Abstract
Suprachiasmatic nucleus neuroanatomy has been a subject of intense interest since the discovery of the SCN’s function as a brain clock and subsequent studies revealing substantial heterogeneity of its component neurons. Understanding the network organization of the SCN has become increasingly relevant in the context of studies showing that its functional circuitry, evident in the spatial and temporal expression of clock genes, can be reorganized by inputs from the internal and external environment. Although multiple mechanisms have been proposed for coupling among SCN neurons, relatively little is known of the precise pattern of SCN circuitry. To explore SCN networks, we examine responses of the SCN to various photic conditions, using in vivo and in vitro studies with associated mathematical modeling to study spatiotemporal changes in SCN activity. We find an orderly and reproducible spatiotemporal pattern of oscillatory gene expression in the SCN, which requires the presence of the ventrolateral core region. Without the SCN core region, behavioral rhythmicity is abolished in vivo, whereas low-amplitude rhythmicity can be detected in SCN slices in vitro, but with loss of normal topographic organization. These studies reveal SCN circuit properties required to signal daily time.
INTRODUCTION
The Problem
Studies of the suprachiasmatic nucleus (SCN) and its function as the “master circadian pacemaker” offer an opportunity to examine multiple temporal and spatial scales so as to explore how complex responses emerge from cellular elements. Breakthroughs in molecular biology and in computer-based storage and analysis of enormous amounts of data have made it possible to measure brain structure, gene expression, and electrical activity simultaneously at many locations with high spatial and temporal resolution. The SCN is an unusual model system in the mammalian brain in that its function is well delineated and can be studied at cellular, tissue, and whole-organism levels. Finally, joint use of experimental and modeling approaches has led to new experimental studies that help to explore and explain seemingly unintuitive results.
The SCN and Its Function
The SCN is an endogenous master circadian pacemaker, whose phase is reset by photic cues via the retino-hypothalamic tract (RHT) and which controls daily rhythms of behavior and physiology (Klein et al. 1991). Ablation of the SCN produces a loss of circadian rhythmicity at the whole-animal level (Moore and Eichler 1972; Stephan and Zucker 1972), with no recovery of function irrespective of the age at which the lesion is made (Mosko and Moore 1979). The SCN oscillates in the absence of input from the rest of the brain both in vivo (Inouye and Kawamura 1979) and in vitro (Green and Gillette 1982; Groos and Hendriks 1982; Shibata et al. 1982). Within the SCN, circadian oscillations can be measured in gene expression, electrical activity, metabolic activity, and neuropeptide release (Reppert and Weaver 2001). Furthermore, fetal SCN grafts can restore behavioral rhythms in SCN-lesioned animals (Lehman et al. 1987; Ralph et al. 1990). The SCN produces both synaptic and diffusible output signals (Hakim et al. 1991; Silver et al. 1996). Neural efferents appear to be necessary for SCN control of neuroendocrine responses but not for activity-dependent rhythms such as drinking, gnawing, and temperature regulation (Lehman et al. 1987; Hakim et al. 1991; Meyer-Bernstein et al. 1999). Finally, individual SCN neurons display circadian rhythmicity upon dispersion (Welsh et al. 1995). These experiments suggest the concept of the SCN as a brain clock comprising a feed-forward circuit of similar cellular elements that together produce a coherent circadian rhythm output that signals time of day information for the rest of the body (Fig. 1A).
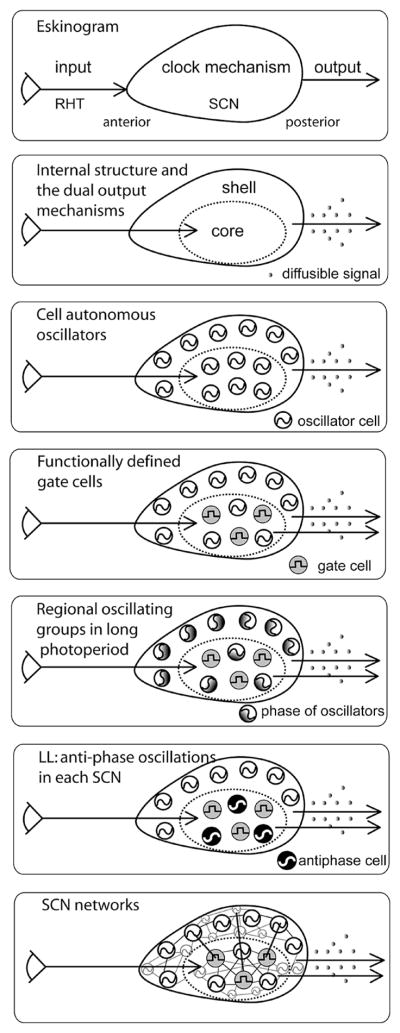
Figure 1.
Building the brain clock: A historical perspective. (A) An early depiction of the circadian system involved a feed-forward mechanism with three components, namely, input, clock mechanism, and output (Pittendrigh 1961; Eskin 1979). This simple heuristic captures several key concepts of circadian timekeeping. (B) Delineation of the distinct core and shell compartments of the SCN began the effort of identifying components of the clock in the black box (Moore 1973; van den Pol and Tsujimoto 1985). Discovery that a diffusible signal was sufficient to maintain coherent activity rhythms (Silver et al. 1996) led to the search for such a signal. (C) In dispersed cell culture, individual SCN neurons show circadian rhythms in their firing rate with different phases, suggesting that circadian oscillation is a property of individual SCN neurons, rather than an emergent network property (Welsh et al. 1995). (D) The SCN is composed of functionally distinct cells. In addition to oscillator cells, some cells in the “core” region lack detectable oscillation of clock gene expression and electrical activity (Hamada et al. 2001; Jobst and Allen 2002; Karatsoreos et al. 2004). Cells of the core and shell region each project to SCN targets sites (Abrahamson and Moore 2001; Kriegsfeld et al. 2004a). (E) Photoperiod induces both temporal and spatial changes in the circadian phase of oscillators within the SCN. In short photoperiod, the oscillators are mostly in-phase; in long photoperiods, the phases of the oscillators change along the rostrocaudal axis. (F) In the behaviorally split hamster, new networks of SCN oscillators are seen under unusual environmental conditions. In the behaviorally split animal, the left and right SCN are in antiphase, and within each SCN, the core and shell of each SCN is in antiphase (Tavakoli-Nezhad and Schwartz 2005; Yan et al. 2005). (G) SCN network organization may resemble a small-world network with mostly local connections between nodes and with a few long-distance connections. On the other hand, it may consist of a locally connected network in which signals spread slowly from one layer of cells to the next.
The Tissue Is the Issue
The initial view of the SCN assigned to it nearly magical properties. Here, within the mammalian brain was a hypothalamic nucleus that was necessary and sufficient for a single function, and upon damage (even neonatally), this function could not be assumed by any other tissue. The early view of the SCN as a black box was a very useful heuristic in drawing attention to inputs and outputs of the master clock (see Fig. 1 in Pittendrigh and Bruce 1959; Eskin 1979). Since that time, numerous advances in the analysis of SCN organization have contributed to understanding the component cellular elements and neural circuits inside this black box (summarized in Fig. 1B–C). The investigation of SCN responses to naturally occurring and experimentally induced challenges has made it clear that robust function of the brain clock is dependent on its intricate neural circuitry (Fig. 1D–F). We know relatively little of SCN circuits, although some general principles of network organization may be useful in conceptualizing available data (Fig. 1G).
Wiring economy or optimization has been suggested as an important general principle for brain organization because larger volumes raise metabolic cost, delay signal transduction, and attenuate signals (Ramón y Cajal et al. 1999). According to this principle, among the various functionally equivalent arrangements of neurons, the one having minimum wiring cost is most evolutionarily fit and therefore is most likely to be selected (Chklovskii and Koulakov 2004). The SCN is small, with small (8–10-μm diameter) densely packed neurons. Even though brain weight and total neuron number differ more than a 1000-fold between human and mouse (human vs. mouse, 1350 g vs. 450 mg; 85 billion vs. 75 million neurons, respectively; Williams and Herrup 1988), the number of neurons in the human SCN is only a fewfold greater (human vs. mouse, 45,000 vs. 10,500; Hofman and Swaab 1989; Abrahamson and Moore 2001). The slight change in the number of SCN neurons during evolution suggests computational performance of the SCN has reached an optimal level at a small size. (This observation also suggests that the SCN does not scale with brain size.) How does the SCN solve the conflict between wiring economy and the need to coordinate long-term, i.e., circadian, rhythmicity among neurons? If the SCN has properties of a small-world network (Watts and Strogatz 1998), one would expect to find local connections between nodes with a few long-distance connections (Fig. 1G). Restated, it would have a small number of connections per node but also a small total distance between any two nodes. Signals in a small-world network tend to propagate very rapidly through the whole network. The advantage of the small-world arrangement for a neural network is not only for wiring economy, but also to increase clustering, and some aspects of SCN circuits are consistent with these properties. For other aspects, particularly the function of the SCN in representing the time of day and season of the year, the evidence of spatiotemporal organization reviewed in this chapter is more consistent with a locally connected network in which signals spread rather slowly from one layer of connected nodes to the next. Such a locally connected network exploits the inherent periodicity of the oscillator cells so as to establish the timescale of the circadian timekeeping. (A “cellular automaton” approach to modeling a locally connected network oscillator is presented below.) In view of regional heterogeneity of the SCN, it is possible that both types of networks occur within the nucleus.
It is our thesis that “the tissue is the issue,” i.e., SCN function can best be understood by delineating circuit properties of the SCN that render it a brain clock. Our objective is to explore how the individual cells that comprise the SCN produce its rhythmic output signal(s). The view that rhythmicity of the SCN is a product of individual, coupled oscillators has set the basis for physiological and modeling studies of the clock, as well as its inputs and outputs. We explore how the cells of the bilateral SCN are organized in space and time, and how unique clock functions are achieved by this organization of SCN cellular elements. The cells that constitute SCN circuits include diverse elements: cell-autonomous oscillators, slave oscillators, and cells that do not appear to oscillate at all (Lee et al. 2003; Hastings and Herzog 2004; Antle and Silver 2005). Thus, within the SCN, heterogeneous, independently phased cellular oscillators occur in a spatially and temporally ordered pattern, and this underlies the timekeeping function of the nucleus. Our goal is to review SCN neuroanatomy briefly, to describe changes in its organization following exposure to various photic stimuli, and to deconstruct its neural circuitry using in vivo and in vitro studies and associated modeling analyses.
SECTION THEMES
Brief Anatomy of SCN Focusing on Core and Shell Organization
Numerous features of SCN organization are consistent among mammals. Each side of this bilaterally symmetrical nucleus of rodents contains approximately 10,000 neurons, with characteristic topography of peptide content, afferent/efferent connections, and clock gene expression. From the time of the earliest studies, it was clear that the SCN is composed of two fundamentally different regions based on cell size and morphology, afferent and efferent connections, and neuropeptide phenotype (Moore 1983). Largely based on the rat SCN, these two regions were designated ventrolateral and dorsomedial. As comparative data emerged, the limited generality of these two “geographic” descriptors came into better focus. Because the topography of SCN organization differs among species, it became more useful to think of the SCN as composed of “core” and “shell” regions, based on physiological and functional criteria (Miller et al. 1996; Antle and Silver 2005). This brought the advantage of conceptually differentiating two regions but lacked precise localization. Because the anatomical loci suggested by the terms core and shell do not delineate SCN regions very precisely, these designations can lead to confusion (for discussion, see Morin and Allen 2006). Nevertheless, anatomical specializations are generally associated with functional specializations, and the core-shell distinction usefully sets the stage for exploring intra-SCN organization (Abrahamson and Moore 2001). As long it is clear which part of the nucleus is being described, and the criteria being used, the core and shell terminology helps to analyze SCN organization and to conceptualize and communicate the results of such analyses.
Distribution of peptidergic phenotypes within the SCN
The study of core-shell functional specializations has been hampered by the fact that there is significant diversity among species in the peptidergic phenotypes of SCN cells (Morin and Allen 2006). The SCN shell contains neurons expressing arginine vasopressin (AVP) in most species, including mouse, rat, hamster, lemur and humans, although not in mink (Larsen and Mikkelsen 1993) or mole rat (Rosen et al. 2007). The SCN core is rich in vasoactive intestinal polypeptide (VIP) and gastrin-releasing peptide (GRP) in mouse, rat, and hamster (Morin et al. 1992; Moore 1996; Abrahamson and Moore 2001). Other peptides in the SCN core, however, are more variable among species (Card and Moore 1984; Hartwich et al. 1994; Silver et al. 1999; Abrahamson and Moore 2001), likely reflecting functional species specializations. For example, the hamster core contains calbindin (CalB) and substance P (SP), whereas the mouse core contains calretinin and neurotensin.
There is a broad association between peptidergic phenotype and clock function. The SCN shell region is delineated by AVP neurons and by rhythmic Per mRNA and AVP expression (Schwartz et al. 1983; Jin et al. 1999). The core region, bearing VIP and GRP cells, lacks detectable rhythmicity or shows very low-amplitude changes in clock gene expression (Fig. 2) (Antle and Silver 2005). In contrast to AVP, neither VIP nor GRP is rhythmically expressed in the SCN under constant conditions, although in light/dark (LD) cycles, GRP is higher in the day and VIP is higher at night (Inouye and Shibata 1994).
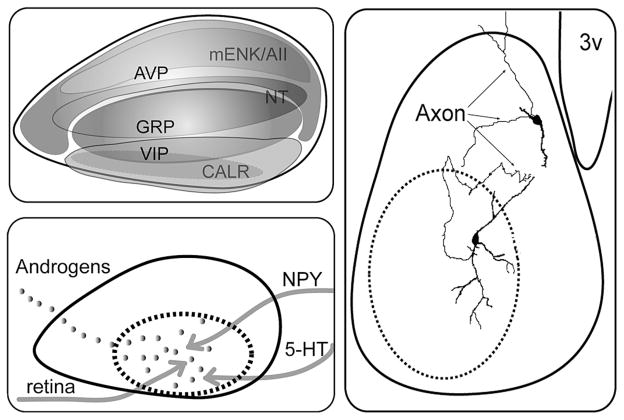
Figure 2.
Anatomical organization of the SCN. (A) Distribution of the neuropeptides in the mouse SCN reconstructed from data in Abrahamson and Moore (2001). (B) Schematic summarizing the neuronal (gray line) and the hormonal (androgen, gray dots) input to the core and shell SCN. (Dashed black line) Boundary of the core SCN. The core region receives both hormonal and neural (retinal input, 5-HT from raphe and NPY from IGL) inputs. The shell region receives GAL and TH afferent fibers. (C) Biotin-filled GRP cell in the SCN core (Drouyer et al. 2005) and AVP cell into the shell (adapted from Pennartz et al. 1998). The GRP cell has an axon and dendrite extending into the shell.
Inputs to SCN
The SCN core and shell have distinct afferent inputs indicative of fundamental differences in function. The core receives direct input from the retina through the retinohypothalamic tract (RHT), secondary visual input from the intergeniculate leaflet (IGL), and the lateral geniculate complex through the geniculohypothalamic tract (GHT), as well as input from the pretectal nuclei and from the midbrain raphe nucleus (Moore 1973; Moga and Moore 1997; Abrahamson and Moore 2001). The shell receives input from the basal forebrain, the cerebral cortex, the hippocampus, brainstem cholinergic nuclei, medullary noradrenergic areas, and several hypothalamic nuclei.
In some instances, the peptidergic phenotype of cells receiving afferent inputs is known. RHT fibers synapse onto VIP cells and GRP cells (rat; Ibata et al. 1989; Tanaka et al. 1997) or on calbindin and GRP cells (hamster; Bryant et al. 2000). Although some retinal fibers may be seen throughout the SCN, the greatest density is in the core region. Functional analysis of Per gene or c-Fos expression reveals that light-induced activation of SCN cells occurs first within the SCN core and later in the rest of the SCN, indicating a multistep propagation of a signal through the SCN (Fig. 1B and 4) (Yan and Silver 2002, 2004; Karatsoreos et al. 2004). The core is also a site of hormone action. In mouse, androgen receptors (AR) are concentrated in GRP cells of the core SCN, among other cell types, and SCN AR expression is modulated by circulating androgen (Karatsoreos et al. 2007).
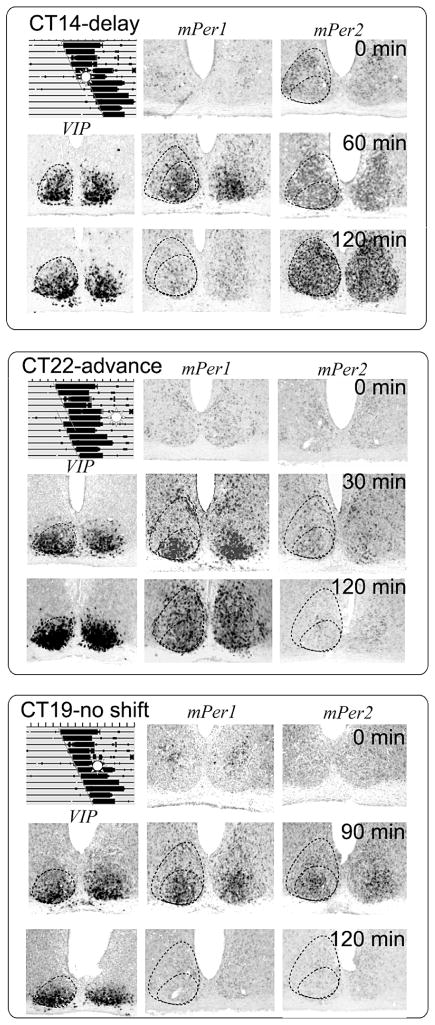
Figure 4.
The photic response is gated in SCN. (A) After a light pulse at CT14, mPer1 is induced only in the core SCN, whereas mPer2 is initially induced in the core and then spreads throughout the SCN. This is correlated with a behavioral phase delay. (B) After a light pulse at CT22, mPer1 is initially induced in the core and then spreads throughout the SCN. There is no mPer2 induction. This is correlated with a behavioral phase advance. (C) After a light pulse at CT19, both mPer1 and mPer2 are transiently induced in the core SCN, without spreading into the shell. This produces no phase-shift. (Modified, with permission, from Yan and Silver 2002 [Wiley-Blackwell].)
Intra-SCN connections: Core and shell specializations
There is very little work done on intra-SCN connections of specific neuronal phenotypes, as the effort is hampered by the small size of the nucleus and of its neurons. Although the phenotype of particular cellular elements is unclear, it is known from electron microscopy studies that the SCN contains large numbers of local circuit axons or local collaterals of long projection axons (van den Pol 1991). Some of these axons appear to be devoted to communication among cells of the SCN, whereas others are axon collaterals of neurons that project outside the SCN and also terminate within the SCN. Indeed, about 80% of synaptic endings within the SCN survive transection of long efferent connections, indicating that SCN axons form extensive axonal circuits within the SCN. The results of the three studies available on known peptidergic phenotypes are consistent with the foregoing. Morphologically, AVP neurons have few axon collaterals, most of which remain inside the boundaries of the SCN but occasionally project to SCN target areas. Their dendritic branches are compact and bear numerous varicosities (Fig. 2C) (Pennartz et al. 1998). In studies using similar methods to study GRP cells in mouse, we find that biocytin-filled GRP cells have axon projections and dendritic arbors in the shell, and GRP dendrites make appositions with AVP neurons and their processes (Fig. 2C) (Drouyer et al. 2005). Furthermore, these intra-SCN circuits are directional. Trans-synaptic labeling of neuronal circuits indicates dense projections from core to shell but little reciprocal innervation (Leak et al. 1999), and this directionality is also seen in biotin-filled core cells (Jobst et al. 2004; Drouyer et al. 2005).
Gap junctions
In addition to synaptic connections, gap junctions also contribute to intra-SCN circuits. In the rat SCN, cells are coupled (Jiang et al. 1997) either within dorsomedial or within ventrolateral SCN, but cells in one region are not coupled to cells in the other region (Colwell 2000). Data from our lab are consistent with these findings: The majority (67%) of GRP cells in mouse SCN are dye-coupled to other cells in the core (Drouyer et al. 2005). In connexin-36 (gap junction protein) knockout mice, behavioral circadian rhythmicity is damped and the onset of activity at the transition to darkness is delayed, suggesting an important functional role for electrical coupling among SCN neurons (Long et al. 2005).
SCN outputs
SCN efferents have been amply reviewed, and we focus here on the question of core/shell differences/similarities. SCN outputs are densest to adjacent hypothalamic nuclei, especially to the subparaventricular zone (SPVZ), the preoptic area (POA), and the dorsomedial hypothalamus (DMH) (Watts et al. 1987; Leak and Moore 2001; Kriegsfeld et al. 2004a), with limited projections to the forebrain, thalamus, and periaqueductal grey. It has been proposed that the former nuclei serve as relays for SCN projections to cell groups throughout the brain (Moore 1996; Deurveilher and Semba 2005). On the basis of track-tracing studies, it appears that core and shell SCN neurons project to all the same targets, although the relative densities of these projections differ (Leak and Moore 2001; Kriegsfeld et al. 2004a). For example, it seems that the medial SPVZ, DMH, and POA receive most of their input from the shell, whereas the lateral SPVZ receives most of its input from the core. This differential pattern of efferent connectivity permits distinct signals from core and shell to reach target sites in the brain.
Diffusible signals represent another SCN output. Polymer-encapsulated SCN fetal tissue grafts, producing diffusible output signals, are sufficient to restore behavioral but not endocrine rhythms (Fig. 1B) (Lehman et al. 1987; Silver et al. 1996; LeSauter and Silver 1998; Meyer-Bernstein et al. 1999). In addition, SCN2.2 cell lines induce rhythms of metabolic activity and clock gene expression in cocultured fibroblasts via a diffusible signal (Allen et al. 2001). A few candidate diffusible factors have been proposed including prokineticin 2 (Cheng et al. 2002; Prosser et al. 2007), transforming growth factor-α (TGF-α) (Kramer et al. 2001), and cardiotrophin-like cytokine (Kraves and Weitz 2006). It is not clear whether these diffusible signals emanate from core or shell, whether they are produced by particular subtypes of SCN cells, and whether additional diffusible factors remain to be identified.
Function of SCN Subregions
Distinct functions of core and shell
Use of immediate-early genes and clock genes as markers indicates that there are two fundamentally different responses of SCN neurons and that these are well aligned with “core” and “shell” compartments. In constant darkness (DD), rhythmic oscillation of Per1 and Per2 expression occurs in the shell (mouse, Shigeyoshi et al. 1997; rat, Yan et al. 1999; hamster, Hamada et al. 2001), but depending on the methods used, such oscillation is very low or not detectable in the core (hamster, Hamada et al. 2001; mouse, Karatsoreos et al. 2004). On the other hand, light-induced Per1 and Per2 expression is largely limited to the core SCN. Similarly, endogenous circadian rhythms of c-FOS and FRA-2 expression occur in the shell, but not in the core, whereas light-induced expression of c-FOS, FRA-2, and FOS-B occurs in the core, but not in the shell (Schwartz et al. 2000). These regional differences may be difficult to detect in some preparations as the VIP and GRP cells that delineate the core have very dense efferent projections that cover the entire SCN and extend beyond its borders (Fig. 3).
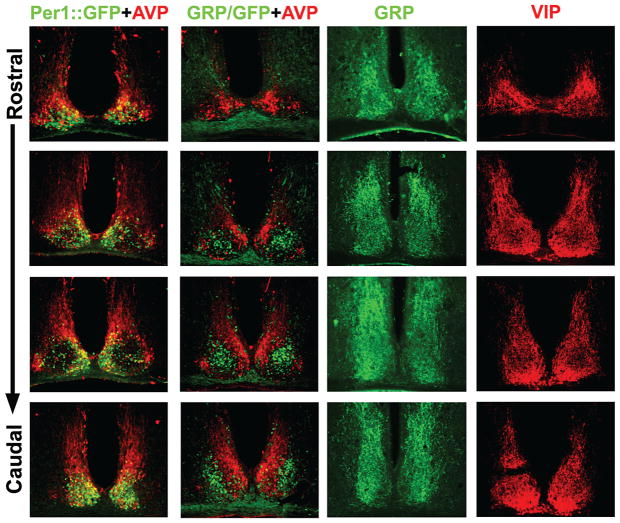
Figure 3.
Photomicrographs showing immunostaining of SCN sections through the rostrocaudal extent of the SCN. (Column 1) Both Per1-GFP (green) and AVP (red) are colocalized in the shell region; (column 2) GRP-GFP (green) cells occur in the core SCN but not in the shell region, delineated by AVP (red); (columns 3 and 4) GRP (column 3) and VIP (column 4) fibers project extensively throughout the SCN, obscuring visualization of GRP somata. In a GRP-GFP transgenic mouse, GRP cell bodies can be visualized. (Data in column 2 is taken, with permission, from Karatsoreos et al. 2004 [Society for Neuroscience].)
This core/shell difference is also seen in electrical activity rhythms. Both rat AVP cells (Schaap et al. 1999) and neurons lying in the AVP-rich region of mouse SCN (delineated by Per1-GFP) (Kuhlman and McMahon 2004) have a higher firing rate during the day than at night. In contrast, calbindin cells of the hamster core SCN (Jobst and Allen 2002) or GRP cells in a transgenic green fluorescent protein (GFP)-reporter mouse (Cloues et al. 2001) show no day/night rhythm in firing rate. In summary, at least some core cells are not detectably rhythmic in gene expression or electrical activity but are light responsive during the night. The light responsiveness of these cells is said to be “gated” (opened or closed) by the circadian clock because it occurs at night but not during the day. Although the gating mechanism is not known, the discovery of these cells sets the stage for exploring the functional importance of cellular diversity within the SCN (Figs. 1D and 3).
Role of core in SCN oscillation
Lesions of the SCN core region eliminate circadian rhythms of locomotor activity, drinking, gnawing, body temperature, and hormone secretion, even when substantial SCN tissue survives ablation (LeSauter and Silver 1999; Kriegsfeld et al. 2004b). The spared SCN tissue contains AVP cells, which are known cellular oscillators, capable of self-sustained oscillation in SCN slices (Noguchi et al. 2004) and even when cultured at low density in vitro (Murakami et al. 1991; Watanabe et al. 1993). In organotypic cultured SCN slices, surgical separation of the ventral and dorsal SCN region causes the neurons in the dorsal shell SCN to desynchronize, whereas the ventral core neurons retain coherent rhythmicity (Yamaguchi et al. 2003). These results suggest that cells of the SCN shell are not sufficient to maintain synchrony among the oscillator neurons and that core-to-shell communication is a requirement for rhythmicity of the tissue as a whole.
Several other lines of evidence support the suggestion that core cells are involved in synchronization and entrainment of SCN oscillators. Intercellular signaling through VIP and its receptor VPAC2 is critical in maintaining the synchrony of SCN cellular oscillators and circadian function within the SCN (Aton et al. 2005; Maywood et al. 2006). In addition, GRP cells are retinorecipient, they communicate with cells in the shell region through GRP receptors, and they function to keep SCN neurons synchronized during entrainment (McArthur et al. 2000; Aida et al. 2002; Karatsoreos et al. 2004; Antle et al. 2005).
Gating of light in the core
One of the fundamental features of circadian oscillation is that resetting by environmental signals depends on the phase at which the stimuli are applied. Thus, a light pulse applied in the early night (or early subjective night) produces phase-delays, whereas the same stimulus presented during the late night (or late subjective night) produces phase-advances. A light pulse applied during the day (or subjective day) typically has little or no effect. This circadian gating of responsiveness (or sensitivity) to photic stimulation is a central feature of all current models of entrainment, yet the underlying mechanisms are not known.
Several lines of evidence suggest that diurnal and circadian gating occur at the level of SCN neurons. Electrical stimulation of the RHT causes light-like changes in the circadian system with phase shifts at night but not during the day in both behavior (de Vries et al. 1994) and electrical activity in vitro (Shibata and Moore 1993). Similarly, light-induced clock gene expression occurs at night or subjective night but not during subjective day (Shigeyoshi et al. 1997). It appears that part of the gating mechanism involves N-methyl-D-aspartic acid (NMDA) receptors on SCN neurons. Application of NMDA causes light-like phase-shifts of the circadian rhythm in neuronal activity in the SCN in vitro (Ding et al. 1994; Shibata et al. 1994). The NMDA component of the evoked excitatory response is larger at night than during the day (Pennartz et al. 2001), and the circadian system gates the magnitude of NMDA-induced currents and Ca2+ transients (Colwell 2001). A hindrance in understanding these gated responses lies in our ignorance of which SCN neurons are involved.
Calbindin-containing cells of the hamster SCN receive direct RHT input (Bryant et al. 2000) and may participate in the gating of photic input. There is a circadian rhythm of subcellular localization of calbindin; whereas the protein is detected at all times in the cytoplasm, it is low or absent in the nucleus during the night. Under normal circumstances, light-induced behavioral phase-shifts and Period (Per) gene expression in the SCN occur only during the subjective night. Surprisingly, after administration of calbindin antisense oligodeoxynucleotides, both behavioral phase-shifts and light-induced Per are blocked during the subjective night and enhanced during the subjective day (Hamada et al. 2003). This evidence suggests that in hamsters, calbindin-containing cells of the SCN may participate in photic gating.
The core is important in gating-light responsiveness, but information from photic signals must reach the shell if phase-shifts are to occur. After an acute light exposure, induction of the clock genes Per1 and Per2 initially occurs in the ventrolateral/core SCN and then spreads into the dorsomedial/shell region, with the exact pattern depending on the timing of light exposure (Yan and Silver 2002). During the delay zone of the phase-response curve, a light pulse induces both mPer1 and mPer2 in the core, but only mPer2 expression spreads to the shell. In contrast, during the advance zone of the phase-response curve, when light causes advances of behavioral rhythms, a light pulse induces mPer1 in the core which then spreads to the shell, but there is no significant induction of mPer2 (Fig. 4).
SCN Plasticity
Whereas the foregoing describes the SCN response to a light pulse, patterns of gene expression within the SCN are also altered by exposure to various other lighting conditions, including long versus short days, constant light, 12-hour days, and other artificial lighting conditions. These phenomena point to the nature of the SCN circuitry underlying plasticity.
Spatiotemporal changes in various photoperiods
Regulation of clock gene expression and neuronal activity in the SCN is dependent on the photoperiod (for review, see Sumova et al. 2004; Johnston 2005). The durations of elevated clock gene expression and neuronal activity are expanded in long photoperiods but compressed in short photoperiods. Electrophysiological data show that in freely moving mice, rhythms in SCN multiunit activity (MUA) (high during subjective day) become compressed in short days and more distributed in long days, and these changes persist even after animals have been transferred into constant darkness (VanderLeest et al. 2007). Corresponding changes are observed in vitro in SCN slices taken from animals under these conditions (Mrugala et al. 2000; Inagaki et al. 2007; VanderLeest et al. 2007).
A model of two separate, but mutually coupled, circadian oscillators has been proposed to explain photoperiodic responses of behavioral rhythms in nocturnal rodents: an evening oscillator, which drives the activity onset and entrains to dusk, and a morning oscillator, which drives the end of activity and entrains to dawn (Pittendrigh and Daan 1976). Measurements of clock gene expression indicate that circadian oscillation in the posterior SCN is phase-locked to the end of activity, whereas oscillations of some cells in the anterior SCN are phase-locked to the onset of activity (Hazlerigg et al. 2005; Johnston 2005; Inagaki et al. 2007).
Photoperiod also produces changes in the SCN core, acting on calbindin expression in the core of hamster SCN. Calbindin expression is negatively correlated with the day length: The number of calbindin-immunopositive neurons and calbindin mRNA levels are markedly increased in hamsters exposed to short photoperiods (light:dark, 6:18). Thus, calbindin neurons may be involved in the encoding of seasonal information by the SCN (Menet et al. 2003).
Spatiotemporal changes in response to constant light
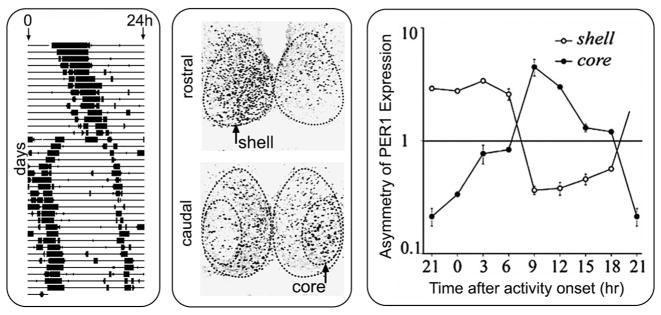
In hamsters, constant light causes “splitting” of circadian rhythms, such that a single daily bout of activity separates into two components, 12 hours apart (Pittendrigh and Daan 1976), with antiphase circadian oscillations of gene expression in the left and right SCN (de la Iglesia et al. 2000). Given the functional heterogeneity of the SCN, in which ventrolateral but not dorsomedial neurons are retinorecipient, we studied how the two compartments respond to the constant lighting conditions that produce splitting. In the rostral and mid level of the hamster SCN, a region containing AVP cells, levels of FOS and PER1 expression are high in one unilateral SCN and low in the other, as previously reported (de la Iglesia et al. 2000). In addition, we found a novel network of oscillators in the core SCN that was not recognized by previous studies of split animals. In a core subregion of the caudal SCN, FOS and PER1 expression are high at phases when expression levels in rostral and mid SCN (on the same side) are low (Fig. 5) (Yan et al. 2005); i.e., the core and the shell regions within each SCN oscillate in antiphase. This suggests that the two antiphase oscillators postulated to underlie splitting (Daan and Berde 1978) may be the product of a new pattern of coherent rhythmicity in a newly emergent SCN network.
Figure 5.
Constant light (LL) produces behavioral “splitting” (A) in which the animal’s single daily bout of activity separates into two components, and the two component free-run until they become stably coupled 12 hours apart. LL also induces antiphase oscillation within each side of the SCN (B). PER1 is not uniform on either side of the SCN, but it is high in the left shell and low in the left core. In contrast, PER1 is low in the right shell and high in the right core. The asymmetry of the bilateral core and shell SCN was quantified in split animals killed every 3 hours (C). The y axis is in log scale. Error bars indicate S.E. (Modified, with permission, from Yan et al. 2005 [Society for Neuroscience].)
Spatiotemporal changes in response to bimodal (7:5:7:5) light/dark cycles
Desynchronized oscillations in different subregions of the SCN have also been reported under other artificial lighting conditions. Bimodal (LDLD) lighting cycles, in which there are two light periods and two dark periods in a 24-hour cycle, induce and entrain split circadian activity rhythms in rodents (Gorman and Elliott 2003). Under such a lighting cycle (LDLD 7:5:7:5), the mPer1 level in the core SCN is higher than in the shell during one of the two light periods (morning photophase, M) and lower in the other light period (evening photophase; E) (Watanabe et al. 2007). In contrast, the mBmal1 level in the core SCN is lower than in the shell in the M phase and higher in the E phase. In yet another variation on this theme, when housed in an 11:11-hour LD cycle, the ventral and dorsal regions of the rat SCN show separate circadian oscillations in the expression of clock genes, with different periods, that reflect two desynchronized components of locomotor activity (de la Iglesia et al. 2004).
Gates and Oscillators in the SCN
The discovery that the SCN is composed of functionally distinct cells, differentiated by their patterns of clock gene expression, forces a paradigm shift in understanding the cellular basis of plasticity and coherent rhythmicity of the master circadian pacemaker. The observation that SCN rhythmicity is not uniform or fixed, but it is plastic in the face of environmental challenges, begs the question of its circuit organization. We now consider how SCN core cells maintain phase synchronization in the oscillator tissue. Importantly, in the absence of some mechanism of synchronization, oscillator cells with different periods will drift out of phase with each other, resulting in a loss of circadian rhythm for the SCN as a whole, despite the continuing oscillation of individual cells. Initial work on this problem centered on the idea that the cells could maintain synchronization through direct cell-to-cell coupling (Liu et al. 1997; Kunz and Achermann 2003; Bernard et al. 2007) or global coupling through a diffusible secreted factor (Garcia-Ojalvo et al. 2004; Gonze et al. 2005). However, the fact that the animal does not sustain circadian rhythms when the SCN core is ablated (LeSauter and Silver 1999; Kriegsfeld et al. 2004b) suggests that such direct coupling among the oscillator cells, if it has a role in synchronization, is not sufficient by itself to maintain synchronization.
Antle et al. (2003, 2007) proposed an alternative model for the maintenance of synchronization in SCN tissue, which gives a central role to the core cells acting as a “gate.” The idea is that the core cells under certain conditions send a phase-resetting signal to the shell oscillators. This signal retards the phase of shell oscillators with advanced phase and advances the phase of shell oscillators with retarded phases, resulting in a reduction in the phase dispersion of the shell oscillators. The resetting signal in this model is triggered either by light or by a feedback signal (direct or indirect) from the circadian system. Computer simulations demonstrate the effectiveness of this gating mechanism in maintaining shell oscillator synchronization and show that the gating model is capable of explaining both entrainment of the SCN to daily light variations (when the gate is triggered by light) and free-running circadian behavior under conditions of constant darkness (when the gate is triggered by feedback from the shell). These simulations also produce phase-response curves to simulated light pulses that are similar to those observed in animals. The model makes the testable prediction that in the absence of the core, the shell oscillators will drift out of phase, much as is seen in peripheral tissues.
Spatiotemporal Oscillations in the SCN
Phase dispersion of SCN oscillators
It is not the case that in vivo, the SCN as a whole has a single phase of clock gene expression (Fig. 6). Instead, each cellular SCN oscillator has a characteristic phase, and the individual oscillators are temporally and spatially organized within the nucleus. During a circadian cycle in vivo, the clock genes Per1 and Per2 are first expressed in cells lying in the dorsomedial, periventricular part of the SCN (near the third ventricle) and then spread out to the central region and finally to the ventral region (Yan and Okamura 2002; Quintero et al. 2003; Yamaguchi et al. 2003). Although the majority of the SCN cellular oscillators (~50–60%) have similar phases, others do not (Quintero et al. 2003; Yamaguchi et al. 2003). Application of the protein synthesis inhibitor cycloheximide (CHX) stops the molecular clock and sets the SCN neurons to the same phase. However, once the CHX is removed, the cells gradually reestablish their original phase relationships (Yamaguchi et al. 2003). This suggests that the phase relationships among the cellular oscillators are an intrinsic property of SCN circuitry.
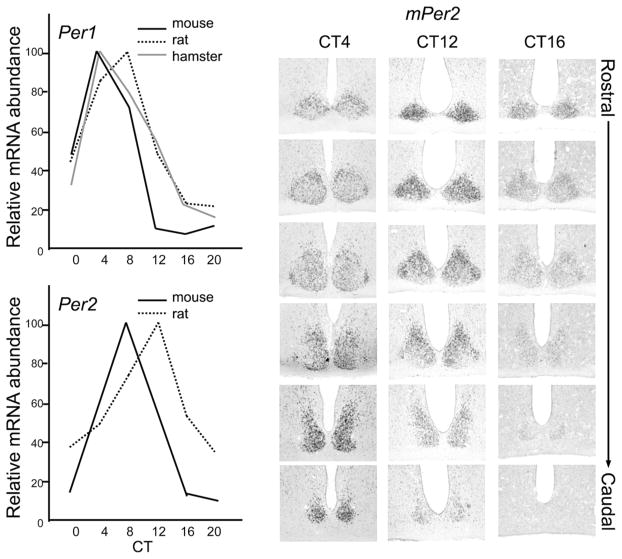
Figure 6.
SCN organization: Temporal and spatial changes in clock gene expression. (A) Temporal profile of clock genes Per1 (A) and Per2 (B) in the SCN (mouse, Takumi et al. 1998; rat, Yan et al. 1999; hamster, Yamamoto et al. 2001). (B) Spatial changes of mPer2 expression in mouse SCN. Representative photomicrographs showing digoxigenin in situ hybridization signals of mPer2 at CT4, CT12, and CT16 from the rostral to caudal end of the SCN. At CT4, mPer2 expression level is moderate in the rostral half and strong in the caudal half of the SCN. At CT12, mPer2 expression is strong in the rostral half and moderate in the caudal half. At CT16, mPer2 is moderate in the rostral half but very low in the caudal half of the SCN. This indicates that the SCN oscillators are spatially and temporally organized.
To examine more precisely the spatiotemporal dynamics of SCN oscillations, we performed bioluminescence imaging of coronal SCN slices cultured from PER2::LUC mice, in which the endogenous Per2 gene is replaced by a fusion of Per2 with the firefly luciferase reporter (Yoo et al. 2004; Liu et al. 2007). This permitted real-time longitudinal monitoring of rhythmic expression of the PER2::LUC fusion protein, with high spatial and temporal resolution (~1 μm, 30 minutes), in various subregions of the SCN. Our most rostral or caudal slices lacked an obvious core in the bioluminescence images (n = 2). In these “core-less” slices, PER2::LUC expression was rhythmic but with low amplitude, and the relative phasing of expression among SCN regions was not consistent over time.
In the coreless slice shown in Figure 7A, bioluminescence is initially elevated in the dorsal, central, and dorsomedial SCN compared to other regions. Bioluminescence then expands to the whole SCN slice before regressing back to the dorsal, central, and dorsomedial regions. In the other core-less slice (not shown), at the trough time, there is more bioluminescence in the center of the SCN than elsewhere; this increases and expands to the whole SCN slice and then regresses back to the center. These data are in accordance with the Per1-luc data of Yamaguchi et al. (2003), who found that dorsal cells were no longer synchronous when isolated from the ventral core SCN by a knife cut.
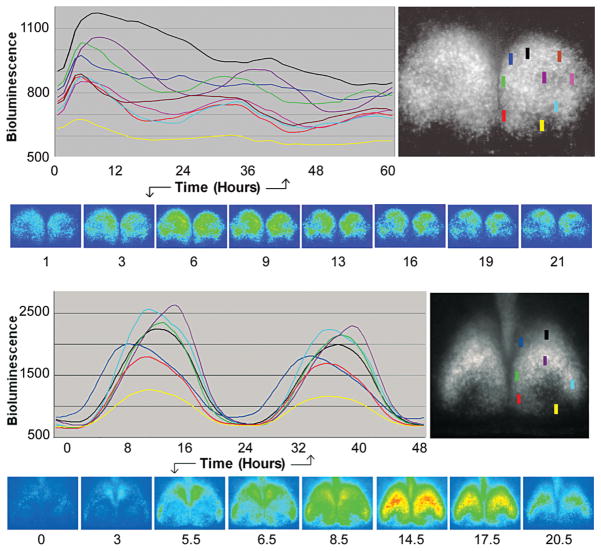
Figure 7.
Spatiotemporal order of PER2::LUC bioluminescence in cultured SCN slices of a wild-type mouse. The time course of these changes, based on measurements of small SCN regions (colored rectangle, each 15 × 50 μm), can be seen in the colored line drawings and visualized in the associated image of slices. (A) Slice in the rostral SCN lacking a core; (B) slice containing both a shell and a core. The presence of the core SCN within the slice is based on the shape of the nucleus and optic tract and on the presence of a region of low-amplitude or nondetectable clock gene expression.
In our other SCN slices, which contained both core and shell (n = 3), we observed strong PER2::LUC rhythms in all subregions, with relative phases that remained consistent over several circadian cycles and across all slices (Fig. 7B). Previous studies have also found differential phasing of cells in SCN slices. Quintero et al. (2003), using a Per1-GFP reporter in acute slices, found that lateral cells tended to peak earlier than medial cells. Yamaguchi et al. (2003), using a Per1-luc reporter in cultured slices, found a “dorsomedial-to-ventral spread” of activation. Careful examination of our high-resolution data, using a PER2::LUC reporter in cultured slices, revealed a more complex pattern. In each circadian cycle, PER2::LUC bioluminescence appears first in a small dorsomedial region (dark blue) near the third ventricle and then in lateral and ventromedial SCN (light blue, red). These three regions of early activation delineate the most peripheral aspects of the SCN shell, the “outer shell.” This is followed by a modest but widespread activation throughout the SCN (with greater intensity in the shell than in the core) and, finally, a stronger activation of the central “inner shell,” or “cap” (purple), just dorsal to the core. Overall, the weakest activation is in the core (yellow). While activation is peaking in the cap (purple), other regions are gradually declining in brightness. In contrast to this complex pattern of activation, the deactivation phase is less complex and appears as a contraction toward the central cap, which is the last area showing bioluminescence. However, we also observed a striking relationship between the time of peak activation of these regions and their rate of deactivation: PER2::LUC expression declines most gradually in the early-peaking outer shell and most steeply in the late-peaking cap. In summary, there appears to be a complex wave of PER2::LUC activation in a series of steps emanating from the most dorsomedial SCN, spreading ventrally in the outer shell, and then centrally to the inner shell (cap); deactivation is simpler and entails a contraction towards the central cap.
Longitudinal in vitro studies using luminescent reporters thus nicely complement our extensive cross-sectional in vivo studies. The convergence of these studies indicates that despite its relatively weak circadian oscillations, the core region is important for maintaining an organized pattern of synchronized oscillations in SCN tissue, which has complex spatiotemporal dynamics.
Space and Time in SCN Circuits: Tides and Waves
The emergent property of coherent oscillation in SCN tissue is not reducible to its cellular elements alone. Mathematical models of coupling in multioscillator systems provide the basis for understanding coherent oscillation of SCN cellular oscillators (Kuramoto 1975) but lack a spatial dimension. The experimental evidence points to complex highly reproducible spatiotemporal patterns of SCN activity on a circadian basis. The gate oscillator model treats this problem at a high level of aggregation, distinguishing only the two main functional SCN compartments (Antle et al. 2003, 2007). The phase-resetting function used to represent the interaction of the gate and the oscillators summarizes what is a complex system of cell-level interactions. In particular, the phase-resetting function of the gating model cannot represent the temporal changes in individual oscillator states with respect to their connections in space, i.e., the spatiotemporal dimension of the SCN between core and shell and within the shell. Anatomically detailed studies of the SCN make it obvious that there is an orderly spatial pattern of activity over the circadian cycle. Depending on which data one examines, it appears that the spatial change can be described as either a wave (Figs. 6 and 7) (Yan and Okamura 2002) or a tide (Figs. 7 and 8) (Hamada et al. 2004).
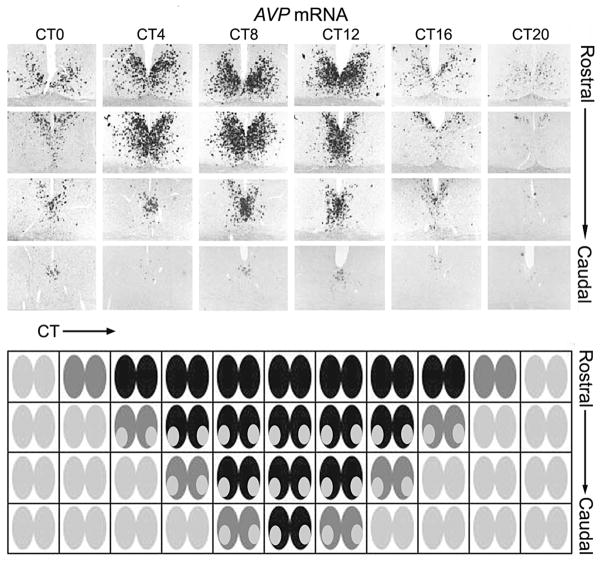
Figure 8.
In vivo evidence and tidal model of the SCN oscillator organization. (A) AVP mRNA expression in hamster SCN detected by digoxigenin in situ hybridization. (B) Model showing expanding tide of a signal that lasts longer in the dorsal aspect than in the ventral aspect of the SCN. (A, Modified, with permission, from Hamada et al. 2004 [Wiley-Blackwell].)
Waves and tides are significantly different. A wave begins with the activation of one region of oscillator cells, which continue their cycles as the adjacent region activates, continuing progressively to the most distal region. In a wave, cells in the first activated region are the first to deactivate, and all regions remain active for the same proportion of the cycle. In contrast, in a tide, the population of cells in the first activated region are the last to deactivate, so that this area is active for a longer proportion of the cycle than are other areas. Waves and tides imply different underlying mechanisms of interoscillator interaction and require different mathematical models. The distinction between waves and tides is important because it reveals the spatiotemporal organization of the SCN oscillation, shows how its functional connectivity arises from the SCN’s circuitry, and imposes constraints on the analysis of SCN data derived from slices and single cells.
One way to visualize and model the spatial changes in the SCN is in the expression of AVP mRNA. The data shown in Figure 8A indicate that expression starts in one region of the SCN tissue, which activates neighboring cells, and these in turn activate their neighbors. But the region of initial activity does not turn off, as one would expect if those cells continued to advance their phase in a regular fashion, but remains active while the tide of activity spreads to the whole SCN. In fact, it is the last activated regions of the SCN that are the first to turn off, so that the region of activity retreats (like a tide) back to the initially active region, which is the last to turn off. A similar tide of expression occurs in PER::LUC expression in the SCN slice and is most obvious in the regression of bioluminescence to the “cap” (Fig. 7B). A testable implication of this pattern is that the population of cells in the initiating region remain longer in the expressing phase of their cycle than do the last cells to be activated. These observations suggest a simple, conceptual model of the circadian oscillation of the SCN, based on the tidal rather than the wave approach. The gate (triggered either by light or by a feedback signal) sends a (excitatory or inhibitory) signal to the oscillator cells. In the case of an excitatory signal, the gate triggers the first group of oscillator cells, which stimulates their expression. The first group of oscillator cells stimulates the next to express AVP mRNA, and so on, continuing out to the furthest SCN region. But the expression of mRNA in the second group of cells appears to slow down the oscillation of the first, so that the first continues to express AVP mRNA until the second group stops expressing AVP mRNA. The last group of cells have no neighbors to activate, nor to slow down their expression, so they turn off first, allowing the next-to-last group of cells to turn off. In this way, the AVP mRNA expression recedes back to the start point. If the effect of a downstream activated group of cells is to slow, but not stop, oscillation in the upstream cells, over a long “day,” the initial group of cells might actually start to turn off before the ebbing tide reaches them.
Although there is no direct evidence for this type of inhibition by downstream cells, an interaction of this kind could explain the observed tidal pattern of activation of the shell. Our preliminary investigation shows that it is possible to create simple “cellular automata” that oscillate over space in a tidal-like pattern similar to the SCN (Fig. 8B). These models require that the cells be oscillators, but the connectivity of the system distorts the shapes of their oscillations. The point is that the tidal mechanism needs to know whether the upstream cells are starting up” or “closing down” in order to function. Because the model represents connections as a directed graph, it is extremely flexible and scalable. It is possible to represent tissue with many connections in complex three-dimensional configurations with this approach. One prediction from this conceptual model is that bioluminescence imaging of a sagittal section that retains the connectivity of the tissue in the rostrocaudal axis will reveal a tidal pattern of cell activation, rather than wavelike patterns. Another testable prediction is that this tide might be the product of increased dispersion of phase in cells of one region compared to another; alternatively, it could be the result of some cells having a longer duration of gene expression than others. The focus of future research will be the calibration of this model to experimentally observed spatiotemporal changes in the SCN.
OVERVIEW AND CONCLUSIONS
On the basis of anatomical, functional, and modeling studies, we propose that SCN circuit organization is key to understanding oscillator synchrony and that the connectivity among the functionally different neurons accounts for the coherent circadian expression pattern of the nucleus and its role as a master pacemaker. The analyses of spatial and temporal changes in SCN point to an important role for the core SCN region and indicate that the daily cycle of SCN activity involves a signal that spreads and then retracts in a wave or tidal pattern, such that the the “on” time of various oscillators is regionally organized. We further point to an important role of cells in the core area for maintaining the orderly pattern of activity and hypothesize that changes in the phase relationships among oscillator networks underlie the ability of the SCN to measure time and to respond in a robust manner to its internal and external environment.
Acknowledgments
This work was supported by National Institutes of Health grants NS37919 and MH075045 to R.S. and K08 MH067657 to D.K.W. We are grateful to Tina Tong and David Vernon for assistance in preparation of the data analysis and manuscript.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol. 2002;61:26. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Allen G, Rappe J, Earnest DJ, Cassone VM. Oscillating on borrowed time: Diffusible signals from immortalized suprachiasmatic nucleus cells regulate circadian rhythmicity in cultured fibroblasts. J Neurosci. 2001;21:7937. doi: 10.1523/JNEUROSCI.21-20-07937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Silver R. Orchestrating time: Arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Antle MC, Kriegsfeld LJ, Silver R. Signaling within the master clock of the brain: Localized activation of mitogen activated protein kinase by gastrin-releasing peptide. J Neurosci. 2005;25:2447. doi: 10.1523/JNEUROSCI.4696-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: A network model of the brain clock. J Biol Rhythms. 2003;18:339. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Foley NC, Foley DK, Silver R. Gates and oscillators. II. Zeitgebers and the network model of the brain clock. J Biol Rhythms. 2007;22:14. doi: 10.1177/0748730406296319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S, Gonze D, Cajavec B, Herzel H, Kramer A. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput Biol. 2007;3:e68. doi: 10.1371/journal.pcbi.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DN, LeSauter J, Silver R, Romero MT. Retinal innervation of calbindin-D28K cells in the hamster suprachiasmatic nucleus: Ultrastructural characterization. J Biol Rhythms. 2000;15:103. doi: 10.1177/074873040001500204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Moore RY. The suprachiasmatic nucleus of the golden hamster: Immunohistochemical analysis of cell and fiber distribution. Neuroscience. 1984;13:415. doi: 10.1016/0306-4522(84)90240-9. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- Chklovskii DB, Koulakov AA. Maps in the brain: What can we learn from them? Annu Rev Neurosci. 2004;27:369. doi: 10.1146/annurev.neuro.27.070203.144226. [DOI] [PubMed] [Google Scholar]
- Cloues RK, Silver R, Sather WA. Abstract Viewer/Itinerary Planner. San Diego: Society for Neuroscience; 2001. Electrophysiological characterization of neurons expressing Calbindin-D28k in the mouse suprachiasmatic nucleus. Program No. 180.3; p. 2001. (online) [Google Scholar]
- Colwell CS. Rhythmic coupling among cells in the suprachiasmatic nucleus. J Neurobiol. 2000;43:379. doi: 10.1002/1097-4695(20000615)43:4<379::aid-neu6>3.0.co;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. NMDA-evoked calcium transients and currents in the suprachiasmatic nucleus: Gating by the circadian system. Eur J Neurosci. 2001;13:1420. doi: 10.1046/j.0953-816x.2001.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Berde C. Two coupled oscillators: Simulations of the circadian pacemaker in mammalian activity rhythms. J Theor Biol. 1978;70:297. doi: 10.1016/0022-5193(78)90378-8. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Meyer J, Carpino A, Jr, Schwartz WJ. Antiphase oscillation of the left and right suprachiasmatic nuclei. Science. 2000;290:799. doi: 10.1126/science.290.5492.799. [DOI] [PubMed] [Google Scholar]
- de Vries MJ, Treep JA, de Pauw ESD, Meijer JH. The effects of electrical stimulation of the optic nerves and anterior optic chiasm on the circadian activity rhythm of the Syrian hamster: Involvement of excitatory amino acids. Brain Res. 1994;642:206. doi: 10.1016/0006-8993(94)90923-7. [DOI] [PubMed] [Google Scholar]
- Deurveilher S, Semba K. Indirect projections from the suprachiasmatic nucleus to major arousal-promoting cell groups in rat: Implications for the circadian control of behavioural state. Neuroscience. 2005;130:165. doi: 10.1016/j.neuroscience.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Ding JM, Chen D, Weber ET, Faiman LE, Rea MA, Gillette MU. Resetting the biological clock: Mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266:1713. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- Drouyer E, LeSauter J, Silver R. Abstract Viewer/Itinerary Planner. Washington, D.C: Society for Neuroscience; 2005. Phenotype matters: Anatomical characteristics of gastrin-releasing peptide cells of the suprachiasmic nucleus. Program No. 312.9; p. 2005. (online) [Google Scholar]
- Eskin A. Identification and physiology of circadian pacemakers. Introduction. Fed Proc. 1979;38:2570. [PubMed] [Google Scholar]
- Garcia-Ojalvo J, Elowitz MB, Strogatz SH. Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing. Proc Natl Acad Sci. 2004;101:10955. doi: 10.1073/pnas.0307095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonze D, Bernard S, Waltermann C, Kramer A, Herzel H. Spontaneous synchronization of coupled circadian oscillators. Biophys J. 2005;89:120. doi: 10.1529/biophysj.104.058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MR, Elliott JA. Entrainment of 2 subjective nights by daily light:dark:light:dark cycles in 3 rodent species. J Biol Rhythms. 2003;18:502. doi: 10.1177/0748730403260219. [DOI] [PubMed] [Google Scholar]
- Green DJ, Gillette R. Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 1982;245:198. doi: 10.1016/0006-8993(82)90361-4. [DOI] [PubMed] [Google Scholar]
- Groos G, Hendriks J. Circadian rhythms in electrical discharge of rat suprachiasmatic neurones recorded in vitro. Neurosci Lett. 1982;34:283. doi: 10.1016/0304-3940(82)90189-6. [DOI] [PubMed] [Google Scholar]
- Hakim H, DeBernardo AP, Silver R. Circadian locomotor rhythms, but not photoperiodic responses, survive surgical isolation of the SCN in hamsters. J Biol Rhythms. 1991;6:97. doi: 10.1177/074873049100600201. [DOI] [PubMed] [Google Scholar]
- Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:1741. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: Rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci. 2001;21:7742. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Lokshin M, Romero MT, Yan L, Venuti JM, Silver R. Calbindin influences response to photic input in suprachiasmatic nucleus. J Neurosci. 2003;23:8820. doi: 10.1523/JNEUROSCI.23-26-08820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwich M, Kalsbeek A, Pevet P, Nurnberger F. Effects of illumination and enucleation on substance-P-immunoreactive structures in subcortical visual centers of golden hamster and Wistar rat. Cell Tissue Res. 1994;277:351. doi: 10.1007/BF00327783. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Herzog ED. Clock genes, oscillators, and cellular networks in the suprachiasmatic nuclei. J Biol Rhythms. 2004;19:400. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- Hazlerigg DG, Ebling FJ, Johnston JD. Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr Biol. 2005;15:R449. doi: 10.1016/j.cub.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Hofman MA, Swaab DF. The sexually dimorphic nucleus of the preoptic area in the human brain: A comparative morphometric study. J Anat. 1989;164:55. [PMC free article] [PubMed] [Google Scholar]
- Ibata Y, Takahashi Y, Okamura H, Kawakami F, Terubayashi H, Kubo T, Yanaihara N. Vasoactive intestinal peptide (VIP)-like immunoreactive neurons located in the rat suprachiasmatic nucleus receive a direct retinal projection. Neurosci Lett. 1989;97:1. doi: 10.1016/0304-3940(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci. 2007;104:7664. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye SIT, Kawamura H. Persistence of circadian rhythmicity in a hypothalamic ‘island’ containing the suprachiasmatic nucleus. Proc Natl Acad Sci. 1979;76:5962. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye ST, Shibata S. Neurochemical organization of circadian rhythm in the suprachiasmatic nucleus. Neurosci Res. 1994;20:109. doi: 10.1016/0168-0102(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Jiang ZG, Yang YQ, Allen CN. Tracer and electrical coupling of rat suprachiasmatic nucleus neurons. Neuroscience. 1997;77:1059. doi: 10.1016/s0306-4522(96)00539-8. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic nucleus. Cell. 1999;96:57. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Jobst EE, Allen CN. Calbindin neurons in the hamster suprachiasmatic nucleus do not exhibit a circadian variation in spontaneous firing rate. Eur J Neurosci. 2002;16:2469. doi: 10.1046/j.1460-9568.2002.02309.x. [DOI] [PubMed] [Google Scholar]
- Jobst EE, Robinson DW, Allen CN. Potential pathways for intercellular communication within the calbindin subnucleus of the hamster suprachiasmatic nucleus. Neuroscience. 2004;123:87. doi: 10.1016/j.neuroscience.2003.08.059. [DOI] [PubMed] [Google Scholar]
- Johnston JD. Measuring seasonal time within the circadian system: Regulation of the suprachiasmatic nuclei by photoperiod. J Neuroendocrinol. 2005;17:459. doi: 10.1111/j.1365-2826.2005.01326.x. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Wang A, Sasanian J, Silver R. A role for androgens in regulating circadian behavior and the suprachiasmatic nucleus. Endocrinology. 2007 doi: 10.1210/en.2007-0775. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: Identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci. 2004;24:68. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DC, Moore RY, Reppert SM, editors. The mind’s clock. Oxford University Press; New York: 1991. Suprachiasmatic nucleus. [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9:212. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- Kriegsfeld LJ, LeSauter J, Silver R. Targeted microlesions reveal novel organization of the hamster suprachiasmatic nucleus. J Neurosci. 2004a;24:2449. doi: 10.1523/JNEUROSCI.5323-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegsfeld LJ, Leak RK, Yackulic CB, LeSauter J, Silver R. Organization of suprachiasmatic nucleus projections in Syrian hamsters (Mesocricetus auratus): An anterograde and retrograde analysis. J Comp Neurol. 2004b;468:361. doi: 10.1002/cne.10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, McMahon DG. Rhythmic regulation of membrane potential and potassium current persists in SCN neurons in the absence of environmental input. Eur J Neurosci. 2004;20:1113. doi: 10.1111/j.1460-9568.2004.03555.x. [DOI] [PubMed] [Google Scholar]
- Kunz H, Achermann P. Simulation of circadian rhythm generation in the suprachiasmatic nucleus with locally coupled self-sustained oscillators. J Theor Biol. 2003;224:63. doi: 10.1016/s0022-5193(03)00141-3. [DOI] [PubMed] [Google Scholar]
- Kuramoto A. Self-entrainment of a population of coupled nonlinear oscillators. Lect Notes Phys. 1975;39:420. [Google Scholar]
- Larsen PJ, Mikkelsen JD. The suprachiasmatic nucleus of the mink (Mustela vison): Apparent absence of vasopressin-immunoreactive neurons. Cell Tissue Res. 1993;273:239. doi: 10.1007/BF00312825. [DOI] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433:312. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Leak RK, Card JP, Moore RY. Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res. 1999;819:23. doi: 10.1016/s0006-8993(98)01317-1. [DOI] [PubMed] [Google Scholar]
- Lee HS, Nelms JL, Nguyen M, Silver R, Lehman MN. The eye is necessary for a circadian rhythm in the suprachiasmatic nucleus. Nat Neurosci. 2003;6:111. doi: 10.1038/nn1006. [DOI] [PubMed] [Google Scholar]
- Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, Bittman EL. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7:1626. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Silver R. Output signals of the SCN. Chronobiol Int. 1998;15:535. doi: 10.3109/07420529808998706. [DOI] [PubMed] [Google Scholar]
- LeSauter J, Silver R. Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J Neurosci. 1999;19:5574. doi: 10.1523/JNEUROSCI.19-13-05574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, III, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: Period determination in the suprachiasmatic nucleus. Cell. 1997;91:855. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Long MA, Jutras MJ, Connors BW, Burwell RD. Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat Neurosci. 2005;8:61. doi: 10.1038/nn1361. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- McArthur AJ, Coogan AN, Ajpru S, Sugden D, Biello SM, Piggins HD. Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro. J Neurosci. 2000;20:5496. doi: 10.1523/JNEUROSCI.20-14-05496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J, Vuillez P, Pevet P. Calbindin expression in the hamster suprachiasmatic nucleus depends on day-length. Neuroscience. 2003;122:591. doi: 10.1016/j.neuroscience.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Meyer-Bernstein EL, Jetton AE, Matsumoto SI, Markuns JF, Lehman MN, Bittman EL. Effects of suprachiasmatic transplants on circadian rhythms of neuroendocrine function in golden hamsters. Endocrinology. 1999;140:207. doi: 10.1210/endo.140.1.6428. [DOI] [PubMed] [Google Scholar]
- Miller JD, Morin LP, Schwartz WJ, Moore RY. New insights into the mammalian circadian clock. Sleep. 1996;19:641. doi: 10.1093/sleep/19.8.641. [DOI] [PubMed] [Google Scholar]
- Moga MM, Moore RY. Organization of neural inputs to the suprachiasmatic nucleus in the rat. J Comp Neurol. 1997;389:508. doi: 10.1002/(sici)1096-9861(19971222)389:3<508::aid-cne11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Moore RY. Retinohypothalamic projection in mammals: A comparative study. Brain Res. 1973;49:403. doi: 10.1016/0006-8993(73)90431-9. [DOI] [PubMed] [Google Scholar]
- Moore RY. Organization and function of a central nervous system circadian oscillator: The suprachiasmatic hypothalamic nucleus. Fed Proc. 1983;42:2783. [PubMed] [Google Scholar]
- Moore RY. Entrainment pathways and the functional organization of the circadian system. Prog Brain Res. 1996;111:103. doi: 10.1016/s0079-6123(08)60403-3. [DOI] [PubMed] [Google Scholar]
- Moore MM, Eichler VB. Loss of circadian adrenal corticosterone rhythm following suprachiasmic nucleus lesion in the rat. J Comp Neurol. 1972;146:1. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- Morin LP, Allen CN. The circadian visual system, 2005. Brain Res Rev. 2006;51:1. doi: 10.1016/j.brainresrev.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Morin LP, Blanchard J, Moore RY. Intergeniculate leaflet and suprachiasmatic nucleus organization and connections in the golden hamster. Vis Neurosci. 1992;8:219. doi: 10.1017/s095252380000287x. [DOI] [PubMed] [Google Scholar]
- Mosko SS, Moore RY. Neonatal suprachiasmatic nucleus lesions: Effects on the development of circadian rhythms in the rat. Brain Res. 1979;164:17. doi: 10.1016/0006-8993(79)90003-9. [DOI] [PubMed] [Google Scholar]
- Mrugala M, Zlomanczuk P, Jagota A, Schwartz WJ. Rhythmic multiunit neural activity in slices of hamster suprachiasmatic nucleus reflect prior photoperiod. Am J Physiol Regul Integr Comp Physiol. 2000;278:R987. doi: 10.1152/ajpregu.2000.278.4.R987. [DOI] [PubMed] [Google Scholar]
- Murakami N, Takamura M, Takahashi K, Utunomiya K, Kuroda H, Etoh T. Long-term cultured neurons from rat suprachiasmatic nucleus retain the capacity for circadian oscillation of vasopressin release. Brain Res. 1991;545:347. doi: 10.1016/0006-8993(91)91312-o. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Watanabe K, Ogura A, Yamaoka S. The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur J Neurosci. 2004;20:3199. doi: 10.1111/j.1460-9568.2004.03784.x. [DOI] [PubMed] [Google Scholar]
- Pennartz CM, Hamstra R, Geurtsen AM. Enhanced NMDA receptor activity in retinal inputs to the rat suprachiasmatic nucleus during the subjective night. J Physiol. 2001;532:181. doi: 10.1111/j.1469-7793.2001.0181g.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennartz CM, Bos NP, Jeu MT, Geurtsen AM, Mirmiran M, Sluiter AA, Buijs RM. Membrane properties and morphology of vasopressin neurons in slices of rat suprachiasmatic nucleus. J Neurophysiol. 1998;80:2710. doi: 10.1152/jn.1998.80.5.2710. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. Circadian rhythms and the circadian organization of living things. Cold Spring Harbor Symp Quant Biol. 1961;25:159. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Bruce VG. Daily rhythms as coupled oscillator systems and their relation to thermoperiodism and photoperiodism. In: Withrow RB, editor. Photoperiodism and related phenomena in plants and animals. AAAS; Washington, D.C: 1959. p. 475. [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Neurol. 1976;106:355. [Google Scholar]
- Prosser HM, Bradley A, Chesham JE, Ebling FJ, Hastings MH, Maywood ES. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci. 2007;104:648. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: A multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal S, Pasik P, Pasik T. Texture of the nervous system of man and the vertebrates. Springer; New York: 1999. [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Rosen GJ, de Vries GJ, Goldman SL, Goldman BD, Forger NG. Distribution of vasopressin in the brain of the eusocial naked mole-rat. J Comp Neurol. 2007;500:1093. doi: 10.1002/cne.21215. [DOI] [PubMed] [Google Scholar]
- Schaap J, Bos NP, de Jeu MT, Geurtsen AM, Meijer JH, Pennartz CM. Neurons of the rat suprachiasmatic nucleus show a circadian rhythm in membrane properties that is lost during prolonged whole-cell recording. Brain Res. 1999;815:154. doi: 10.1016/s0006-8993(98)01025-7. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Coleman RJ, Reppert SM. A daily vasopressin rhythm in rat cerebrospinal fluid. Brain Res. 1983;263:105. doi: 10.1016/0006-8993(83)91205-2. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Carpino A, Jr, de la Iglesia HO, Baler R, Klein DC, Nakabeppu Y, Aronin N. Differential regulation of fos family genes in the ventrolateral and dorsomedial subdivisions of the rat suprachiasmatic nucleus. Neuroscience. 2000;98:535. doi: 10.1016/s0306-4522(00)00140-8. [DOI] [PubMed] [Google Scholar]
- Shibata S, Moore RY. Neuropeptide Y and optic chiasm stimulation affect suprachiasmatic nucleus circadian function in vitro. Brain Res. 1993;615:95. doi: 10.1016/0006-8993(93)91118-c. [DOI] [PubMed] [Google Scholar]
- Shibata S, Oomura Y, Kita H, Hattori K. Circadian rhythmic changes in neuronal activity in the suprachiasmatic nucleus of the rat hypothalamic slice. Brain Res. 1982;247:154. doi: 10.1016/0006-8993(82)91041-1. [DOI] [PubMed] [Google Scholar]
- Shibata S, Watanabe A, Hamada T, Ono M, Watanabe S. N-methyl-D-aspartate induces phase shifts in circadian rhythm of neuronal activity of rat SCN in vitro. Am J Physiol. 1994;267:R360. doi: 10.1152/ajpregu.1994.267.2.R360. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Silver R, Sookhoo AI, LeSauter J, Stevens P, Jansen HT, Lehman MN. Multiple regulatory elements result in regional specificity in circadian rhythms of neuropeptide expression in mouse SCN. Neuroreport. 1999;10:3165. doi: 10.1097/00001756-199910190-00008. [DOI] [PubMed] [Google Scholar]
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity are eliminated by suprachiasmatic lesions. Proc Natl Acad Sci. 1972;54:1521. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumova A, Bendova Z, Sladek M, Kovacikova Z, Illnerova H. Seasonal molecular timekeeping within the rat circadian clock. Physiol Res. 2004;53(suppl 1):S167. [PubMed] [Google Scholar]
- Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, Yagita K, Yan L, Young ML, Okamura H. A light independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Hayashi S, Tamada Y, Ikeda T, Hisa Y, Takamatsu T, Ibata Y. Direct retinal projections to GRP neurons in the suprachiasmatic nucleus of the rat. Neuroreport. 1997;8:2187. doi: 10.1097/00001756-199707070-00020. [DOI] [PubMed] [Google Scholar]
- Tavakoli-Nezhad M, Schwartz WJ. c-Fos expression in the brains of behaviorally “split” hamsters in constant light: Calling attention to a dorsolateral region of the suprachiasmatic nucleus and the medial division of the lateral habenula. J Biol Rhythms. 2005;20:419. doi: 10.1177/0748730405278443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN. Glutamate and aspartate immunoreactivity in hypothalamic presynaptic axons. J Neurosci. 1991;11:2087. doi: 10.1523/JNEUROSCI.11-07-02087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN, Tsujimoto KL. Neurotransmitters of the hypothalamic suprachiasmatic nucleus: Immunocytochemical analysis of 25 neuronal antigens. Neuroscience. 1985;15:1049. doi: 10.1016/0306-4522(85)90254-4. [DOI] [PubMed] [Google Scholar]
- VanderLeest HT, Houben T, Michel S, Deboer T, Albus H, Vansteensel MJ, Block GD, Meijer JH. Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol. 2007;17:468. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Koibuchi N, Ohtake H, Yamaoka S. Circadian rhythms of vasopressin release in primary cultures of rat suprachiasmatic nucleus. Brain Res. 1993;624:115. doi: 10.1016/0006-8993(93)90067-w. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Naito E, Nakao N, Tei H, Yoshimura T, Ebihara S. Bimodal clock gene expression in mouse suprachiasmatic nucleus and peripheral tissues under a 7-hour light and 5-hour dark schedule. J Biol Rhythms. 2007;22:58. doi: 10.1177/0748730406295435. [DOI] [PubMed] [Google Scholar]
- Watts AG, Swanson LW, Sanchez-Watts G. Efferent projections of the suprachiasmatic nucleus. I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J Comp Neurol. 1987;258:204. doi: 10.1002/cne.902580204. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small world’ networks. Nature. 1998;393:440. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Williams RW, Herrup K. The control of neuron number. Annu Rev Neurosci. 1988;11:423. doi: 10.1146/annurev.ne.11.030188.002231. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Shigeyoshi Y, Ishida Y, Fukuyama T, Yamaguchi S, Yagita K, Moriya T, Shibata S, Takashima N, Okamura H. Expression of the Per1 gene in the hamster: Brain atlas and circadian characteristics in the suprachiasmatic nucleus. J Comp Neurol. 2001;430:518. [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur J Neurosci. 2002;16:1531. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Silver R. Resetting the brain clock: Time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur J Neurosci. 2004;19:1105. doi: 10.1111/j.1460-9568.2004.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Takekida S, Shigeyoshi Y, Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: Circadian profile and the compartment-specific response to light. Neuroscience. 1999;94:141. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- Yan L, Foley NC, Bobula JM, Kriegsfeld LJ, Silver R. Two antiphase oscillations occur in each suprachiasmatic nucleus of behaviorally split hamsters. J Neurosci. 2005;25:9017. doi: 10.1523/JNEUROSCI.2538-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCI-FERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci. 2004;101:5339. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]