Abstract
Personalized genomic medicine and surgery (PGMS) represents a new approach to health care that customizes patients’ medical treatment according to their own genetic information. This new approach is the result of increased knowledge of the human genome and ways this information can be applied by physicians in the medical and surgical management of their patients. A patient’s genotype can yield important information concerning disease susceptibility and the effectiveness of medications, therefore guiding specific, targeted imaging and treatment therapies. This review summarizes major achievements of human genomic studies and applications of genomics in health care. Five years ago we developed a model for the development of PGMS in which genomic profile guides choice of therapy. In this article we discussed our progress, including an updating of the model, and a future vision of PGMS.
Introduction
The Genomic and Personalized Medicine Act of 2007 was intended to provide funding for the advancement of this new healthcare initiative [1] and by 2008, the president’s Council of Advisors on Science and Technology (PCAST), provided a comprehensive definition of personalized medicine stating that it:
…refers to the tailoring of medical treatment to the individual characteristics of each patient. It does not literally mean the creation of drugs or medical devices that are unique to a patient, but rather the ability to classify individuals into subpopulations that differ in their susceptibility to a particular disease or their response to a specific treatment. Preventive or therapeutic interventions can then be concentrated on those who will benefit, sparing expense and side effects for those who will not [2].
The United States Department of Health and Human Services (HHS) also has acknowledged the importance of this new method of health care through the issuance of their Personalized Health Care directive aimed at increasing the usefulness and application of genomic knowledge [3]. Ginsburg and Willard have described genomic medicine as being a core element for the creation of a personalized health care in which physicians use information from an individual’s genome and its components to influence the decision-making process regarding patient care [4], thus, minimizing side effects and/or tailoring a treatment for a more successful outcome. Genomic information also can indicate the potential risk for certain diseases before the patient is symptomatic, prompting a proactive strategy of observation and screening.
Personalized genomic medicine will further classify many complex diseases and lead to the discovery of more specific treatments based on the genomic profiling, eliminating some traditional treatments that could prove ineffective or have undesirable side effects in certain patient populations. For example, 20–75% of patients could fail to respond to many commonly used drugs [5]. Just as the effectiveness of a particular medication is often based on characteristics specific to an individual, such as the rate at which the medication is metabolized or the causal factors of the disease, it is clear that differences in the genomes of individuals would likewise affect the effectiveness of medication. Based on the analysis of an individual genome, medication selection tailored to the patient’s genomic indications would improve clinical outcomes. The goal of personalized genomic medicine is to ensure that patients receive a personalized diagnosis and treatment, and to accomplish it at an efficient, cost-effective manner.
This review summarizes major achievements of human genomic studies and applications of genomics in health care. This new health care model is of critical importance to surgeons, who have access to human disease tissues for genomic analysis, as well as the need to apply genomic information to improve the outcomes of their operations. We have therefore updated our model, now renamed the GIFT (genomic, imaging, function, and therapy) Model for Personalized Genomic Medicine and Surgery (PGMS). The vision of the development of PGMS is discussed here.
Studies of the human genome
While glimpses of the revolutionary scientific work to come were revealed in a White House ceremony in the millennial year 2000, it was not until 2003 that the Human Genome Project was complete [6, 7]. In the10-year period following that initiating ceremony the study of genomic science experienced rapid expansion and discovery [8]. Building on the success of the Genome Project, the International HapMap (Haplotype Map) Project was begun [9–11] in an effort to discover single nucleotide polymorphisms (SNPs) and copy number variations (CNVs). Data from this project were released in three stages over a period of seven years (2002–2009). Researchers compiled a list of data revealing frequently-found SNPs and CNVs in four human populations. Single nucleotide polymprphisms are present when a single variation of alleles, usually two, occurs in the DNA sequence on one of two paired chromosomes. When sets of these SNPs are found in close proximity to one another on the same chromosome, they are termed haplotypes; groups of haplotypes are further classified into haplogroups. The presence of certain SNPs can identify someone as having a greater propensity for developing certain diseases, thus underscoring the importance of this mapping project to the advancement of personalized genomic health care. The HapMap Project was designed to uncover these common genetic variants that have become extensively applied to the investigation of varying diseases in what are termed genome-wide association studies (GWAS). For example, SNPs in the gene, apolipoprotein E (ApoE), are associated with varying alleles (ε2, ε3, and ε4) that, depending on which allele has been inherited, either increases (ε4) or decreases (ε2) an individual’s chance for developing Alzheimer’s disease [12–15]. While research into SNPs continues to expand, there are presently over 500 identified genomic regions linked to an increased risk for various diseases [16].
While similar to SNPs in that they have been linked to disease vulnerability or resistance, CNVs encompass a more extensive area of the genome (nearly 10 times that of SNPs) [17] that has been either removed or strengthened, resulting in a variation of the copy number among individuals or between normal and diseased tissues. Discovered after completion of the Human Genome Project [18, 19], the causes of CNVs have been established as either heredity or de novo mutation [20]. Because CNVs often contain entire genes or even more than one gene, they could greatly influence a number of healthcare-related factors including the likelihood that a given person will develop certain diseases or gain benefit from certain medications [21, 22]. Indeed, CNVs have been associated with an increasing number of diseases [23]. For example, individuals with a higher copy number of CCL3L1 showed lower susceptibility to human immunodeficiency virus (HIV) infection [24]; whereas individuals with a low copy number of FCGR3B (the CD16 cell surface immunoglobulin receptor) can have increased susceptibility to systemic lupus erythematosus and similar inflammatory autoimmune disorders [25].
Advances in DNA sequencing technology have increased efficiency and lowered costs. These rapid advances have allowed sequencing of whole human genomes such as that of Archbishop Desmond Tutu of South African [7]; James Watson [26], Nobel laureate and co-discoverer of the DNA double helix; Craig Venter [27], co-author of the human genome sequence, and several others [28–30].
In 2008 a new genetic mapping consortium project was set underway and called the 1000 Genomes Project [16]. This project aims to sequence the genome of 1,000 individuals to identify the most common (≥1% of populations studied) genetic variants that will then be compiled into a comprehensive database available to researchers. Any two humans are at least 99.9% identical in DNA sequence (excluding the copy number variation mentioned above). This is remarkable given the uniqueness experienced among the human species and it underscores the importance of the differing genetic material, regardless of its implied numeric value. A primary goal of the 1000 Genomes Project is to create a detailed map of common SNPs as well as rare SNPs occurring in the range of 1–5% of the population.
Genomics and clinical risk factors
In 2006, scientists predicted that the cost of sequencing an individual genome would fall to a price of around $1,000, commonly referred to as “the $1,000 genome” with the advent of new DNA sequencing platforms [31]. The high cost of personalized genomic medicine and surgery remains an impediment to its widespread clinical application for the general population; however, as new sequencing technologies, termed “next-gen” platforms, continue to improve, associated costs are falling. It is not beyond the realm of possibility that a personal genome sequence will become a future standard component of every patient’s medical records. As previously discussed, genomic information can affect health care in many ways. Genomic variability could signal the risk of certain diseases before clinical signs and symptoms appear. Accordingly, strategies of prevention and early intervention can be planned. For example, Farmer and colleagues and Venkitaraman have shown that two genes (BRCA1 & 2) are of critical importance to fundamental cell processes [32, 33] and that mutations of these genes contribute to tumorigenesis in breast and ovarian tissues. In a study by Roukos and Briasoulis the mean lifetime risk for breast cancer in women ranges between 10 and 14% in Europe and the United States [34], whereas women with germline BRCA mutations have a 36–85% lifetime chance of developing breast cancer (BRCA1 or BRCA2) [35–37]. Additionally, variants of these genes increase a woman’s chance of developing ovarian cancer to between 25 and 60%; a stark contrast to a reported 1.7% risk among women who do not carry these variants [34, 38]. These genomic risk analyses currently guide clinical decision making about aggressive surveillance programs such as increased frequency of mammography or even prophylactic surgery, including mastectomy and removal of the ovaries. Similarly, the protein products of MLH1 and MSH2 genes help to repair mistakes made in DNA replication. People with mutations in MLH1 and MSH2 genes may have a high risk for colon cancer. Therefore, Wiesner et al. have recommended that these patients undergo early and regular screening colonoscopy in an effort to increase the chances for early detection of the disease [39]. Although recent GWASs have linked many chronic diseases and prevalent cancers to genetic indicators [40], further studies are needed to determine the extent to which the data obtained will translate into practical clinical contributions for patients.
Genomics and drug metabolism
Drug selection is a major benefit of personal genome data. Having prior knowledge regarding the suspected efficacy, or lack thereof, of a particular drug for a particular patient gives physicians a marked advantage in selecting appropriate therapies and improving outcomes while minimizing side effects and toxicity. The National Center for Biotechnology Information (NCBI) defines the term pharmacogenomics as “a science that examines the inherited variations in genes that dictate drug response and explores the ways these variations can be used to predict whether a patient will have a good response to a drug, a bad response to a drug, or no response at all” [41]. For example, warfarin (Coumadin), a widely used anticoagulant, is also known to have unfavorable side effects. There are genetic variants in two genes that are known to influence the turnover of warfarin. One of these genes (CYP2C9*) encodes an enzyme that is responsible for metabolic clearance of the drug. Three alleles of this gene (CYP2C9*1, CYP2C9*2, and CYP2C9*3) contain genetic variants causing a reduced clearance rate [42] and are thought to contribute to associated bleeding complications. The second gene (VKORC1) contains specific SNPs that make it less susceptible to suppression by warfarin (warfarin resistance) [43, 44]. Armed with this knowledge, the U.S. Food and Drug Administration (FDA) has encouraged physicians to use genetic testing when estimating “reasonable warfarin” dosing for their patients [45]. Another example of applied pharmacogenomics can be seen with the use of tamoxifen, an antagonist of the estrogen receptor in breast tissue used for over 30 years as the standard anti-estrogen treatment for breast cancer [46]. As a prodrug, tamoxifen requires several cytochrome P450 enzymes to produce the active metabolite endoxifen. CYP2D6, the primary enzyme responsible for this conversion, is associated with several known SNPs, some of which yield a reduced capacity for conversion, while others are characterized by an increased capacity [46]. Patients with reduced-capacity SNPs convert lesser amounts of the active metabolite, therefore, leading to underdosing and the possibility of a lessened response to treatment [47]. CYP2D6, along with another P450 enzyme, CYP2C19, are known to be responsible for the metabolism of nearly 25% of all prescription drugs [48], therefore, the FDA has approved the Roche-AmpliChip CYP450 test to help physicians determine adequate dosing strategies. Additionally, the FDA has approved inserts extolling the benefits of genetic testing for certain pharmaceuticals in which known genetic variants may bear weight on dosing or therapy recommendations [49].
Genomics and molecular targets
Molecular targets discovered from genomics and gene expression levels have been successfully translated into clinical use. For example, patients with breast cancer often undergo testing to ascertain the expression levels of estrogen and progesterone receptors in an effort to better determine the individual’s response to tamoxifen [50]. About 30% of breast cancer cases showing overexpression of human epidermal growth factor receptor 2 (ERBB2) also had less favorable outcomes [51, 52]. In 1998 a diagnostic test for ERBB2 overexpression was developed, along with a humanized monoclonal antibody, trastuzumab (Herceptin), against the extracellular domain of ERBB2. Together, these have been successfully used to treat patients with HER2-positive breast cancer [53]. Further clinical studies have shown that trastuzumab therapy also is effective in an ERBB2-positive subpopulation of gastric cancers [54, 55].
The Philadelphia chromosome is a translocation between chromosomes 9 and 22 that has been linked to 95% of chronic myelogenous leukemias (CML) [56, 57]. The translocation results in the BCR-ABL oncogene, encoding a membrane-associated tyrosine kinase. An inhibitor Imatinib (Gleevec) was discovered to block BCR-ABL and specifically target CML cells [58]. Since 2001, Gleevec has been the principal treatment for CML [59].
Gefitinib (Iressa) is an epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor that is characterized by its sensitivity to non-small-cell lung cancers with mutations in the EGFR tyrosine kinase domain [60–63]. Reserved for only non-small-cell lung carcinoma (NSCLC) patients with at least one previous chemotherapy regimen, Erlotinib also inhibits EGFR tyrosine kinase, and two anti-EGFR monoclonal antibodies [Erbitux (cetuximab) and Vectibix (panitumumab)] have been introduced as therapies for metastatic colorectal cancer [64]. These two antibodies are more effective in treating to the cancer with wild-type KRAS [65, 66]. A randomized trial assessment of response to wild-type p53 gene replacement in patients with recurrent squamous cell cancer of the head and neck region who had low expression of a mutant p53 gene or no evidence of a p53 gene mutation resulted in significantly improved survival rates compared to with the survival seen after p53 gene replacement in patients who had demonstrated higher expression of a mutant p53 gene [67]. Thus, the genotype of several genes can predict response to therapy.
Targeted therapies are beginning to be developed for other, non-cancerous diseases [68]. For example, a chemokine receptor CCR5 inhibitor called maraviroc (Selz-entry) has been developed for the treatment of HIV infection [69]. Human immunodeficiency virus can use either CCR5 or CXCR4 as a co-receptor to bind and enter a human macrophage and/or T lymphocyte [70], but maraviroc is limited to CCR5 tropism, which is determined by diagnostic assay (Trofile).
Personalized genomic medicine and surgery
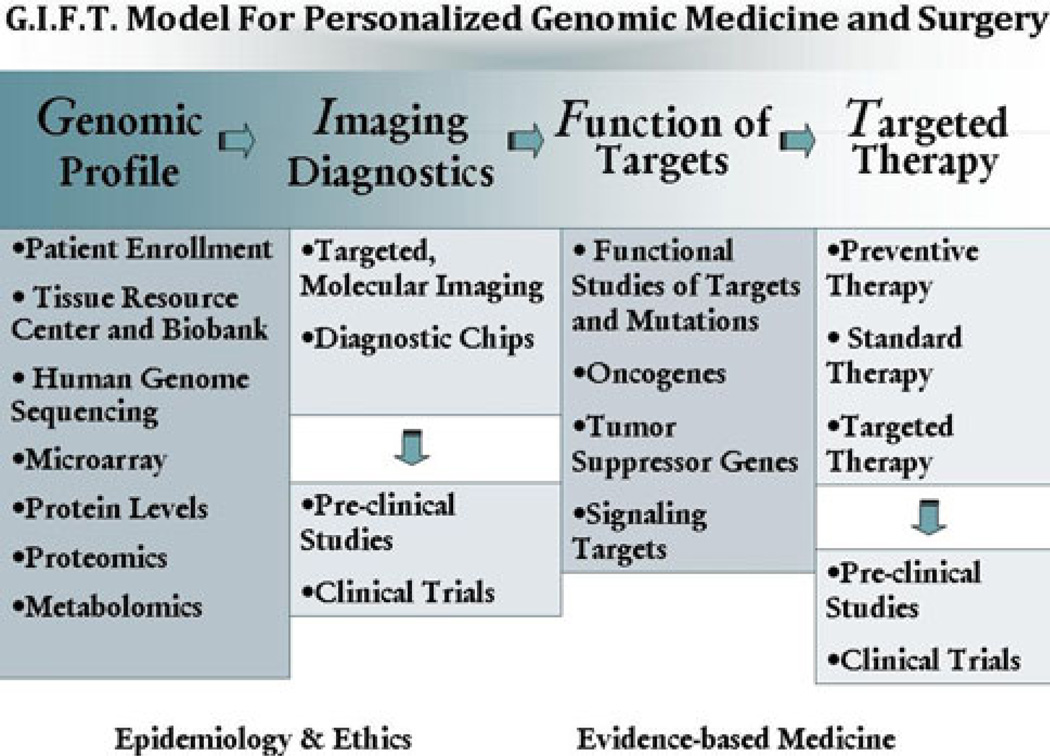
While remarkable discoveries have been made through genomic sequencing, its progress was hindered by the lack of a practical model of personalized genomic medicine and surgery in which the information gleaned from genomics could be readily translated into clinical care. Therefore, the leadership of the Human Genome Sequencing Center and the Michael E. DeBakey Department Surgery at Baylor College of Medicine (BCM) previously developed its model for Personalized Genomic Medicine and Surgery (PGMS) and began implementing its phases. Since its debut approximately five years ago, the model has undergone some revision and restructuring of its phases (Fig. 1). Our restructured GIFT model for PGMS consists of the following four phases: (1) genomic profiling to identify molecular/genomic targets, (2) the development of imaging diagnostics, (3) the examination of the function of selected molecular targets, and (4) therapies (see Fig. 1).
Fig. 1.
The G.I.F.T. model for personalized genomic medicine and surgery. The model is rolled out in four phases: genomic profile, imaging diagnostics, function of targets, and targeted therapy
While the key phrase for the model is that “the genomic profile guides choice of therapy,” our team began implementation of the model by focusing on phase 1 with the building of a genomic Biobank. To review our progress, we hosted the first “Molecular Surgeon Symposium on Personalized Genomic Medicine and Surgery (PGMS)” on April 12, 2008 [71]. The development of our PGMS model was discussed, with emphasis on the creation of a pipeline of tissues from the operating rooms of the BCM-affiliated hospitals into the BCM, as well as establishing our goals for the next 2 years [72, 73]. While we have made substantial advances in the development of the model of PGMS, we hosted the second symposium on May 7, 2010, to review our progress and re-establish goals for the next two years. This symposium brought together 19 surgeons, physicians, and scientists from varying disciplines determined to improve the translation of genomic applications to clinical therapies (Table 1). An update regarding our progress related to all four phases of the PGMS model was presented, with the most noteworthy accomplishment being the establishment and funding of a biological specimen tissue repository (a Biobank) in the Human Genome Sequencing Center.
Table 1.
Presentations given at the 2010 Molecular Surgeon Symposium on Personalized Genomic Medicine and Surgery (PGMS)
| Speaker | Presentation topic |
|---|---|
| F. Charles Brunicardi, MD, FACS | PDX-1 as an archetype for PGMS |
| Richard Gibbs, PhD | Genomics in personalized medicine |
| John Nemunaitis, MD | Personalized therapeutics using gene based technology |
| Gustavo Ayala, MD | From nerves to gene regulation |
| David Wheeler, PhD | Mutation profiling in human cancer using second generation sequencers |
| William E. Fisher, MD, FACS, | Can genomic studies overcome the universal failure of clinical trials in pancreatic cancer? |
| Kenneth L. Scott, PhD | Oncogenomics-guided functional screening for metastasis determinants |
| Qizhi (Cathy) Yao, MD, PhD | Trop2: expression, DNA sequence and functions in pancreatic cancer |
| Christian Marin-Muller | Mesothelin and miR-198: Expression, DNA sequence and functions in pancreatic cancer |
| Sarah M. Weakley, MD | Loss of XIST enhances the tumor growth and pathogenesis of female human pancreatic cancer |
| Changyi (Johnny) Chen, MD, PhD | miR-196a: Expression, DNA sequence and functions in pancreatic cancer |
| Min Li, PhD | ZIP4 is a novel therapeutic target in pancreatic cancer |
| Guisheng Zhou, PhD | SSTR5 P335L acts as a hypofunctional single-nucleotide polymorphism (SNP) by upregulating PDX-1 expression |
| Amy L. McGuire, PhD, JD | Genome research, biobanking, and shared resources |
| Courtney J. Balentine, MD | Flipping the cancer master switch |
| Dolores J. Lamb, PhD | Identification of de novo copy number variants associated with human disorders of genitourinary development |
| Jacfranz Guiteau, MD | Development and maintenance of an HCC bioresource bank: practical issues |
| Ronald T. Cotton, MD | Liver cancer genome: Mutation profiling for viral hepatitis-associated hepatocellular carcinoma |
| Seth P. Lerner, MD | p53 targeted therapy trial for organ confined invasive urothelial bladder cancer |
Large numbers of patients with different diseases have been enrolled with informed consent. Special consents were designed with Internal Review Board (IRB) approval for patients consenting to the study of their tissues for genomic analyses, including DNA sequencing, epigenetic analysis, RNA microarray, and proteomics. The genomic data generated from these studies led to the identification of genomic targets for patients, as well as the discovery of disease-specific molecular targets. The greatest example was the historic sequencing of the whole genome of a patient with Charcot-Marie-Tooth disease, resulting in the discovery of the gene causing the disease, SH3TC2. This magnificent achievement opened up the era for PGMS [30]. With the generation of enormous amounts of patient-specific genomic information, we must now determine how their genomic profiles guide the choice of therapy using the GIFT model. This update on the progress of PGMS and the means by which to quickly achieve it has resulted in the collaboration of genomic scientists, basic scientists, and clinical scientist–surgeons on coordinating the different phases of the model. It is our hope that the genomic analyses will result in practical diagnostic genomic tests that will guide choice of preventive, standard, and targeted therapies for the positive application to the care of our patients.
Summary
The past decade has brought about an exponential evolution in the sequencing field. These achievements include the original sequencing of the human genome, the sequencing of the whole genome of several individuals, and the successful search for a patient’s disease-causing gene using whole genome sequencing. With such a dramatic drop in the cost of entire genome sequencing (now approximately $6,000) it is anticipated that the overall cost will eventually be close to $1,000 within the next few years. Continued cost reduction will make whole genome information readily available to our patients and to us as clinicians. Genomic information can be used to influence patient care in many ways, including prediction of disease susceptibility, risk analysis, determining the effectiveness of medications, and specific targeted imaging and therapy. While it is clear that the progress in DNA sequencing has been extensive over a short period of time, the great challenge before us is to utilize genomic information to guide the choice of therapy for our patients. In order to further develop PGMS, we will continue in our efforts to develop the highest quality BioBank for genomic studies. Whole genome sequencing of patients’ blood and tissues will be continued. Several molecular targets will be advanced to preclinical and clinical trials. We will promote educational programs in PGMS and encourage more basic scientists, clinical scientists, surgeons, pathologists, and physicians to participate in PGMS in the different phases of the model, with the ultimate goal being to significantly improve healthcare quality by utilizing personal genomics to guide choice of therapy.
Acknowledgments
The authors are grateful to Katie Elsbury for editorial assistance. This study was partially supported by grants from the following sources: National Institutes of Health (NIH) grants NIDDK R01-DK46441 and NCI R01-CA095731 (to F.C. Brunicardi); U54-HG003273; U54-HG004973 (to R. A. Gibbs); Cancer Prevention & Research Institution of Texas (CPRIT) grant RP101353-P01/P07 (to R. A. Gibbs); the Vivian L. Smith Foundation, the MD Anderson Foundation, the Elkins Pancreas Center at the Baylor College of Medicine, the MacDonald General Research Fund Award, St. Luke’s Episcopal Hospital (09RDM006, to C. Chen), and the Dan L. Duncan Cancer Center at the Baylor College of Medicine (DLDCC PILOT PROJECT 09-10 to C. Chen).
Contributor Information
F. Charles Brunicardi, Email: cbrunica@bcm.tmc.edu, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, 1709 Dryden Street, Suite 1500, Houston, TX 77030, USA.
Richard A. Gibbs, Human Genome Sequencing Center, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, MSC-226, Houston, TX 77030, USA
David A. Wheeler, Human Genome Sequencing Center, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, MSC-226, Houston, TX 77030, USA
John Nemunaitis, Mary Crowley Cancer Research Center, Medical City, 7777 Forest Lane, Suite 707, Dallas, TX 75230, USA.
William Fisher, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, 1709 Dryden Street, Suite 1500, Houston, TX 77030, USA.
John Goss, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, 1709 Dryden Street, Suite 1500, Houston, TX 77030, USA.
Changyi Chen, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, 1709 Dryden Street, Suite 1500, Houston, TX 77030, USA.
References
- 1.Obama B. Genomics and personalized medicine act of 2007. Washington, DC: U.S. Congress; 2007. [Google Scholar]
- 2.President’s Council of Advisors on Science Technology. Priorities for Personalised Medicine. [Accessed Feb 12, 2011];2008 http://www.ostp.gov/galleries/PCAST/pcast_report_v2.pdf.
- 3.U.S. Department of health and Human Services (HHS) Personalized health care: opportunities, pathways, resources. http://www.hhs.gov/myhealthcare/
- 4.Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res. 2009;154:277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 8.Collins FS. Has the revolution arrived? Nature. 2010:674–675. doi: 10.1038/464674a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 10.International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artiga MJ, Bullido MJ, Frank A. Risk for Alzheimer’s disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet. 1998;7:1887–1892. doi: 10.1093/hmg/7.12.1887. [DOI] [PubMed] [Google Scholar]
- 13.Holmes C. Genotype and phenotype in Alzheimer’s disease. Br J Psychiatry. 2002;180:131–134. doi: 10.1192/bjp.180.2.131. [DOI] [PubMed] [Google Scholar]
- 14.Belbin O, Dunn JL, Ling Y, et al. Regulatory region single nucleotide polymorphisms of the apolipoprotein E gene and the rate of cognitive decline in Alzheimer’s disease. Hum Mol Genet. 2007;16:2199–2208. doi: 10.1093/hmg/ddm171. [DOI] [PubMed] [Google Scholar]
- 15.van Vliet P, Westendorp RG, Eikelenboom P, et al. Parental history of Alzheimer disease associated with lower plasma apolipoprotein E levels. Neurology. 2009;73:681–687. doi: 10.1212/WNL.0b013e3181b59c2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Human Genome Research Institute; [Accessed Feb 12, 2011]. International Consortium Announces the 1000 Genomes Project. http://www.genome.gov/26524516. [Google Scholar]
- 17.Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 19.Iafratel AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 20.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 21.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman JL, Perry GH, Feuk L, et al. Copy number variation: new insights into genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Gu W, Hurles ME, et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez E, Kulkarni H, Bolivar H. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 25.Aitman TJ, Dong R, Vyse TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 27.Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto A, Nakagawa H, Hosono N, et al. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat Genet. 2010;42:931–936. doi: 10.1038/ng.691. [DOI] [PubMed] [Google Scholar]
- 29.Sobreira NLM, Cirulli ET, Avramopoulos D, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a mendelian disease gene. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mardis ER. Anticipating the $1, 000 genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 33.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 34.Roukos DH, Briasoulis E. Individualized preventive and therapeutic management of hereditary breast ovarian cancer. Nat Clin Pract Oncol. 2007;4:578–590. doi: 10.1038/ncponc0930. [DOI] [PubMed] [Google Scholar]
- 35.National Cancer Institute. BRCA1 and BRCA2: cancer risk and genetic testing. [Accessed Feb 12, 2011]; http://www.cancer.gov/cancertopics/factsheet/risk/brca.
- 36.Struewing JP, Hartge P, Wacholder S, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 37.Ries LAG, Harkins D, Krapcho M, et al. SEER cancer statistics review, 1975–2003. Bethesda: National Cancer Institute; 2006. [Google Scholar]
- 38.Narod SA, Offit K. Prevention and management of hereditary breast cancer. J Clin Oncol. 2005;23:1656–1663. doi: 10.1200/JCO.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 39.Wiesner G, Slavin T, Barnholtz-Sloan J. Colorectal cancer. In: Willard H, Ginsburg GS, editors. Genomic it personalized medicine. Durham: Elsevier; 2009. pp. 879–897. [Google Scholar]
- 40.Manolio T, Brooks L, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Center for Biotechnology Information (NCBI) One size does not fit all: the promise of pharacogenomics. [Accessed Feb. 12, 2011];2011 http://www.ncbi.nlm.nih.gov/About/primer/pharm.html.
- 42.Schwarz UI. Clinical relevance of genetic polymorphisms in the human CYP2C9 gene. Eur J Clin Invest. 2003;33:23–30. doi: 10.1046/j.1365-2362.33.s2.6.x. [DOI] [PubMed] [Google Scholar]
- 43.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 44.Oldenburg J, Watzka M, Rost S, et al. VKORC1: molecular target of coumarins. J Thromb Haemost. 2007;5(1):1–6. doi: 10.1111/j.1538-7836.2007.02549.x. [DOI] [PubMed] [Google Scholar]
- 45.FDA Approves Updated Warfarin (Coumadin) Prescribing Information. [Accessed Apr. 8, 2009]; http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2007/ucm108967.htm.
- 46.Higgins MJ, Stearns V. Pharmacogenetics of endocrine therapy for breast cancer. Annu Rev Med. 2011;62:281–293. doi: 10.1146/annurev-med-070909-182545. [DOI] [PubMed] [Google Scholar]
- 47.Dezentjé VO, Guchelaar HJ, Nortier JW, et al. Clinical implications of CYP2D6 genotyping in tamoxifen treatment for breast cancer. Clin Cancer Res. 2009;15:15–21. doi: 10.1158/1078-0432.CCR-08-2006. [DOI] [PubMed] [Google Scholar]
- 48.Jain KK. Applications of AmpliChip CYP450. Mol Diagn. 2005;9:119–127. doi: 10.2165/00066982-200509030-00002. [DOI] [PubMed] [Google Scholar]
- 49.FDA. Table of valid genomic biomarkers in the context of approved drug labels, 2009. [Accessed Feb 12, 2011];2009 http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm.
- 50.Dowsett M, Dunbier AK. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin Cancer Res. 2008;14:8019–8026. doi: 10.1158/1078-0432.CCR-08-0974. [DOI] [PubMed] [Google Scholar]
- 51.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 52.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Herceptin adjuvant (HERA) trial study team. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Eng J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 53.Ross JS, Slodkowska EA, Symmans WF, et al. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14:320–368. doi: 10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 54.Bang Y, Chung H, Xu J, et al. Pathological features of advanced gastric cancer (GC): relationship to human epidermal growth factor receptor 2 (HER2) positivity in the global screening programme of the ToGA trial. J Clin Oncol. 2009;27 abstract 4556. [Google Scholar]
- 55.Van Cutsem E, Kang Y, Chung H, et al. Efficacy results from the ToGA trial: a phase III study of trastuzumab added to standard chemotherapy (CT) in first-line human epidermal growth factor receptor 2 (HER2)-positive advanced gastric cancer (GC) J Clin Oncol. 2009;27 abstract 2409. [Google Scholar]
- 56.Joske DJ. Chronic myeloid leukaemia: the evolution of gene-targeted therapy. Med J Aust. 2008;189:277–282. doi: 10.5694/j.1326-5377.2008.tb02027.x. [DOI] [PubMed] [Google Scholar]
- 57.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 58.Schindler T, Bornmann W, Pellicena P. A structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1857–1859. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 59.National Comprehensive Cancer Network. clinical practice guidelines in oncology chronic myelogenous leukemia. 2009 doi: 10.6004/jnccn.2009.0065. http://www.nccn.org/professionals/physician_gls/PDF/cml.pdf. [DOI] [PubMed] [Google Scholar]
- 60.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers“ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 62.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 63.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 64.Mack GS. FDA holds court on post hoc data linking KRAS status to drug response. Nat Biotechnol. 2009;27:110–112. doi: 10.1038/nbt0209-110c. [DOI] [PubMed] [Google Scholar]
- 65.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 66.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 67.Nemunaitis J, Clayman G, Agarwala SS, et al. Biomarkers predict p53 gene therapy efficacy in recurrent squamous cell carcinoma of the head and neck. Clin Cancer Res. 2009;15:7719–7725. doi: 10.1158/1078-0432.CCR-09-1044. [DOI] [PubMed] [Google Scholar]
- 68.U.S. Food and Drug Administration. Table of valid biomarkers in the context of approved drug labels. 2009 http://www.fda.gov/cder/genomics/genomic_biomarkers_table.htm.
- 69.Levy JA. HIV pathogenesis: 25 years of progress and persistent challenges. AIDS. 2009;23:147–160. doi: 10.1097/QAD.0b013e3283217f9f. [DOI] [PubMed] [Google Scholar]
- 70.Biswas P, Tambussi G, Lazzarin A. Access denied? The status of co-receptor inhibition to counter HIV entry. Expert Opin Pharmacother. 2007;8:923–933. doi: 10.1517/14656566.8.7.923. [DOI] [PubMed] [Google Scholar]
- 71.Brunicardi FC, Gibbs RA, Fisher W, et al. Overview of the molecular surgeon symposium on personalized genomic medicine and surgery. World J Surg. 2009;33:612–614. doi: 10.1007/s00268-008-9861-9. [DOI] [PubMed] [Google Scholar]
- 72.Voidonikolas G, Kreml SS, Chen C, et al. Basic principles and technologies for deciphering the genetic map of cancer. World J Surg. 2009;33:615–629. doi: 10.1007/s00268-008-9851-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voidonikolas G, Gingras MC, Hodges S, et al. Developing a tissue resource to characterize the genome of pancreatic cancer. World J Surg. 2009;33:723–731. doi: 10.1007/s00268-008-9877-1. [DOI] [PMC free article] [PubMed] [Google Scholar]