Abstract
The mechanism whereby brief light exposure resets the mammalian circadian clock in a phase dependent manner is not known, but is thought to involve Per gene expression. At the behavioural level, a light pulse produces phase delays in early subjective night, phase advances in late subjective night, and no phase shifts in mid-subjective night or subjective day. To understand the relationship between Per gene activity and behavioural phase shifts, we examined light-induced mPer1 and mPer2 expression in the suprachiasmatic nucleus (SCN) of the mouse, in the subjective night, with a view to understanding SCN heterogeneity. In the VIP-containing region of the SCN (termed `core'), light-induced mPer1 expression occurs at all times of the subjective night, while mPer2 induction is seen only in early subjective night. In the remaining regions of the SCN (termed `shell'), a phase delaying light pulse produces no mPer1 but significant mPer2 expression, while a phase advancing light pulse produces no mPer2 but substantial mPer1 induction. Moreover, following a light pulse during mid-subjective night, neither mPer1 nor mPer2 are induced in the shell. The results reveal that behavioural phase shifts occur only when light-induced Per gene expression spreads from the core to the shell SCN, with mPer1 expression in shell corresponding to phase advances, and mPer2 corresponding to phase delays. The results indicate that the time course and the localization of light-induced Per gene expression in SCN reveals important aspects of intra-SCN communication.
Keywords: circadian rhythm; light pulse; mouse, suprachiasmatic nucleus
Introduction
In mammals, the suprachiasmatic nuclei (SCN) of the hypothalamus contain the master circadian clock that controls the timing of physiological and behavioural activities (van Esseveldt et al., 2000). The circadian clock has two distinct functions: generation of rhythms, and synchronization (or entrainment) of these rhythms to the environment. The molecular mechanism underlying rhythm generation involves interlocked positive and negative transcription/translation-based feedback loops, in which the expression of putative `clock genes' is suppressed periodically, by their protein products (Dunlap, 1999; Harmer et al., 2001; Reppert & Weaver, 2001).
Light is the most salient cue for the entrainment of the circadian clock. Light exposure during the subjective night produces phase shifts of locomotor activity rhythms (Pittendrigh & Daan, 1976). It is widely accepted that resetting of the mammalian circadian clock by light involves acute induction of Per1 and Per2 genes (Dunlap, 1999; Lowrey & Takahashi, 2000). However, the mechanisms whereby a light pulse causes phase delays in early subjective night and phase advances in late subjective night in mammals is not well understood. Also not yet established is the distinct role of per1 and per2 in this photic entrainment. Finally, there are discrepancies reported in the consequences of light pulses on Per expression. Thus, Albrecht et al. (1997) found that mper1, unlike mper2, is expressed throughout the SCN after exposure to light. On the other hand, Shigeyoshi et al. (1997) reported that mper1 induction is restricted to the ventral or core SCN. The goal of the present study was to delineate the cellular and the molecular mechanism mediating photic entrainment by examining the time course and localization of light-induced mper1 and mper2 expression in the mouse SCN. One hypothesis to be explored is that light-induced mper1 and mper2 expression initially occurs in a limited subregion of the SCN, and subsequently spreads to the remaining SCN. A related question is the determination of whether induction of clock genes in distinct SCN regions is phase-dependent and associated with the direction of the behavioural changes. To explore this relationship, we applied light pulses which resulted in either phase delays, phase advances or no phase shifts, and then examined mPer1 and mPer2 expression in the SCN, with attention to the regional distribution of these genes.
Materials and methods
Animals housing and experimental groups
Male C57BL/6 mice (Charles River Laboratories, Wilmington, MA) 5 weeks of age, were housed in a 12-h light : 12-h dark (LD) cycle (light, 300 lux) for 2–4 weeks, before being used in the experiment. Food and water were available ad libitum. A white noise generator (91dB sound pressure level) masked environmental noise.
For measurement of phase shifts in this strain of mice, wheel-running behaviour was monitored using DataQuest III (Data Science Int., St. Paul, MN) on a PC computer. The activity of the animals was consulted daily and data were analysed using Clocklab (Actimetrics, Evanston, IL). After 2 weeks in LD, mice (n = 16) were transferred to constant dark (DD). On the third day in DD, animals were exposed to a light pulse (900 lux, 30 min) at the delay phase – circadian time (CT) 14, the advance phase – CT22, or in the transition zone – CT19, 19.5 and 20 (Schwartz & Zimmerman, 1990). Each animal was tested at each of the 3 time zones, using a random order of light pulse application. Phase shifts of wheel-running activity were measured after a stable phase had been established (Daan & Pittendrigh, 1976).
For mRNA analysis, animals were divided into 3 groups, defined by the circadian time of light pulse administration. Animals were exposed to light at CT14, 19.5 or 22 on the third day in DD (as in the behavioural study). To characterize the time course of light-induced mper1 and mper2 mRNA expression, animals (n = 4/time point) were killed at 0, 30, 60, 90, 120, 180 and 240 min after the beginning of the light pulse. Control animals (n = 2/time point) were treated identically, but were not exposed to the light pulse. Mice were deeply anaesthetized (pentobarbital, 200 mg/kg) under a red safe light (< 1 lux, Delta 1, Dallas, TX), and perfused intracardially with 10 mL of autoclaved ice-cold saline and 20 mL of a fixative (4% paraformaldehyde in 0.1 m phosphate buffer (PB), pH 7.4). The brains were removed, postfixed for 16 h at 4 C, and cryoprotected in 0.1 m PB with 20% sucrose for 48 h.
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Columbia University.
In situ hybridization using Digoxigenin-labelled cRNA probes
The mper1, mper2 and VIP cDNA fragment-containing plasmids were the gift of Dr H. Okamura; Kobe University, Japan. The mper1, mper2 and VIP cDNA fragment-containing vectors were linearized with restriction enzymes and then used as templates for sense or antisense complementary RNA (cRNA) probes. Digoxigenin (Dig)-labelled probes for mper1, mper2 and VIP were made using Dig-UTP (Boehringer Mannheim, Mannheim, Germany) with a standard protocol for cRNA synthesis (Yan et al., 1999). Sense mper1, mper2 and VIP cRNA probes revealed no specific hybridization signals in brain sections.
Serial coronal sections (30 μm) were made from the rostral to the caudal end of the SCN using a cryostat (Reichert-Jung, Heidelburg, Germany). Every third section was collected for each of the following probes: mper1, mper2, and VIP. To minimize technical variations in the hybridization procedure, sections from each temporal series across different experimental conditions were processed simultaneously.
The in situ hybridization histochemistry was performed as described previously (Yan & Okamura, 2002). Briefly, tissue sections were processed with proteinase K (1 mg/mL, 0.1 m Tris buffer pH 8.0; 50 mM EDTA; 10 min) at 37 °C and 0.25% acetic anhydride in 0.1 m triethanolamine for 10 min. The sections were then incubated in hybridization buffer [60% formide, 10% dextran sulphate, 10 mM Tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0), 0.6 m NaCl, 0.2% N-laurylsarcosine, 500 mg/mL, 200 mg/mL tRNA, 1 × Denhardt's, 0.25% SDS and 10 mM dithiothreitol (DTT)] containing the Diglabelled mper1, mper2 and VIP antisense cRNA probes (0.1 mg/mL) for 16 h at 60 °C. After a high-stringency posthybridization wash, sections were treated with RNase A, and then were further processed for immunodetection with a nucleic acid detection kit (Boehringer Mannheim). The sections were incubated in 1.0% of blocking reagent in buffer 1 (100 mM Tris-HCl buffer, 150 mM NaCl, pH 7.5) for 1 h at room temperature. Sections were then incubated at 4 °C in alkaline phosphatase-conjugated Dig antibodies diluted 1 : 5000 in buffer 1 for 3 days. On the following day, sections were washed in buffer 1 twice (5 min each), and incubated in buffer 3 (100 mM Tris-HCl buffer, pH 9.5, containing 100 mM NaCl and 50 mM MgCl2) for 3 min. They were then incubated in a solution containing nitroblue tetrazolium salt (0.34 mg/mL) and 5-bromo-4-chrolo-3-indolyl phosphate toluidinium salt (0.18 mg/mL) (Boerhringer Mannheim) for 16 h. The colourimetric reaction was stopped by immersing the sections in buffer 4 (10 mM Tris-HCl containing 1 mM EDTA, pH 8.0).
Delineation of the SCN regions and quantitative analysis
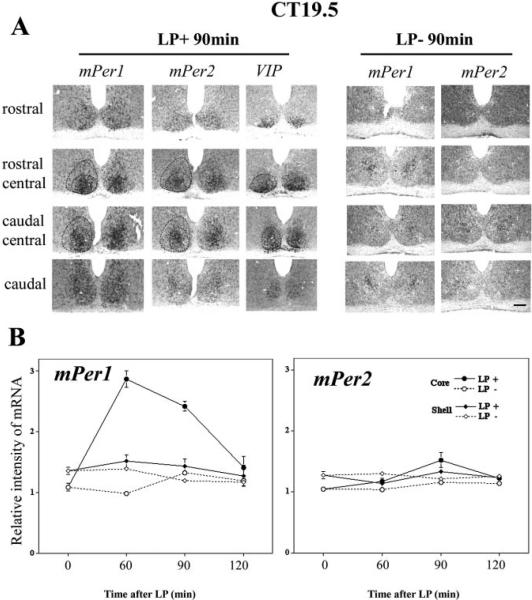
To delineate distinct SCN regions, cells positive for vasoactive intestinal polypeptide (VIP) mRNA were used as a marker for the core of the SCN, with the non-VIP expressing region representing the shell. The outer border of the SCN was defined by light-induced mPer2 in the delay phase (Fig. 3A) and light-induced mPer1 in the advance phase (Fig. 5A), and confirmed by comparing mPer1, mPer2 and cresyl violet staining using alternate sections in a non-experimental animal. To accurately define the SCN in animals with low Per staining (e.g. control animals), we created a template of this hypothalamic region using hypothalamic markers.
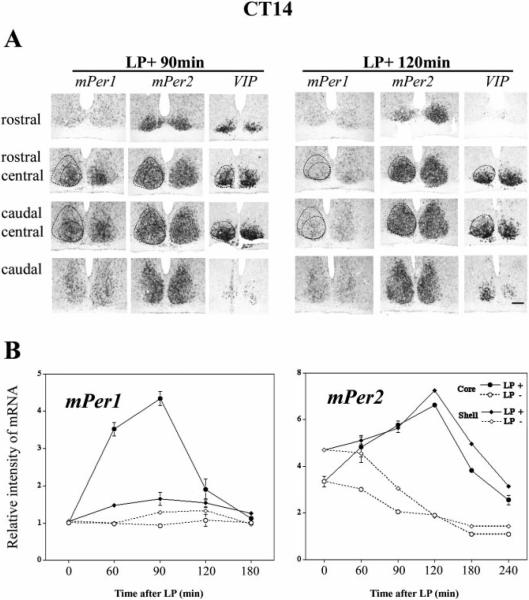
Fig. 3.
Topographic and quantitative analysis of light-induced mPer1 and mPer2 in the SCN after a light pulse (900 lux, 30 min) at CT14. (A), mPer1, mPer2 and VIP expression in adjacent sections from rostral to caudal SCN are shown in two representative animals from the LP+ 90-min and LP+ 120-min groups. VIP was used as the marker for the core SCN, and light-induced mPer2 staining was used to delineate the border of the SCN. Quantitative analysis was done in the core region (VIP-containing cells region) and shell region (no VIP-containing SCN region) independently.(B), Quantitative analysis was for the relative intensity of mPer1 (left) and mPer2 (right) in core and shell SCN with (LP+) or without (LP−) the light pulse. For mPer1, the mean effect of light pulse is significant in the core region (two-way anova, P < 0.0001), but not in the shell region (P > 0.05). For mPer2, the mean effect of light-pulse is significant in both the core and the shell region (P < 0.0001). The data are presented as Mean ± SEM. Scale bar = 100 μm.
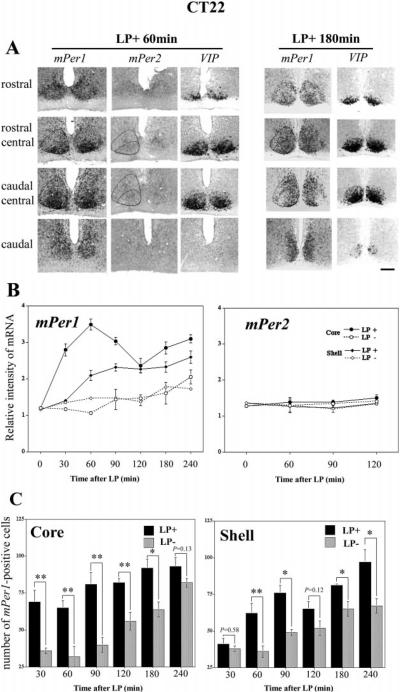
Fig. 5.
Topographic and quantitative analysis of light-induced mPer1 in the SCN after a light pulse (900 lux, 30 min) at CT22. (A), mPer1, mPer2 and VIP staining in adjacent sections from rostral to caudal SCN are shown two representative animals from the LP+ 60-min and LP+ 180-min groups. VIP was used as a marker of the core SCN, and mPer1 staining was used to delineate the border of the SCN. Quantitative analysis was done in the core region (VIP-containing cells region) and shell region (no VIP-containing SCN region) independently. (B), The relative intensity of mPer1 (left) and mPer2 (right) in core and shell SCN with (LP+) or without (LP−) the light pulse. For mPer1, the effect of light pulse is significant in both the core and the shell SCN (P < 0.0001). While for mPer2, there is no significant effect of light pulse in either the core or the shell SCN (P > 0.05). (C), Number of mPer1-positive cells in core (left) and shell (right) at each time point following the light pulse. *P < 0.05, **P < 0.01, student t-test. The data are presented as Mean ± SEM. Scale bar = 100 μm.
Various terms have been used to describe SCN subregions (Moore, 1996). This has resulted in substantial confusion. In this paper, we use the following descriptive terminology as the basis of quantitative analysis. Mouse SCN is approximately 550 μm in rostro-caudal extent. We used 30 μm sections and analysed the mid-SCN (about 180 μm) as follows. The region of cells expressing VIP mRNA was designated `the core', while the remaining SCN region was designated the `shell'. We recognize that this definition differs from that used by Moore and colleagues (Moore, 1973; Abrahamson & Moore, 2001). Our purpose was to establish a consistent measurement system, rather than map the present results onto previously identified SCN zones (e.g. dorsomedial vs. ventrolateral) established on the basis of peptidergic maps.
For quantification of optical density (OD), images of serial sections of mid-SCN were captured using a CCD video camera (Sony XC77) attached to light microscope (BH-2; Olympus Optical, Tokyo, Japan). mRNA expression was quantified using the NIH Image program (version 1.61). Relative optical density (ROD), assessing the mean grey value per pixel was used to quantify the intensity of the signal in the SCN compared to the adjacent hypothalamus area. The ratio of the SCN density to the background was the value for each section. The average ratio of the sections from each brain was used as the value of one animal.
To confirm the results of ROD measurements in the phase advance groups, cell counting was carried out. In the two middle sections of the SCN, which were analysed using ROD measures, the number of mPer1 positive cells was counted in each SCN, and the average between left and right SCN was obtained. The average of two adjacent sections was then calculated. This analysis was undertaken at each time point after the CT22 light pulse, as well as the respective control groups by an experimenter blind to the experimental condition of the animals.
A two-way ANOVA was used to analyse the difference between light pulsed (LP+) and control groups that were killed on the same schedule but were not exposed to a light pulse (LP−). To analyse the effect of a light pulse at specific time points, unpaired Student's t-test was applied.
Results
Light phase shifts circadian behaviour in a time-dependent manner
The phase shifting behaviour of C57BL/6 mice, which display both phase advances and phase delays, have previously been described (Schwartz & Zimmerman, 1990). These studies were repeated in the conditions of our laboratory (Fig. 1). Light pulses resulted in the following phase shifts: delay of 91 ± 8 min at CT14 (early subjective night; Fig. 1A and D) and advance of 56 ± 9.5 min at CT22 (late subjective night; Fig. 1B and D). One animal shortened its free-running circadian period following a light pulse at CT22, and was not included in the analysis. In the transition zone (mid-subjective night), no phase shifts were observed at any of the three time points examined (Fig. 1C and D). The results are: 3.5 ± 7.2 min at CT19, 0 ± 4.5 min at CT19.5 (Fig. 1C), and 0 ± 6.1 min at CT20.
Fig. 1.
Actograms showing phase shifts of wheel-running activity after a light pulse (900 lux, 30 min) during subjective night. Mice maintained in a 12-h LD cycle (indicated by the white-black bar on the top) were placed in constant darkness (DD) as indicated by the label to the right of each figure. Light pulses were given on the third day in DD. The magnitude of the phase shifts was calculated by comparing eye-fitted lines drawn according to the onset of the circadian behaviour before and after the light pulses. (A) A light pulse at CT14 results in phase delay of circadian rhythm; (B) a light pulse at CT22 results in phase advance of circadian rhythm; (C) a light pulse at CT19.5 reduces no-phase shift; (D), Mean ± SEM of the phase shifting effects of light at CT14, 19, 19.5, 20 and 22. Numbers above or under bars denote sample sizes for each condition. ▽ Indicates the light pulse.
MPer1 and mPer2 expression following phase delaying light pulses
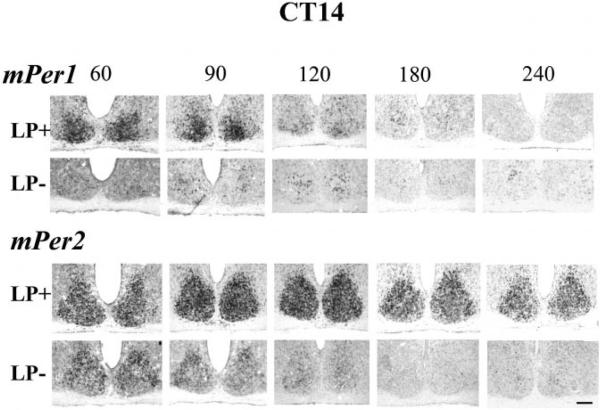
The time course of mPer1 and mPer2 expression was examined after a light pulse (LP+) at CT14 and in control animals (LP−) killed at the same times, but not exposed to light (Fig. 2). In LP+ animals, robust mPer1 expression was observed at 60 and 90 min, which then decreased from 120 min onwards (Fig. 2, row1). In LP− animals (Fig. 2, row 2), mPer1 expression was low at all time points. Analysis of mPer2 in LP+ animals indicated very strong expression at 90 and 120 min, and moderate to strong expression at all other time points (Fig. 2, row3). In LP− animals, mPer2 expression was initially weak at CT15 (60 min), and decreased steadily to undetectable levels by CT18 (240 min; Fig. 2, row4). It is important to note that the SCN sections in Fig. 2, rows 1 + 3 and rows 2 + 4, are adjacent sections, allowing comparison of mPer1 and mPer2 staining in the same animal. This analysis suggests that mPer2 staining occurs in the whole SCN, while mPer1 staining was restricted to the ventral, core SCN. This was assessed in the next part of the results using VIP as a marker for the core SCN.
Fig. 2.
Light-induced mPer1 and mPer2 expression in the SCN after a light pulse at CT14 as shown by in situ hybridization. Animals were given a light pulse (900 lux, 30 min) at CT14, then killed at the following the time points: 60, 90, 120, 180 and 240 min after the beginning of the light pulse. Control animals that were not exposed to the light were killed on the same schedule. Every third SCN section from each animal was processed with mPer1, mPer2 and VIP in situ probes. mPer1 LP+ (row 1) and mPer2 LP+ (row 3) are shown in adjacent sections. mPer1 LP− (row 2) and mPer2 LP− (row 4) are also adjacent sections. LP, light pulse; the numbers across the top indicate time in minutes after the beginning of the light pulse. Scale bar = 100 μm.
Figure 3A shows rostral-to-caudal SCN processed for mPer1, mPer2 and VIP in serial adjacent sections, from the LP+ 90-min and LP+ 120-min groups, respectively. VIP expressing cells were used as markers for delineation of the core and shell SCN (see details in materials and methods section). For LP+ 90-min animals, light-induced mPer1 was observed in the core SCN, while light-induced mPer2 was observed in both the core and shell SCN. For LP+ 120-min animals, light-induced mPer1 was weak in the core SCN, while light-induced mPer2 was robust throughout the SCN.
Quantification of mPer1 and mPer2 expression in LP+ and LP− groups was done in the core and shell SCN, respectively (Fig. 3B). For mPer1 (Fig. 3B, left panel), there was a significant effect of light pulse in the core region (core, LP+ vs. LP−; F1,14 = 167.971, P < 0.0001, two-way ANOVA), but not in the shell region (shell, LP+ vs. LP−; F1,14 = 4.198, P > 0.05, two-way ANOVA). For mPer2 (Fig. 3B, right panel), the main effect of the light pulse was significant in each region (core, LP+ vs. LP−; F1,16 = 299.383, P < 0.0001; shell, LP+ vs. LP−; F1,16 = 486.091, P < 0.0001, two-way ANOVA). In the core, a comparison of LP+ and LP− groups (compare open and closed circles, Fig. 3B, right panel) indicates that light induces mPer2 starting at 60 min (core, 60 min LP+ vs. LP−; P < 0.05, unpaired t-test). In the shell (compare open and closed diamonds in Fig. 3B, right panel), the increase of mPer2 was first detected at 90 min (60 min LP+ vs. LP−; P > 0.05, 90 min LP+ vs. LP−; P < 0.001, unpaired t-test), indicating a 30-min time lag between responses in the core and shell SCN.
MPer1 and mPer2 expression following phase advancing light pulses
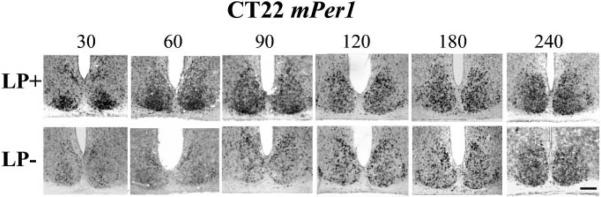
After a phase advancing light pulse at CT22, expression of mPer1 and mPer2 were examined. For mPer1, in LP+ animals (Fig. 4, row 1), strong expression was observed in the ventral core of the SCN at 30–60 min, with moderate to strong expression seen throughout the SCN from 90 to 240 min. In LP− control animals (Fig. 4, row 2), weak mPer1 expression was observed in the SCN. In contrast to the data for mPer1, there was no light-induced expression of mPer2 at 60–120 min (see below).
Fig. 4.
Light-induced mPer1 expression in the SCN after a light pulse (900 lux, 30 min) at CT22 as shown by in situ hybridization. Animals were given a light pulse at CT22, then killed as described in Fig. 2. Control animals that were not exposed to the light pulse were killed on the same schedule. Quantification of the results is shown in Fig. 5. LP, light pulse; the numbers along the top indicate time in minutes after the beginning of the light pulse. Scale bar = 100 μm.
Figure 5A shows rostral-to-caudal SCN processed for mPer1, mPer2 and VIP in serial adjacent sections, from animals in the LP+ 60-min and LP+ 180-min groups, respectively. As above, VIP expression cells were used to delineate the core SCN. In LP+ 60-min animals, light-induced mPer1 was strong in the core region, and weak in the shell region, with no detectable light-induced mPer2 in both core and shell SCN. In LP+ 180-min animals, light-induced mPer1 was strong in both core and shell SCN. mPer2 expression was not detected at 60, 90 120 min (CT 23, 23.5, 24) and was not examined at LP+ 180 min (CT 1 in next subjective day).
Quantification of these results is shown in Fig. 5B. The effect of a light pulse on Per1 expression was significant in both core and shell SCN (core, LP+ vs. LP−; F1,26 = 220.987, P < 0.0001; shell, LP+ vs. LP−; F1,26 = 77.529, P < 0.0001, two-way ANOVA). At CT22, there was no significant effect of a light pulse on Per2 expression in either core (LP+ vs. LP−; F1,10 = 1.261, P > 0.05, two-way ANOVA) or shell (LP+ vs. LP−; F1,10 = 1.709, P > 0.05, two-way ANOVA). In the core region (compare open and closed circles in Fig. 5B, left panel), an increase of light-induced mPer1 was observed starting at 30 min (core, 30 min LP+ vs. LP−; P < 0.05, unpaired t-test). In the shell region (compare open and closed diamonds in Fig. 5B, left panel), the increase of mPer1 was not seen at 30 min (shell, 30 min LP+ vs. LP−; P > 0.05, unpaired t-test), but was observed from 60 min (shell, 60 min LP+ vs. LP−; P < 0.05, unpaired t-test). Thus, there is also a 30-min time lag of light-induced mPer1 spreading from the core to shell SCN.
Because endogenous levels of mPer1 are increasing at CT 22, we wanted to further evaluate the effect of the light pulse using an independent measure of mPer1 induction. To this end, cell counting of the LP+ and LP− groups was done in both core and shell regions (Fig. 5C). A significant effect of light exposure on the number of mPer1 positive cells was observed in both core and shell region (P < 0.0001, two way ANOVA). The significance of the effect of light at each time point was assessed by student t-test (Fig. 5C), and the results were consistent with the earlier ROD analysis.
MPer1 and mPer2 expression following light pulse at CT 19.5: a nonshifting light pulse
Expression of light-induced mPer1 and mPer2 were examined after a light pulse at CT19.5, when light has no effect on the phase of locomotor behaviour (Fig. 6A). At LP 90 min, light-induced mPer1 and mPer2 was observed in LP+ animals, but not in LP− 90-min animals. Quantification of the results for all animals in these groups is shown in Fig. 6B. For mPer1 (Fig. 6B, left panel), the LP+ increase was seen at 60 and 90 min in the core SCN, but not in the shell SCN (core, LP+ vs. LP−; F1,14 = 153.188, P < 0.0001; shell, LP+ vs. LP−; F1,14 = 2.379, P > 0.05, two-way ANOVA). For mPer2 (Fig. 6B, right panel), in the core region, a weak effect of the light pulse was detected (two-way ANOVA, core, LP+ vs. LP−; F1,14 = 5.456, P = 0.47), with a significant light-induced increase of mPer2 at 90 min (core, 90 min LP+ vs. LP−; P < 0.05, unpaired t-test). In shell region, no effect of light on mPer2 was detected (shell, LP+ vs. LP−; F1,14 = 0.751, P > 0.05, two-way ANOVA). Consistent results were observed after the light pulses at CT19 and CT20 (data not shown).
Fig. 6.
Topographic and quantitative analysis of light-induced mPer1 and mPer2 in the SCN after a light pulse (900 lux, 30 min) at CT19.5. (A), 90 min after the beginning of the light pulse (LP+ 90 min), mPer1, mPer2 and VIP staining in adjacent sections from rostral to caudal SCN, compared with the no light pulse control (LP− 90 min). (B), Relative intensity of mPer1 and mPer2 in core and shell SCN with (LP+) or without (LP−) the light pulse. For mPer1, the mean effect of light pulse is significant in the core region (P < 0.0001), but not in the shell region (P > 0.05). While for mPer2, there is a small, but significant increase in the core SCN (P < 0.05), which was confirmed at 90 min (t-test, P < 0.05), and not significant increase in the shell region (P > 0.05). The data are presented as Mean ± SEM. Scale bar = 100 μm.
Discussion
Based on afferent input and neuronal phenotype, it has long been suggested that the SCN of rodents has two anatomically defined regions (Moore, 1996; Leak et al., 1999). The core or ventral region of the SCN, which receives directly retinal input, is delineated by VIP and/or GRP (gastrin-releasing peptide) containing cells, the shell or dorsomedial zone is defined by arginine vasopressin (AVP)-containing cells (van den Pol & Tsujimoto, 1985; Abrahamson & Moore, 2001). By taking into account the anatomical and functional organization of the SCN, the present results provide new insights into the role of Per genes in phase shifting (Fig. 7). Substantial light induced mPer1 expression occurs in the core region of the SCN throughout the subjective night, even at times of day when behavioural phase shifts do not occur (Fig. 7, upper left panel). In contrast, strong expression of mPer2 in the core SCN occurs only after a phase-delaying light pulse (Fig. 7, upper right panel). These results eliminate the possibility that light-induced mPer1 or mPer2 expression in the core region of the SCN is itself sufficient to produce phase shifts.
Fig. 7.
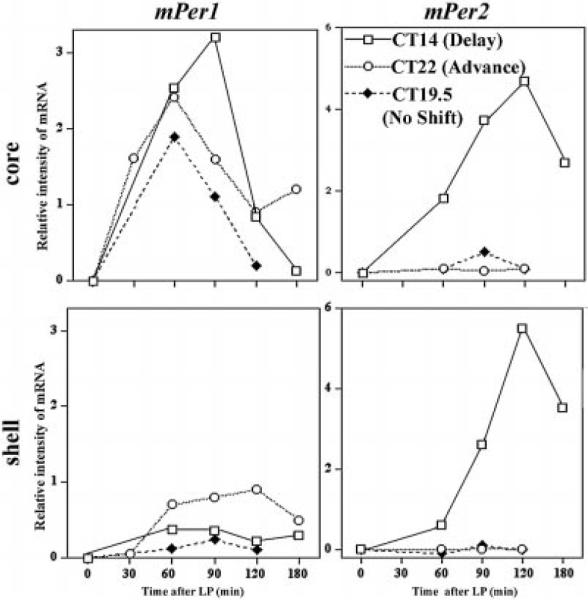
Summary of light-induced mPer1 and mPer2 in core and shell SCN, respectively. Solid line and open squares show the result of a phase delaying pulse at CT14, dotted line and open circles show the result of a phase advancing light pulse at CT22, and dashed line and solid diamonds show the results of a nonphase shifting light pulse at CT19.5. The data are presented as the difference between the Mean of LP+ and LP− at each distinct condition.
In contrast to the pattern seen in the core SCN, light-induced Per gene expression in the shell SCN is related to the occurrence of behavioural phase shifts (Fig. 7, lower panels). Following a light pulse that results in behavioural phase delays (CT14), significant mPer2, but little mPer1 expression is observed in the shell. Following a light pulse that does result in behavioural phase advances (CT22), substantial induction of mPer1 but not mPer2 is observed in the shell. Finally, following a light pulse administered in the middle of the subjective night (CT19.5) where no phase shifts occur, neither mPer1 nor mPer2 are induced in the shell SCN. These results indicate that behavioural phase shifts occur only when light-induced Per gene expression spreads from the core to the shell region of the SCN. Furthermore, in the shell region of the SCN, light-induced mPer1 expression is associated with phase advances, and light-induced mPer2 expression in the shell is associated with phase delays.
The amplitude of the Per2 mRNA response in the shell is about 500% above baseline, while it is about 100% over baseline for Per1. This may account for the difference in the rapidity and amplitude of phase shifts seen between delays and advances in mice. Concomitantly, phase delays in our mice were about 1.5 times larger than phase advances. Moreover, phase delays, are complete by the day after the light pulse, while phase advances reach their new phase after 2 or 3 days of transient phase shifts (Daan & Pittendrigh, 1976; Schwartz & Zimmerman, 1990).
The region of VIP mRNA expressing cells was used as the marker for the core of the mid-SCN region. Compared with other rodent species, the anatomical subdivisions of mouse SCN are less distinct. While older studies using autoradiographic tract tracing suggested that retinal projections extend beyond the boundaries of the SCN (Cassone et al., 1988), more recent work using the anterograde tracer choleratoxin (CTB) indicates that retinal efferents are concentrated in the core SCN (Abrahamson & Moore, 2001). Moreover, in the middle third of the SCN, the shell region is devoid of CTB-labelled fibres. It should be pointed out that, in both of these studies, VIP immunoreactive (ir) cells were examined, and the distribution of the VIP-ir cells is concentrated in the ventral region of the core SCN. In contrast to VIP-ir cells, the distribution of the VIP mRNA-positive cells in the present study is much larger, and is similar to the distribution of retinal input in the middle SCN and it is also comparable to the distribution of neuropeptide Y (NPY) fibre staining, which is present throughout the core of the SCN (Abrahamson & Moore, 2001). The differences in the distribution of VIP-ir and VIP mRNA positive cells might be due to peptide release from neuronal perikarya, resulting in an underestimation of the extent of VIP cells.
The present results explain the differences between the reports of Shigeyoshi et al. (1997) and Albrecht et al. (1997) in that the former reported that light-induced mPer1 is restricted to the core SCN while the latter reported a widespread signal following light-induced mPer1 expression. Timing of the light pulse appears to account for the difference in their results, as the former study used a phase-delaying light pulse at CT16, while the latter administered a phase-advancing light pulse at CT22.
The function of Per genes in phase shifting has been studied in Per1 and Per2 mutant animals, as well as in animals treated with Per antisense oligonucleotide. The mutant animals were studied using different testing protocols and reached different conclusions. Using the Aschoff type II protocol (Albrecht et al., 2001), behavioural phase shifts were studied in Per1 (Zheng et al., 2001) or Per2 (Zheng et al., 1999) mutant animals. Per1 mutant animals show a normal phase delay but no phase advance, while Per2 mutant animals show an exaggerated phase advance but no phase delay following an appropriate phase-shifting light pulse. This suggests Per1 plays a role in phase advance and Per2 is essential for phase delay control of the clock. Using the Aschoff type I-testing protocol, Cermakian et al. (2001) also studied the phase shifts in mPer1 mutant mice strain, but saw no difference from the wild type animals on phase shifts to light. This suggests that mPer1 has no effect on light-induced phase shifts. In experiments using antisense oligonucleotide (AS-ODN) probes, intracerebroventricular injection of mPer1 and/or mPer2 AS-ODN attenuated light-induced phase delay of locomotor behaviour, suggesting that acute induction of mPer1 and mPer2 have additive effects on light-induced phase delays (Akiyama et al., 1999; Wakamatu et al., 2001).
Both light-induced mPer1 and mPer2 were observed first in the core SCN. It is well known that light information is transmitted to the SCN via the retinal hypothalamic tract (RHT), and the projection of RHT terminals is primarily to the core SCN (Abrahamson & Moore, 2001). Glutamate is the primary neurotransmitter from the RHT to the SCN (Johnson et al., 1988; Hannibal et al., 2000). A light pulse stimulates the release of glutamate from the RHT, activating multiple glutamate receptors (Colwell & Menaker, 1992; Mintz et al., 1999), and induces phosphorylation of the Ca2+/cAMP response element binding protein (CREB) and CRE-mediated transcription in both early and late subjective night. Light-induced pCREB is also observed in the core SCN (Ding et al., 1997), and overlaps with VIP cells (von Gall et al., 1998), suggesting the pCREB/CRE pathway is critical for the light-induced mPer1 expression in this region. There are two CRE sites in the mPer1 gene and one consensus CRE element in the mPer2 promoter region (Reppert & Weaver, 2001). It is reasonable to speculate that light-induced mPer1 and mPer2 expression in the VIP-containing core region, which was observed in the present study, is directly regulated via these pathways.
There was a 30-min time lag for the light-induced Per expression to spread from the core to the shell region of the SCN. This time lag suggests that temporal information is transmitted from the core to the shell. Data available for the rat, using swine herpesvirus (PRV) for tract tracing, indicates that the core projects densely to the shell SCN, but not vice versa (Leak et al., 1999). Comparable anatomical data is not available for the mouse, although physiological experiments do suggest connections from the core to the shell. It has been suggested that in mouse, GRP neurons, which are localized in the core SCN, convey photic information from the core to the shell SCN (Aida et al., 2002).
The differential Per gene response in the shell region of the SCN suggests a distinct regulatory mechanism underlying phase delays (involving Per2) and advances (involving Per1). Numerous second messenger pathways are known to participate in photic signal transduction, with divergent signalling pathways involved in the light-induced phase delays and advances (Gillette & Tischkau, 1999). In the early night, phase delays require activation of the ryanodine receptors to release intracellular stores of Ca2+. Activators of ryanodine receptors were found to induce phase resetting; whereas inhibitors blocked delays induced by light and glutamate (Ding et al., 1998). In the late night, administration of the specific cGMP-dependent protein kinase (PKG) inhibitor significantly attenuated light-induced advances in the phase of activity, implicating PKG in the signalling pathways responsible for photic phase advances (Weber et al., 1995; Mathur et al., 1996). Different signalling pathways for phase delays and advances likely account for the differential Per1 and Per2 gene expression in the shell of the SCN. It would be interesting to explore how these distinct pathways regulate Per1 and Per2 induction in the core and shell of the SCN. The present results alert us to the fact that attention to the anatomical organization and network properties of the SCN are necessary to understand its functions.
Acknowledgements
We thank Dr Hitoshi Okamura for kindly donating the plasmid for in situ hybridization, Dr Joseph LeSauter, Lance Kriegsfeld, Michael Antle, Toshiyuki Hamada and Ilia Karatsoreos for their comments on an earlier draft. This work was supported by NIH grants NS37919 and NS41069 to RS.
Abbreviations
- AVP
arginine vasopressin
- CREB
cAMP reactive element binding protein
- CT
circadian time
- DD
constant darkness
- DIG
digoxigein
- GRP
gastrin-releasing peptide
- LP
light pulse
- NPY
neuropeptide Y
- RHT
retinal hypothalamic tract
- ROD
relative optical density
- SCN
suprachiasmatic nucleus
- VIP
vasoactive intestinal peptide
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of period gene in mice. Mol. Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J. Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Cassone VM, Speh JC, Card JP, Moore RY. Comparative anatomy of the mammalian hypothalamic suprachiasmatic nucleus. J. Biol. Rhythms. 1988;3:71–91. doi: 10.1177/074873048800300106. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Menaker M. NMDA as well as non-NMDA receptor antagonists can prevent the phase-shifting effects of light on the circadian system of the golden hamster. J. Biol. Rhythms. 1992;7:125–136. doi: 10.1177/074873049200700204. [DOI] [PubMed] [Google Scholar]
- Daan S, Pittendrigh CS. A function analysis of circadian pacemakers in nocturnal rodents. II. The variability of phase response curves. J. Comp. Physiol. 1976;106:253–266. [Google Scholar]
- Ding JM, Buchanan GF, Tischkau SA, Chen D, Kuriashkina L, Faiman LE, Alster JM, McPherson PS, Campbell KP, Gillette MU. A neuronal ryanodine receptor mediates light-induced phase delays of the circadian clock. Nature. 1998;394:381–384. doi: 10.1038/28639. [DOI] [PubMed] [Google Scholar]
- Ding JM, Faiman LE, Hurst WJ, Kuriashkina LR, Gillette MU. Resetting the biological clock: mediation of nocturnal CREB phosphorylation via light, glutamate, and nitric oxide. J. Neurosci. 1997;17:667–675. doi: 10.1523/JNEUROSCI.17-02-00667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- van Esseveldt KE, Lehman MN, Boer GJ. The suprachiasmatic nucleus and the circadian time-keeping system revisited. Brain Res. Brain Res. Rev. 2000;33:34–37. doi: 10.1016/s0165-0173(00)00025-4. [DOI] [PubMed] [Google Scholar]
- von Gall C, Duffield GE, Hastings MH, Kopp MD, Dehghani F, Korf H-W, Stehle JH. CREB in the mouse SCN: a molecular interface coding the phase-adjusting stimuli light, gluatamate, PACAP, and melatonin for clockwork access. J. Neurosci. 1998;18:10389–10397. doi: 10.1523/JNEUROSCI.18-24-10389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette MU, Tischkau SA. Suprachiasmatic nucleus: the brain's circadian clock. Recent Prog. Horm. Res. 1999;54:33–59. [PubMed] [Google Scholar]
- Hannibal J, Moller M, Ottersen OP, Fahrenkrug J. PACAP and glutamate are co-stored in the retinohypothalamic tract. J. Comp. Neurol. 2000;418:147–155. [PubMed] [Google Scholar]
- Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- Johnson RF, Morin LP, Moore RY. Retinohypothalamic projections in the hamster and rat demonstrated using cholera toxin. Brain Res. 1988;462:301–312. doi: 10.1016/0006-8993(88)90558-6. [DOI] [PubMed] [Google Scholar]
- Leak RK, Card JP, Moore RY. Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res. 1999;819:23–32. doi: 10.1016/s0006-8993(98)01317-1. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system: Photic entrainment, circadain pacemaker mechanisms, and postranslational regulation. Annu. Rev. Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- Mathur A, Golombek DA, Ralph MR. cGMP-dependent protein kinase inhibitors block light-induced phase advances of circadian rhythms in vivo. Am. J. Physiol. 1996;270:R1031–R1036. doi: 10.1152/ajpregu.1996.270.5.R1031. [DOI] [PubMed] [Google Scholar]
- Mintz EM, Marvel CL, Gillespie CF, Price KM, Albers HE. Activation of NMDA receptors in the suprachiasmatic nucleus produces light-like phase shifts of the circadian clock in vivo. J. Neurosci. 1999;19:5124–5130. doi: 10.1523/JNEUROSCI.19-12-05124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY. Retinohypothalamic projection in mammals: a comparative study. Brain Res. 1973;49:403–409. doi: 10.1016/0006-8993(73)90431-9. [DOI] [PubMed] [Google Scholar]
- Moore RY. Entrainment pathways and the functional organization of the circadian system. Prog. Brain Res. 1996;111:103–119. doi: 10.1016/s0079-6123(08)60403-3. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan SA. A function analysis of circadian pacemakers in nocturnal rodents. IV. Entrainment: pacemaker as clock. J. Comp. Physiol. 1976;106:291–331. [Google Scholar]
- van den Pol AN, Tsujimoto KL. Neurotransmitters of the hypothalamic suprachiasmatic nucleus: Immunocytochemical analysis of 25 neuronal antigens. Neuroscience. 1985;15:1049–1086. doi: 10.1016/0306-4522(85)90254-4. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Wakamatu H, Takahashi S, Moriya T, Inouye SIT, Okamura H, Akiyama M, Shibata S. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. Neuroreport. 2001;12:127–131. doi: 10.1097/00001756-200101220-00033. [DOI] [PubMed] [Google Scholar]
- Weber ET, Gannon RL, Rea MA. cGMP-dependent protein kinase inhibitor blocks light-induced phase advances of circadian rhythms in vivo. Neurosci. Lett. 1995;197:227–230. doi: 10.1016/0304-3940(95)11961-u. [DOI] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Takekida S, Shigeyoshi Y, Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience. 1999;94:141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]