoG itself is a poor substrate (18, 24, 26–28), which is in contradiction with work that showed that NEIL1 can remove the oxidized guanine (20, 29). In at least one of these cases, the discrepancy may be attributed to a difference in the position of the lesion within the DNA oligonucleotide (29). Among the commercially available substrates tested, MvNei1 recognizes and removes thymine glycol (5,6-dihydro-5,6-dihydroxythymine (Tg)) and 5-hydroxyuracil (5-OHU) and displays negligible activity toward 8-oxoG (26). Tg is the most common oxidation product of thymine (30, 31), and 5-OHU is generated by the oxidative deamination of cytosine (32, 33) (Fig. 1).
FIGURE 1.
Schemes of the oxidation pathways leading to Tg (45) and 5-OHU (62).
We reported previously the crystal structures of MvNei1 unliganded and in complex with a DNA oligomer containing tetrahydrofuran (THF), a stable AP site analog (34). The structure of MvNei1 was found to be very similar to that of hNEIL1, EcoNei, and other Fpg/Nei enzymes. The MvNei1·THF complex illustrated how this enzyme binds DNA, although the question of how it recognizes a lesion was left unanswered. Here, we present the structures of MvNei1 in complex with DNA oligomers containing either Tg or 5-OHU, which are the first crystal structures of any Nei with a damaged base. The structures of MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G show that the damaged base makes few interactions with MvNei1. The paucity of contacts to the damaged base suggests that lesion recognition occurs before base flipping.
EXPERIMENTAL PROCEDURES
Protein Preparation
The MvNei1 variants were created using the primers 5′-gatatacatatgccaCAAggtcctgaagtagc-3′ for E3Q, 5′-cctgaaggtcctGCAgttgctctcac-3′ for E6A, and 5′-gaatttgattttcttgttTTTcgcaaaaagaaagacccc-3′ for Y253F using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Expression and purification of MvNei1 variants were performed using a published protocol (26). For the kinetics experiments, all MvNei1 variants were stored at −20 °C in a storage buffer (20 mm Hepes-KOH, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, and 50% (v/v) glycerol).
DNA Preparation and Complex Formation
For crystallization, a 13-mer oligodeoxynucleotide 5′-CGTCCAXGTCTAC-3′ (X = Tg or 5-OHU) was used. The oligodeoxynucleotides (oligo) containing TG and 5-OHU were purchased from Midland Certified Reagent Co. (Midland, TX) and gel-purified. The commercially available thymine glycol is predominantly the 5R,6S stereoisomer (87%) (Glen Research, Sterling, VA) (35, 36). Annealing of the damage-containing oligodeoxynucleotide with the complementary strand and complex formation with the enzyme were performed as reported previously (34). For the kinetics experiments, a 35-mer 5′-TGTCAATAGCAAGXGGAGAAGTCAATCGTGAGTCT-3′ (X = Tg or 5-OHU) oligonucleotide and its complementary strand were used. An oligonucleotide with uracil at the X position was used to generate a site of base loss (AP site). The modified strand was labeled at the 5′-end using [γ-32P]ATP with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and annealed to the complementary strand in a 1:1 ratio. To minimize the error because of the uncertainty in the DNA concentration after ethanol precipitation, the labeled oligonucleotide for the glycosylase assay was diluted 10-fold with the corresponding unlabeled damaged oligonucleotide (26). For the AP lyase assay, the labeled oligodeoxynucleotide was used undiluted. The double-stranded oligonucleotide containing a uracil was treated with uracil DNA glycosylase (New England Biolabs, Ipswich, MA) for 1 h at 37 °C to generate an AP site.
Kinetics Experiments
Kinetics experiments were performed under single-turnover conditions ([Enzyme] > [DNA]), using the 35-base pair duplex to evaluate the glycosylase and lyase activity of MvNei1 variants. In each case, the total reaction volume was 100 μl with a final duplex DNA concentration of 2.5 nm in the reaction buffer (20 mm Hepes-KOH, pH 7.8, 75 mm KCl, 1 mm EDTA, 0.1 mg/ml BSA, 1 mm DTT, 5% (v/v) glycerol) and 300 nm MvNei1 WT, 600 nm Y253F, or 900 nm E6A at 37 °C. For the Tg:A reaction, aliquots were removed from the reaction at various time points, 1/3, 2/3, 1, 2, 5, 10, 30, and 60 min, and quenched with 2 μl of 1 n NaOH following a heat treatment at 90 °C for 2 min. For the 5-OHU:G reaction, aliquots were removed from the reaction at various time points, 1/10, 2/10, 1/3, 2/3, 1, 2, 5, 10, 20, and 40 min, and quenched with 2 μl of 1 n NaOH following heat treatment at 90 °C for 2 min. Because of the fast lyase reaction, kinetics experiments were performed using an RQF-3 rapid quench flow instrument (KinTek Corp., Austin, TX). The time points for AP:A were 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5, and 20 s. The reaction samples were quenched using a 1% (w/v) SDS solution, and the mixture was incubated on ice for 1 h. After the incubation, the supernatant was taken for quantification. The kinetic parameters for both activity experiments (glycosylase and lyase) were obtained from three independent experiments and fitted using Prism (GraphPad Software, La Jolla, CA).
Crystallization, X-ray Data Collection, and Structure Determination
Crystallization conditions for MvNei1E3Q in complex with Tg or 5-OHU are similar to those reported for the MvNei1·tetrahydrofuran (THF) complex (34). However, sodium formate (0.15 m) was used as a salt instead of 0.15 m magnesium nitrate to crystallize MvNei1E3Q in complex with 5-OHU. The pH of the crystallization solution was measured to be 7.0. The base opposite the lesion is a C for Tg and a G for 5-OHU. Although a Tg:A base pair is more biologically relevant, crystals were obtained readily with C opposite Tg, although attempts to crystallize a complex with an oligonucleotide containing Tg opposite A yielded only small needles. Both crystallographic data sets of the MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes were collected at 100 K at λ = 1.0332 Å at beamline 23-ID-B of the Advanced Photon Source, and additional data for MvNei1E3Q·Tg:C were collected at copper radiation on a Mar345 detector (MarResearch, Hamburg). Both data sets were processed, merged, and scaled with Denzo/Scalepack (37).
Both structures were determined by difference Fourier methods using phases from the MvNei1·THF model devoid of all non-protein atoms (Protein Data Bank code 3A46 (34)). The refinement of the MvNei1E3Q·Tg:C complex was performed using CNS (38). The MvNei1E3Q·5-OHU:G complex was refined using CNS (38) followed by phenix.refine (39). The final models for both structures were found to exhibit good geometry, as determined using Procheck (40). The original FoMvNei1E3Q·Tg:C or MvNeilE3Q·5-OHU:G-FoMvNei1·THF isomorphous difference Fourier maps indicated that the putative lesion recognition loop (residues 217–245) in both MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes was not well ordered throughout this region. After building the damaged bases (Tg and 5-OHU) and further refinement, the electron density map corresponding to the loop was judged adequate for building main-chain atoms only. There are no residues in the disallowed region of the Ramachandran plot. Refinement statistics are shown in Table 1. All structure figures were prepared using PyMOL (41).
TABLE 1.
Data collection and refinement statistics
Rmerge = Σ |I − 〈I〉|/ΣI, where 〈I〉 is the average intensity from multiple observations of symmetry-related reflections. RFriedel = Σ|I + − I−|/Σ 〈I〉; Rwork and Rfree = Σ‖Fo|−|Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 10% of the reflections not used in refinement.
| Thymine glycol·DNA complex | 5-Hydroxyuracil·DNA complex | |
|---|---|---|
| Beamlines | APS 23-ID-B + home source | APS 23-ID-B |
| No. of crystals | 2 | 1 |
| Wavelength | 1.0332 + 1.5418 Å | 1.0332 Å |
| Space group | P21 | P21 |
| Molecules per asymmetric unit | 2 | 2 |
| Data collection statistics | ||
| Resolution | 50 to 2.0 Å (2.07 to 2.00 Å) | 50 to 2.1 Å (2.18 to 2.10 Å) |
| Unit cell dimensions | ||
| a, b, c | 39.7, 121.6 80.8 Å | 39.6, 121.5, 80.7 Å |
| β (°) | 95.5° | 95.5° |
| Unique reflections | 48,426 (3,775) | 41,763 (4,439) |
| Redundancy | 8.0 (2.8) | 2.8 (2.7) |
| Rmergea | 0.133 (0.370) | 0.080 (0.232) |
| RFriedela | 0.07 (0.43) | |
| Completenessa | 94.2% (73.3%) | 92.6% (98.9%) |
| Overall I/σ(I)a | 14.9 (2.1) | 14.6 (3.5) |
| Refinement statistics | ||
| Rwork | 20.0% | 21.3% |
| Rfree | 23.0% | 26.6% |
| Root mean square deviations | ||
| Bond length | 0.005 Å | 0.005 Å |
| Bond angles | 1.16° | 0.99° |
| B-factor | ||
| Protein (molecule A, B) | 17.9, 18.3 Å2 | 23.7, 23.1 Å2 |
| DNA (molecule A, B) | 27.1, 25.1 Å2 | 31.3, 30.8 Å2 |
| Water (all molecules) | 29.4 Å2 | 30.7 Å2 |
| Ramachandran plot | ||
| Most favored | 88.4% | 88.6% |
| Additional allowed | 11.4% | 11.4% |
| Generously allowed | 0.2% | 0.0% |
| Disallowed | 0.0% | 0.0% |
a Values for the highest resolution shell are shown in parentheses.
Protein Data Bank Accession Codes
Coordinates and structure factors for MvNei1E3Q bound to Tg and 5-OHU have been deposited in the Protein Data Bank with accession codes 3VK8 and 3VK7, respectively.
RESULTS
Structure Determination of MvNei1 in Complex with DNA Containing Tg:C or 5-OHU:G
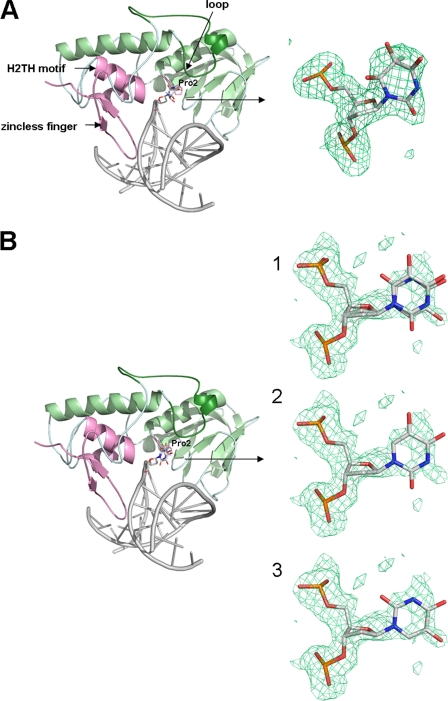
To date, there is no crystal structure of any Nei glycosylase in complex with a damaged base, which prompted our effort to crystallize MvNei1 with DNA containing a lesion. We tested several glycosylase-deficient variants, including P2G and E3Q. The residual glycosylase activity of the P2G and E3Q variants was negligible. The E3Q variant was selected for crystallization trials because it was previously documented that the lack of the catalytic proline could affect the position of the αF-β9/10 loop in Fpg (16, 42). The E3Q variant was crystallized with 5-OHU and Tg, which are commercially available and are good substrates for Nei enzymes, including MvNei1, hNEIL1, and EcoNei (Fig. 1) (24, 26, 43, 44). Although both are oxidized pyrimidines, 5-OHU is planar, and Tg is not (45). Crystals of MvNei1E3Q with Tg-containing DNA were obtained using crystallization conditions similar to those used for the complex of MvNei1 with DNA containing an abasic site analog (THF) (34). Diffraction data were collected to 2.0 Å resolution at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) and at our home x-ray source. Because of the high isomorphism between the MvNei1·THF and MvNei1E3Q·Tg:C complexes (overall cross-R of 0.18 on amplitudes), the initial phases were obtained by using the model of MvNei1 in complex with THF devoid of all non-protein atoms. The structure of MvNei1E3Q with Tg-containing DNA was refined to R-factor and Rfree values of 20.0 and 23.0%, respectively (Table 1 and Fig. 2A).
FIGURE 2.
Overall structures of MvNei1 E3Q in complex with Tg:C (A) and 5-OHU:G (B). The zinc-less finger, helix-two-turn-helix (H2TH), and catalytic proline are highlighted in pink. The DNA is shown in gray. The loop corresponding to αF-β9/10 loop in Fpg is shown in dark green. Close-up views of Tg (A) and 5-OHU (B) are shown on the right-hand side with overlaid simulated annealing omit map contoured at 2.5 σ (green mesh) at 1) both syn and anti conformations, 2) anti conformation, and 3) syn conformation of 5-OHU.
A single crystal of MvNei1E3Q in complex with 5-OHU:G-containing DNA was used to collect a 2.1 Å resolution data set at the Advanced Photon Source synchrotron. The MvNei1E3Q·5-OHU:G data set was also isomorphous with the MvNei1·THF complex (cross-R on amplitudes was 0.28). The initial phases for MvNei1E3Q·5-OHU:G were also obtained using the MvNei1·THF model devoid of all non-protein atoms. The residual Fo − Fc map after several cycles of refinement revealed that two conformations of 5-OHU were bound in the active site with ∼70% occupancy for the syn conformation and ∼30% occupancy for the anti conformation in molecule A. The structure of MvNei1E3Q·5-OHU:G was refined with CNS (37) followed by phenix.refine (39) to R-factor and Rfree values of 21.3 and 26.6%, respectively (Table 1 and Fig. 2B). Although the crystal asymmetric unit comprises two MvNei1E3Q·DNA complexes, the description below will focus mainly on one of the complexes, molecule A, because both molecules have comparable average B-factors.
Overall Structure of MvNei1E3Q in Complex with Tg:C or 5-OHU:G
The overall models of the MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU·G complexes are nearly identical to that of MvNei1·THF (root mean square deviation 0.24 Å (calculated on 288 Cα) and 0.34 Å (calculated on 287 Cα), respectively). The differences between these structures are localized in the active site and are due to slight changes in the position of three residues in the lesion binding pocket (Pro-2, Glu/Gln-3, and Glu-6), and changes in the conformation of the putative lesion recognition loop (residues 217–245 in MvNei1).
In the structures of the MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes, the lesion is flipped out of the DNA double helix and located in the lesion-binding site (Fig. 2). The density observed for Tg indicated that Tg is in the anti conformation in the MvNei1E3Q·Tg:C complex. However, the electron density observed for 5-OHU in the MvNeiE3Q·5-OHU:G complex suggested that both the syn and anti conformations are possible, with the respective occupancies refined to ∼0.70 and ∼0.30. The catalytic amino acid residues Pro-2 and Gln-3 (Glu-3 in the wild-type enzyme) in the MvNei1E3Q·Tg:C and /5-OHU:G complexes adopt positions that are nearly identical to those in the MvNei1·THF:C structure, although there are slight changes in the side chain orientations. The side chain of Gln-3 rotates away from O4′ on the deoxyribose (Fig. 3) The Pro-2 ring also adopts slightly different rotamers. The distance between the amine of the proline and C1′ of the deoxyribose is 4.0 Å in the complex with Tg, 4.4 Å in the complex with 5-OHU, and 3.7 Å in the THF complex. The carboxylate moiety of Glu-6 in MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G is rotated toward the lesion (Tg or 5-OHU) (Fig. 3).
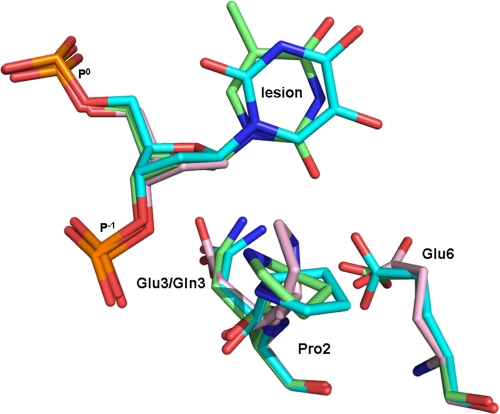
FIGURE 3.
Superposition MvNei1 E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes onto the MvNei1·THF complex. The MvNei1E3Q·Tg:C complex is shown in green, MvNei1E3Q·5-OHU:G syn conformation complex in cyan, and MvNei1·THF (Protein Data Bank code 3A46 (34)) in pink.
Interactions of MvNei1 with Tg or 5-OHU
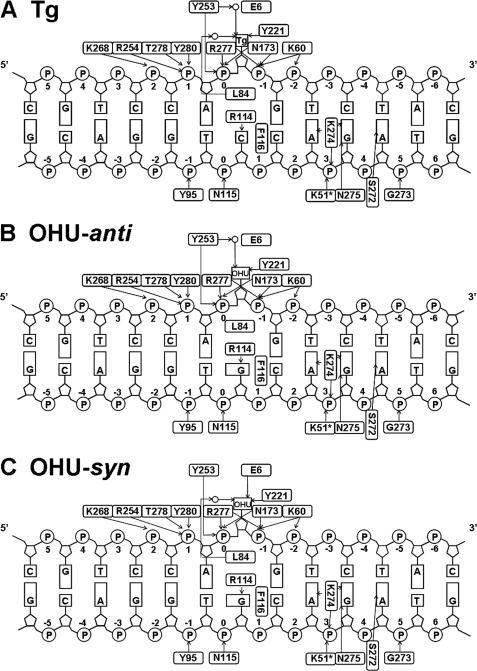
The DNA binds to MvNei1 in the MvNeiE3Q·Tg:C or MvNei1E3Q·5-OHU:G complexes in a manner that is very similar to that described for the MvNei1·THF complex (34), with the exception of the interactions involving the lesions (Tg or 5-OHU) and the estranged base guanine. The 13-mer duplex is bound, as expected, in the DNA binding cleft between the two domains and lies perpendicularly to the long axis of MvNei1 (Fig. 2). Arg-277, which is well conserved among the Fpg/Nei family and plays an important role in the glycosylase but not the lyase activity, contacts the oxygen of two phosphates (P0 and P−1) adjacent to the lesion (Fig. 4) (46).
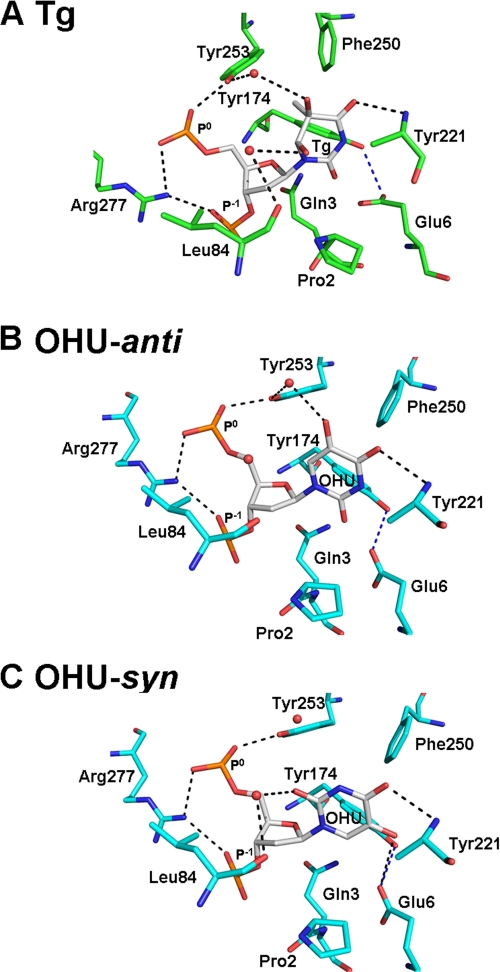
FIGURE 4.
Close-up view of the interactions between MvNei1 residues and lesion. A, Tg; B, anti-5-OHU; C, syn-5-OHU. MvNei1E3Q·Tg:C is shown in green, MvNei1E3Q/5-OHU:G in cyan, and damaged base in gray. Hydrogen bonds are represented by black dashed lines.
In the MvNei1E3Q·Tg:C complex, there are very few hydrogen bonding interactions involving the Tg base. A direct hydrogen bond interaction between Tg and MvNei1 was observed, between the main-chain amide of Tyr-221 and O4 of Tg (Figs. 4A and 5A). (The electron density for the side chain of Tyr-221 was unclear and therefore was not built.) Two additional water-mediated hydrogen bond interactions were also observed, connecting the hydroxyl group of Tyr-253 and O5 of Tg and the main-chain carbonyl group of Leu-84 with O6 of Tg. Three aromatic residues, Tyr-174, Phe-250, and Tyr-253, surround Tg. Tyr-174 and Tyr-253 are within van der Waals distance and may therefore help stabilize the damaged base (Fig. 4A).
FIGURE 5.
Diagram of protein-DNA interactions. Nucleotides are numbered beginning from the damaged base (Tg or 5-OHU) 0, with positive numbers toward the 5′-end. Hydrogen bonds are represented with black arrows pointing toward the acceptors. Open circles represent water molecules. A, Tg; B, 5-OHU-anti; C, 5-OHU-syn.
In the MvNei1E3Q·5-OHU:G complex, 5-OHU adopts two conformations, anti and syn. In both conformations, a hydrogen bond between the main-chain amide of Tyr-221 and the O4 of 5-OHU is maintained, reminiscent of the MvNei1E3Q·Tg:C structure (Figs. 4 and 5). In the anti conformation, a water-mediated hydrogen bond between the hydroxyl group of Tyr-253 and O5 of 5-OHU was also observed in molecule A, as in the MvNei1E3Q·Tg:C structure (Fig. 4, A and B). In the syn conformation, an additional hydrogen bond was observed between the carboxyl group of Glu-6 and the hydroxyl group at position 5 in 5-OHU (Figs. 4C and 5C). A water-mediated hydrogen bond from the hydroxyl group of Tyr-253, which was seen in the MvNei1E3Q·Tg:C complex, was not observed in the syn conformation of the MvNei1E3Q·5-OHU complex. In contrast, a hydrogen bond between the main-chain carbonyl group of Leu-84 and the lesion (O6 of Tg and O2 of 5-OHU) was maintained (Fig. 4C). As seen with Tg, three aromatic residues, Tyr-174, Phe-250, and Tyr-253, are in the vicinity of 5-OHU, and the two tyrosines are within van der Waals distance of the damaged base (Fig. 4, B and C).
Interactions of MvNei1 with the Estranged Base
In both the MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes, Leu-84, Arg-114, and Phe-116 were observed filling the void created by the extrusion of the damaged base, as described before (34). Leu-84 takes the place of the excised damaged base and participates in a hydrogen bond interaction with the damaged base (Tg and 5-OHU-syn) via its main-chain carbonyl (Figs. 4 and 5). Phe-116 is wedged between the estranged base (cytosine or guanine) and its 5′ neighbor. In the MvNei1E3Q·Tg:C complex, Arg-114 interacts with the orphaned cytosine via two hydrogen bonds, involving O2 and N3 of the estranged C and NϵH and NηH of Arg-114 (supplemental Fig. S1A), as described previously (34). In the MvNei1E3Q·5-OHU:G complex, Arg-114 interacts with the orphaned guanine also via two hydrogen bonds (O6 and N7 of estranged G and NϵH and NηH of Arg-114) (supplemental Fig. S1B), as reported in the Bacillus stearothermophilus (Bst) Fpg complex with DNA containing a reduced abasic site (47).
Activity Assays of MvNei1WT, E6A, and Y253F Variants
Very few hydrogen bond interactions were observed between the lesion and MvNei1 in the complexes of MvNei1E3Q with either Tg:C or 5-OHU:G (Fig. 4). Tyr-253 interacts with both Tg and anti-5-OHU via a water molecule (Figs. 4 and 5). We hypothesized that Tyr-253 might thus contribute to either lesion recognition or lesion stabilization after the lesion has flipped out into the glycosylase active site. The tyrosine also contacts phosphate P0 and could therefore play a part in the lyase reaction. In addition, Glu-6 is located within hydrogen bonding distance of Tg and 5-OHU in the anti conformation, although formation of such an H-bond would require the glutamate to be protonated (Fig. 4, A and B). However, Glu-6 interacts with the hydroxyl 5-OH of syn-5-OHU via a weak H-bond (∼3.5 Å). To evaluate the potential role of the only protein side chains seen contacting the lesions in either the glycosylase or lyase reaction, activity assays of MvNei1 with DNA containing Tg:A, 5-OHU:G, or AP:A were performed with enzyme variants mutated at Glu-6 and Tyr-253. The MvNei1 Y253F variant was designed because it lacks the ability to hydrogen bond with a water molecule while maintaining the aromatic ring. The MvNei1 E6A variant forfeits any possibility of a hydrogen bond from its side-chain atoms with the lesion.
An initial glycosylase activity assay showed that the Y253F and E6A variants exhibited a level of activity similar to that of MvNei1 WT (data not shown). This result indicated that Tyr-253 and Glu-6 were not key residues in the recognition of the lesion. However, further kinetics experiments of MvNei1 WT and the two variants (Y253F and E6A) with Tg:A and 5-OHU:G were performed to analyze differences in reaction velocity. When performing pre-steady state kinetics on DNA glycosylases, either a single- or double-exponential model can be used to fit the data depending on the substrates tested. The Tg:A data were fit to a single-exponential model, as observed for Candida albicans Fpg with a double-stranded Tg substrate (48). We note that another group reported that kinetics data obtained with human NEIL1 and Tg-containing DNA were fit to a double-exponential model, presumably because of the interconversion of the cis- and trans-Tg epimers (49). Mutating Tyr-253 and Glu-6 in MvNei1 decreased the velocity of the glycosylase reaction by 6.7- and 3.6-fold, respectively (Table 2). No significant difference in the velocity of the glycosylase reaction was observed with 5-OHU:G (Table 2). Furthermore, the glycosylase reaction using 5-OHU fits a double-exponential model, instead of a single-exponential model as observed with Tg:A. It presumably corresponds to the presence of both syn and anti conformations observed for 5-OHU in the MvNei1E3Q·5-OHU:G complex.
TABLE 2.
Kinetic rates (kobs) per min of MvNei1 with double strand substrates (Tg:A, 5-OHU:G, or AP:A) determined under single-turnover conditions
The kinetic parameters were obtained from three independent experiments. Data obtained with 5-OHU:G and AP:A were fit with a double-exponential model, which yields two rate constants (k1 and k2). Standard deviations are shown. NA means not applicable.
| k1 | k2 | |
|---|---|---|
| WT | ||
| Tg:A | 0.541 ± 0.028 | NA |
| 5-OHU:G | 5.75 ± 2.36 | 0.107 ± 0.0316 |
| AP:A | 898.2 ± 179.2 | 27.3 ± 4.03 |
| E6A | ||
| Tg:A | 0.144 ± 0.006 | NA |
| 5-OHU:G | 6.76 ± 1.07 | 0.081 ± 0.0168 |
| AP:A | 523.1 ± 86.5 | 12.8 ± 2.98 |
| Y253F | ||
| Tg:A | 0.081 ± 0.004 | NA |
| 5-OHU:G | 6.98 ± 1.39 | 0.104 ± 0.0157 |
| AP:A | 1101.6 ± 371.6 | 62.7 ± 8.06 |
To check the effect of mutating Tyr-253 and Glu-6 on the lyase reaction, kinetics experiments with MvNei1 WT and the two variants (Y253F and E6A) with AP:A were performed. The result of the lyase reaction, which fit the double-exponential model presumably because a naturally occurring AP site can adopt open and closed ring conformations, indicated no drastic difference arising from mutating either of these residues. The E6A variant exhibited a kobs that was decreased by almost 2-fold, although the kobs for Y253F was slightly higher than that of wild-type MvNei1.
DISCUSSION
One of the pressing questions regarding Nei glycosylases is how these enzymes locate and recognize a damaged base (50, 51). Unlike Fpg, an enzyme of the same family with a narrow substrate specificity, Nei enzymes efficiently recognize a wide variety of oxidized lesions (12, 13, 26, 52–55). We therefore set out to crystallize MvNei1E3Q in complex with two of its best commercially available substrates, Tg and 5-OHU (26), in an attempt to answer this question. Our work describes the first crystal structure of any Nei in complex with a damaged base. Several structures of Fpg in complex with a lesion (8-oxoG, ring-opened formamidopyrimidine, and 5-hydroxy-5-methylhydantoin) have been reported (15, 16, 42, 56), and these structures were compared with the MvNei1 complexes to gauge how lesions are recognized. Among the Fpg structures, the BstFpgE3Q·8oxoG:C complex was chosen as a reference because the E3Q variant was employed in both BstFpg and MvNei1 structures (15).
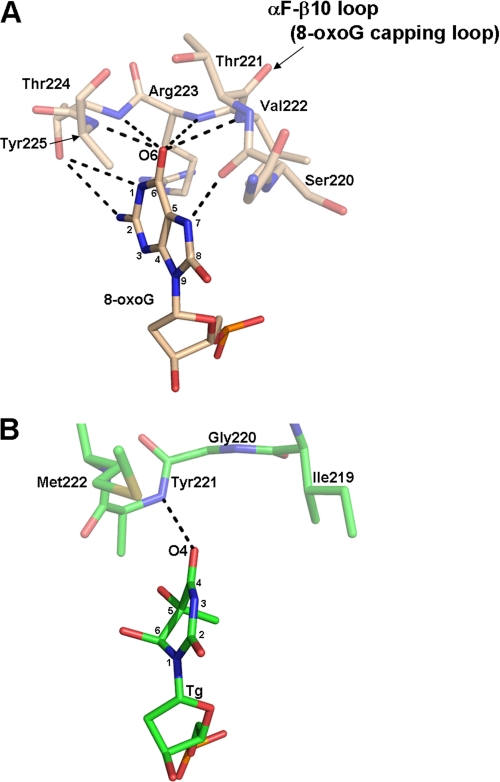
The structures of MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes revealed that the lesion (Tg or 5-OHU) is extruded from the DNA helix and located in the active site of the enzyme. In these structures, the electron density for the tip (residues 219–223) of the putative lesion recognition loop is disordered, and as a result only the main-chain atoms of these residues were built. However, the αF-β10 loop in the BstFpgE3Q·8oxoG:C complex is well ordered. Few hydrogen bond interactions between MvNei1 and the damaged base are observed in the MvNei1E3Q·Tg:C and MvNei1E3Q·5-OHU:G complexes. In contrast, 8-oxoG is stabilized by several strong hydrogen bond interactions in the BstFpgE3Q·8oxoG:C complex (15). In particular, the αF-β10 loop wraps around 8-oxoG providing several important main-chain interactions to O6 and the protonated N7. Verdine and co-workers (17) showed that a BstFpg variant in which the αF-β10 loop was deleted showed no glycosylase activity toward 8-oxoG, underscoring the crucial role of this protein segment in the recognition of the oxidized guanine.
A superposition of MvNei1E3Q·Tg:C onto the BstFpgE3Q·8-oxoG:C complex reveals that O4 and N3 of Tg are located approximately at the same position as O6 and N7 in 8-oxoG. O4 of Tg participates in one H-bond interaction with the main-chain amide of Tyr-221. N3, however, does not partake in any H-bond interaction with the putative lesion recognition loop. The loop is shorter than that of Fpg and as a result it cannot wrap around the lesion (Fig. 6 and supplemental Fig. 2). This finding suggests that the MvNei1 loop does not play a key role in either the recognition of the damaged base nor its stabilization. Moreover, the structure of the MvNei1E3Q·5-OHU:G complex revealed that 5-OHU can adopt both anti and syn conformations in the active site, suggesting that the lesion in the active site of MvNei1 is not well anchored, in contrast to 8-oxoG in the BstFpgE3Q·8-oxoG:C complex. The putative lesion recognition loop in MvNei1 is shorter than the αF-β9/10 loop in Fpg as mentioned before, and this segment in hNEIL1 is even shorter than in MvNei1 (supplemental Fig. S2). Nei enzymes prefer oxidized pyrimidines and the further oxidation products of 8-oxoG, such as guanidinohydantoin and spiroiminodihydantoin, although several groups have shown that 8-oxoG itself is removed very inefficiently (18, 24, 26–28). Taken together, these observations suggest that the long αF-β9/10 loop in Fpg has evolved to stabilize and cap its target lesion, 8-oxoG.
FIGURE 6.
Comparison of the hydrogen bond interactions between BstFpg and 8-oxoG versus MvNei1E3Q and Tg. There are multiple H-bond interactions (shown as black dashed lines) between the αF-β10 loop (8-oxoG capping loop) of BstFpg to 8-oxoG. In contrast, there is only one direct H-bond from the main-chain amide group of Tyr-221 to O4 of Tg in the MvNei1E3Q·Tg:C complex. A, BstFpg·8-oxoG:C complex (Protein Data Bank code 1R2Y (15)). B, MvNei1E3Q·Tg:C complex.
Previous work on E. coli Nei had predicted that the binding pocket for the everted base would be formed by three aromatic residues, namely Phe-228, Phe-230 and Tyr-170 (9). The authors further predicted that the plane of the tyrosine would be perpendicular to that of Tg. This is exactly what we have observed with the corresponding residues (Phe-250, Phe-253, and Tyr-174) in the everted lesion binding pocket of MvNei1 (Fig. 4A). Hydrogen bonding interactions were also predicted between Tg and Pro-2, Glu-6, and Arg-213. Pro-2 and Glu-6 are in close proximity of the lesion in the MvNei1 structure. Arg-213, which was predicted to contact O4 and O5, is not conserved, and we do not observe any residue capable of making these H-bond interactions. Another group also used molecular dynamics calculations to predict which hNEIL1 residues might interact with Tg, i.e. Pro-2, Glu-3, Glu-6, Tyr-177, and Tyr-263 (57). The corresponding residues (Pro-2, Glu-3, Glu-6, Tyr-174, and Tyr-253) are indeed all part of the MvNei1 lesion binding pocket. At the time Jia et al. (57) published their work, mutational data for Glu-6 was not available, and this residue was predicted to be important for glycosylase activity. However, we found in this study that mutating this glutamate to alanine does not adversely affect the glycosylase activity.
Mutating the other residue whose side chain is in close contact to Tg (Tyr-253) does not abolish the glycosylase activity either. In addition, mutating either residue to alanine has little effect on the AP lyase activity. The decreased velocity of the Tyr-253 variant for the Tg:A substrate suggests that Tyr-253 might participate in stabilizing the lesion via its water-mediated hydrogen bond interaction. The decreased velocity of the E6A variant for Tg:A indicates that Glu-6 affects the stability of the lesion. However, H-bond interaction between Glu-6 and O2 of Tg would require the glutamate to be protonated. The pKa of glutamate is 4.5, and the pH of the crystallization solution is 7.0, meaning that Glu-6 should in theory carry a negative charge. There are, however, a number of variables that can alter the pKa of a carboxylate group, including whether it is buried and how many hydrogen bonds with which it participates (58). The exact nature of the interaction between Glu-6 and O2 of Tg remains to be elucidated. No significant change of the kinetic rate for 5-OHU:G was observed after mutating either Tyr-253 or Glu-6. This result suggests that 5-OHU is not significantly stabilized by these protein side chains. This is consistent with the observation that two conformations of 5-OHU (anti and syn) coexist in the MvNei1E3Q·5-OHU:G complex.
Our mutational studies suggest that the two protein residues whose side chains are in close contact to the damaged base are not involved in lesion recognition. The question then of how Nei enzymes recognize their target substrates still remains. In an attempt to answer this question, an unmodified thymine was modeled into the active site of MvNei1 (supplemental Fig. S3). A modeled thymine was substituted for Tg in the MvNei1E3Q·Tg:C complex structure (supplemental Fig. S3). Although two water-mediated H bonds involving the 5- and 6-hydroxyl groups of Tg would be lost, the hydrogen bond interaction between O4 of thymine and the main chain amide of Tyr-221 of MvNei1 would likely remain, as in the MvNei1E3Q·Tg:C and /5-OHU:G complexes (Fig. 4). The hydrophobic interactions with Tyr-253, Phe-250, and Tyr-174 would remain essentially unchanged. Although the superposition suggests that once a normal base finds it way into the active site it would likely be cleaved because no obvious discrimination between the damaged and nondamaged base is observed, cleavage of normal thymine by MvNei1 is not observed (data not shown). This leads to the hypothesis that the lesion might be recognized before the lesion flips out of the DNA helix. The same speculation was originally put forward by Qi et al. (17) for the recognition of 8-oxoG by BstFpg. The authors obtained crystal structures of BstFpg cross-linked to DNA containing G or 8-oxoG within the double helix and reported that the C2′-endo sugar pucker of guanine undergoes a pseudorotation to an alternative pucker (C4′-exo) when G is replaced by 8-oxoG. This localized reorganization of the DNA backbone was proposed to be a key element in the way Fpg discriminates between a target lesion and a regular base. A local conformational perturbation may also be at play here. The presence of (5R,6S)-Tg within double-stranded DNA is known to hinder proper stacking of the 5′ base, especially when that base is a purine (45, 59, 60). Furthermore, the nature of the base opposite the lesion was shown to modulate repair of Tg by human NEIL1 (49); Tg occupies the wobble position when mispaired with G (61) and is excised much more rapidly than Tg:A (49). Importantly the Tg:G mispair in DNA results from oxidative damage to 5-methylcytosine followed by deamination. Similarly the 5-OHU:G mispair arises after oxidative deamination of cytosine. Whether Nei enzymes sense local conformational perturbations in the DNA to recognize their target lesions will require further investigation.
We presented here the first crystal structures of an Nei enzyme in complex with a DNA duplex containing either Tg or 5-OHU. This study showed that Tg in the active site is in the anti conformation, whereas 5-OHU can adopt both anti and syn conformations. Unexpectedly, very few hydrogen bond interactions were observed between the enzyme and either lesion, and mutating two of the residues in contact with the lesion did not affect the glycosylase activity. This finding suggests that lesion recognition is likely performed before the lesion is flipped out from the DNA double helix.
Acknowledgments
We thank Dr. Pierre Aller and Karl Zahn for data collection of the MvNei1·Tg complex at beamline 23-ID-B at the Advanced Photon Source; Minmin Liu for helpful discussions regarding the kinetics experiments; Dr. Matthew Hogg for technical assistance with the rapid quench instrument; and Drs. Peter Orth, Terukazu Nogi, and Mark Rould for helpful crystallographic suggestions. The beamline GM/CA CAT has been funded in whole or in part by National Institutes of Health Grant Y1-CO-1020 from NCI and Grant Y1-GM-1104 from NIGMS. Use of the Advanced Photon Source was supported by United States Department of Energy, Basic Energy Sciences, Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant P01CA098993 from NCI.

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3VK8 and 3VK7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Fpg
- formamidopyrimidine DNA glycosylase
- Tg
- thymine glycol
- 5-OHU
- 5-hydroxyuracil
- hNEIL1
- human Nei-like enzyme 1
- 8-oxoG
- 8-oxoguanine
- Nei
- endonuclease VIII
- THF
- tetrahydrofuran
- AP
- apurinic or apyrimidinic.
REFERENCES
- 1. Wallace S. S. (1998) Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res. 150, S60–S79 [PubMed] [Google Scholar]
- 2. Wallace S. S. (2002) Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 33, 1–14 [DOI] [PubMed] [Google Scholar]
- 3. Wilson D. M., 3rd, Sofinowski T. M., McNeill D. R. (2003) Repair mechanisms for oxidative DNA damage. Front. Biosci. 8, d963–d981 [DOI] [PubMed] [Google Scholar]
- 4. Hegde M. L., Hazra T. K., Mitra S. (2008) Early steps in the DNA base excision/single strand interruption repair pathway in mammalian cells. Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. David S. S., O'Shea V. L., Kundu S. (2007) Base-excision repair of oxidative DNA damage. Nature 447, 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wallace S. S., Bandaru V., Kathe S. D., Bond J. P. (2003) The enigma of endonuclease VIII. DNA Repair 2, 441–453 [DOI] [PubMed] [Google Scholar]
- 7. Zharkov D. O., Shoham G., Grollman A. P. (2003) Structural characterization of the Fpg family of DNA glycosylases. DNA Repair 2, 839–862 [DOI] [PubMed] [Google Scholar]
- 8. Sugahara M., Mikawa T., Kumasaka T., Yamamoto M., Kato R., Fukuyama K., Inoue Y., Kuramitsu S. (2000) Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 19, 3857–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zharkov D. O., Golan G., Gilboa R., Fernandes A. S., Gerchman S. E., Kycia J. H., Rieger R. A., Grollman A. P., Shoham G. (2002) Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 21, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilboa R., Zharkov D. O., Golan G., Fernandes A. S., Gerchman S. E., Matz E., Kycia J. H., Grollman A. P., Shoham G. (2002) Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 277, 19811–19816 [DOI] [PubMed] [Google Scholar]
- 11. Chetsanga C. J., Lozon M., Makaroff C., Savage L. (1981) Purification and characterization of Escherichia coli formamidopyrimidine-DNA glycosylase that excises damaged 7-methylguanine from deoxyribonucleic acid. Biochemistry 20, 5201–5207 [DOI] [PubMed] [Google Scholar]
- 12. Chung M. H., Kasai H., Jones D. S., Inoue H., Ishikawa H., Ohtsuka E., Nishimura S. (1991) An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat. Res. 254, 1–12 [DOI] [PubMed] [Google Scholar]
- 13. Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S. (1991) 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U.S.A. 88, 4690–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu M., Bandaru V., Bond J. P., Jaruga P., Zhao X., Christov P. P., Burrows C. J., Rizzo C. J., Dizdaroglu M., Wallace S. S. (2010) The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 4925–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fromme J. C., Verdine G. L. (2003) DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 278, 51543–51548 [DOI] [PubMed] [Google Scholar]
- 16. Coste F., Ober M., Le Bihan Y. V., Izquierdo M. A., Hervouet N., Mueller H., Carell T., Castaing B. (2008) Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode. Chem. Biol. 15, 706–717 [DOI] [PubMed] [Google Scholar]
- 17. Qi Y., Spong M. C., Nam K., Banerjee A., Jiralerspong S., Karplus M., Verdine G. L. (2009) Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 462, 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., Mitra S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 3523–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bandaru V., Sunkara S., Wallace S. S., Bond J. P. (2002) A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair 1, 517–529 [DOI] [PubMed] [Google Scholar]
- 20. Morland I., Rolseth V., Luna L., Rognes T., Bjørås M., Seeberg E. (2002) Human DNA glycosylases of the bacterial Fpg/MutM superfamily. An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 30, 4926–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takao M., Kanno S., Kobayashi K., Zhang Q. M., Yonei S., van der Horst G. T., Yasui A. (2002) A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 277, 42205–42213 [DOI] [PubMed] [Google Scholar]
- 22. Hickerson R. P., Chepanoske C. L., Williams S. D., David S. S., Burrows C. J. (1999) Mechanism-based DNA protein cross-linking of MutY via oxidation of 8-oxoguanosine. J. Am. Chem. Soc. 121, 9901–9902 [Google Scholar]
- 23. Sugden K. D., Martin B. D. (2002) Guanine and 7,8-dihydro-8-oxo-guanine-specific oxidation in DNA by chromium (V). Environ. Health Perspect. 110, Suppl. 5, 725–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnamurthy N., Zhao X., Burrows C. J., David S. S. (2008) Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 47, 7137–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hazra T. K., Muller J. G., Manuel R. C., Burrows C. J., Lloyd R. S., Mitra S. (2001) Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 29, 1967–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bandaru V., Zhao X., Newton M. R., Burrows C. J., Wallace S. S. (2007) Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair 6, 1629–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hailer M. K., Slade P. G., Martin B. D., Rosenquist T. A., Sugden K. D. (2005) Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair 4, 41–50 [DOI] [PubMed] [Google Scholar]
- 28. Kropachev K. Y., Zharkov D. O., Grollman A. P. (2006) Catalytic mechanism of Escherichia coli endonuclease VIII. Roles of the intercalation loop and the zinc finger. Biochemistry 45, 12039–12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parsons J. L., Zharkov D. O., Dianov G. L. (2005) NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 33, 4849–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teoule R., Bonicel A., Bert C., Cadet J., Polverelli M. (1974) Identification of radioproducts resulting from the breakage of thymine moiety by γ irradiation of E. coli DNA in an aerated aqueous solution. Radiat. Res. 57, 46–58 [PubMed] [Google Scholar]
- 31. Frenkel K., Goldstein M. S., Teebor G. W. (1981) Identification of the cis-thymine glycol moiety in chemically oxidized and γ-irradiated deoxyribonucleic acid by high pressure liquid chromatography analysis. Biochemistry 20, 7566–7571 [DOI] [PubMed] [Google Scholar]
- 32. von Sonntag C., Schuchmann H. P. (1986) The radiolysis of pyrimidines in aqueous solutions: an updating review. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 49, 1–34 [DOI] [PubMed] [Google Scholar]
- 33. Wagner J. R., Hu C. C., Ames B. N. (1992) Endogenous oxidative damage of deoxycytidine in DNA. Proc. Natl. Acad. Sci. U.S.A. 89, 3380–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Imamura K., Wallace S. S., Doublié S. (2009) Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J. Biol. Chem. 284, 26174–26183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iwai S. (2001) Synthesis and thermodynamic studies of oligonucleotides containing the two isomers of thymine glycol. Chemistry 7, 4343–4351 [DOI] [PubMed] [Google Scholar]
- 36. Lustig M. J., Cadet J., Boorstein R. J., Teebor G. W. (1992) Synthesis of the diastereomers of thymidine glycol, determination of concentrations, and rates of interconversion of their cis-trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res. 20, 4839–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 39. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L. W., Jain S., Kapral G. J., Grosse Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C, Richardson J. S., Terwilliger T. C., Zwart P. H. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laskowski R., MacArthur M., Moss D., Thornton J. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 10.1107/S002188982009944 [DOI] [Google Scholar]
- 41. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC, New York [Google Scholar]
- 42. Coste F., Ober M., Carell T., Boiteux S., Zelwer C., Castaing B. (2004) Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J. Biol. Chem. 279, 44074–44083 [DOI] [PubMed] [Google Scholar]
- 43. Parsons J. L., Kavli B., Slupphaug G., Dianov G. L. (2007) NEIL1 is the major DNA glycosylase that processes 5-hydroxyuracil in the proximity of a DNA single strand break. Biochemistry 46, 4158–4163 [DOI] [PubMed] [Google Scholar]
- 44. Dou H., Mitra S., Hazra T. K. (2003) Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684 [DOI] [PubMed] [Google Scholar]
- 45. Aller P., Rould M. A., Hogg M., Wallace S. S., Doublié S. (2007) A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. U.S.A. 104, 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Doublié S., Bandaru V., Bond J. P., Wallace S. S. (2004) The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zinc-less finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fromme J. C., Verdine G. L. (2002) Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 9, 544–552 [DOI] [PubMed] [Google Scholar]
- 48. Kathe S. D., Barrantes-Reynolds R., Jaruga P., Newton M. R., Burrows C. J., Bandaru V., Dizdaroglu M., Bond J. P., Wallace S. S. (2009) Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair 8, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ocampo-Hafalla M. T., Altamirano A., Basu A. K., Chan M. K., Ocampo J. E., Cummings A., Jr., Boorstein R. J., Cunningham R. P., Teebor G. W. (2006) Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis-trans epimerization. DNA Repair 5, 444–454 [DOI] [PubMed] [Google Scholar]
- 50. Zharkov D. O., Grollman A. P. (2005) The DNA trackwalkers. Principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 577, 24–54 [DOI] [PubMed] [Google Scholar]
- 51. Perry J. J., Cotner-Gohara E., Ellenberger T., Tainer J. A. (2010) Structural dynamics in DNA damage signaling and repair. Curr. Opin. Struct. Biol. 20, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiang D., Hatahet Z., Melamede R. J., Kow Y. W., Wallace S. S. (1997) Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 272, 32230–32239 [DOI] [PubMed] [Google Scholar]
- 53. Melamede R. J., Hatahet Z., Kow Y. W., Ide H., Wallace S. S. (1994) Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry 33, 1255–1264 [DOI] [PubMed] [Google Scholar]
- 54. Purmal A. A., Lampman G. W., Bond J. P., Hatahet Z., Wallace S. S. (1998) Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J. Biol. Chem. 273, 10026–10035 [DOI] [PubMed] [Google Scholar]
- 55. Dizdaroglu M., Burgess S. M., Jaruga P., Hazra T. K., Rodriguez H., Lloyd R. S. (2001) Substrate specificity and excision kinetics of Escherichia coli endonuclease VIII (Nei) for modified bases in DNA damaged by free radicals. Biochemistry 40, 12150–12156 [DOI] [PubMed] [Google Scholar]
- 56. Le Bihan Y. V., Angeles Izquierdo M., Coste F., Aller P., Culard F., Gehrke T. H., Essalhi K., Carell T., Castaing B. (2011) 5-Hydroxy-5-methylhydantoin DNA lesion, a molecular trap for DNA glycosylases. Nucleic Acids Res. 39, 6277–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jia L., Shafirovich V., Geacintov N. E., Broyde S. (2007) Lesion specificity in the base excision repair enzyme hNeil1. Modeling and dynamics studies. Biochemistry 46, 5305–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harris T. K., Turner G. J. (2002) Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life 53, 85–98 [DOI] [PubMed] [Google Scholar]
- 59. Brown K. L., Roginskaya M., Zou Y., Altamirano A., Basu A. K., Stone M. P. (2010) Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5′-GTgG-3′ sequence. Destabilization of two base pairs at the lesion site. Nucleic Acids Res. 38, 428–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aller P., Duclos S., Wallace S. S., Doublié S. (2011) A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex. J. Mol. Biol. 412, 22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown K. L., Basu A. K., Stone M. P. (2009) The cis-(5R,6S)-thymine glycol lesion occupies the wobble position when mismatched with deoxyguanosine in DNA. Biochemistry 48, 9722–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thiviyanathan V., Somasunderam A., Volk D. E., Hazra T. K., Mitra S., Gorenstein D. G. (2008) Base-pairing properties of the oxidized cytosine derivative, 5-hydroxy uracil. Biochem. Biophys. Res. Commun. 366, 752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]