Abstract
Aβ (amyloid beta peptide) is an important contributor to Alzheimer’s disease (AD). We modeled Aβ toxicity in yeast by directing the peptide to the secretory pathway. A genome-wide screen for toxicity modifiers identified the yeast homolog of phosphatidylinositol binding clathrin assembly protein (PICALM) and other endocytic factors connected to AD whose relationship to Aβ was previously unknown. The factors identified in yeast modified Aβ toxicity in glutamatergic neurons of Caenorhabditis elegans and in primary rat cortical neurons. In yeast, Aβ impaired the endocytic trafficking of a plasma membrane receptor, which was ameliorated by endocytic pathway factors identified in the yeast screen. These links between Aβ, endocytosis, and human AD risk factors can be ascertained using yeast as a model system.
Yeast cells lack the specialized processes of neuronal cells and the cell-cell communications that modulate neuropathology. However, the most fundamental features of eukaryotic cell biology evolved before the split between yeast and metazoans. Yeast studies of the cell cycle, DNA damage repair and checkpoints produced pivotal advances in cancer biology (1). More recently, the conservation of protein-homeostasis networks, vesicular trafficking, mitochondrial biology, autophagy, and apoptosis facilitated the development of yeast models for protein-misfolding pathologies (1). When human diseases impinge on common features of eukaryotic cell biology, yeast’s unequaled toolkit offers an attractive discovery platform, as established for multiple aspects of α-synuclein toxicity (2–7).
Here, we wanted to create a yeast model of cellular toxicities elicited by the amyloid β (Aβ) peptide. According to the still hotly debated “amyloid cascade” hypothesis, Aβ is causal in both sporadic and familial Alzheimer’s Disease (AD) (8). The oligomeric forms of the peptide appear to be the most toxic (9–12). Similar toxic oligomers, formed by unrelated proteins but all recognized by the same conformation-specific antibody, are associated with other neurodegenerative diseases and with yeast prions (13, 14). Thus, the toxicity of such oligomers is an ancient protein-folding problem.
In addition to Aβ, neurofibrillary tangles (NFTs) of tau, a microtubule-binding protein, are hallmarks of AD pathology (15). Aβ seems to act upstream of tau (16, 17). Genetic AD risk factors are now being identified through genome-wide association studies (GWAS), but their relationship to Aβ is unknown.
A yeast model of Aβ toxicity
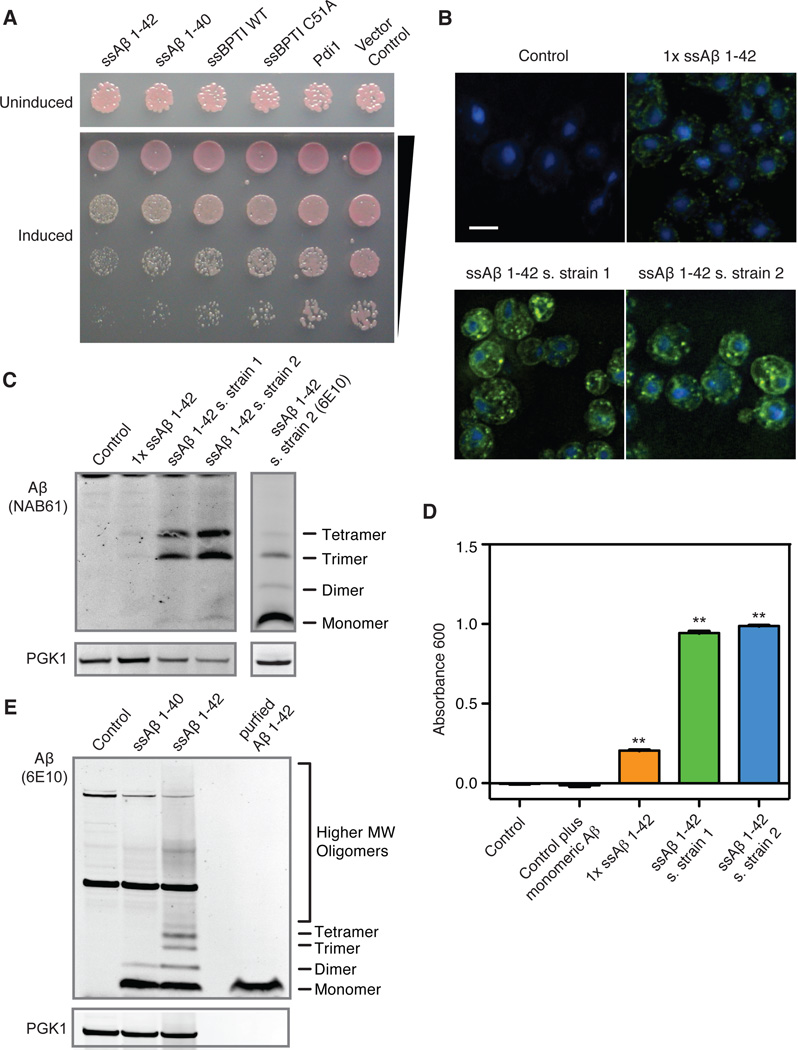
The most toxic form of Aβ, Aβ 1-42, is generated by proteolytic cleavage of APP, the transmembrane amyloid precursor protein (18, 19). APP processing occurs in the secretory pathway, which releases Aβ into the trans-Golgi, endosomal compartments, and extracellular space. Aβ then interacts with the plasma membrane and is subject to endocytosis and further vesicular trafficking (18). To recapitulate this multi-compartment trafficking in yeast, we fused an ER targeting signal to the N-terminus of Aβ 1-42 (referred to as ssAβ 1-42, Fig. 1A). Without an ER retention signal, after cleavage of the signal sequence Aβ 1-42 should simply transit through the secretory pathway to the plasma membrane (20). The yeast cell wall will restrain secreted peptides from diffusing into the culture medium, allowing Aβ to interact with the plasma membrane, undergo endocytosis, and thereby transit through endocytic compartments potentially relevant to AD (Fig. S1A).
Figure 1.
Expression of Aβ in the yeast secretory pathway. (A) Comparison of ssAβ 1-42 toxicity with ssAβ 1-40, ssBPTI (WT and C51A) and Pdi1. Proteins were expressed using the inducible GAL1 promoter and a high copy number plasmid. Strains carrying the plasmids were serially diluted and spotted on inducing (galactose) and non-inducing (glucose) media. (B) Immunostaining for ssAβ 1-42 reveals localization to the ER/secretory pathway. Aβ was detected in the ER (ring surrounding the nucleus, stained blue with DAPI) and in small foci throughout the cell. The scale bar is 5µm. All figures are on the same scale. (C) Immunoblot with NAB61, an antibody specific for soluble Aβ oligomers, detects oligomers in unboiled samples. An immunoblot with the 6E10 Aβ-specific antibody is shown for reference. (D) An indirect ELISA assay using a monoclonal Aβ oligomer-specific antibody detects Aβ oligomers in the 1×ssAβ strain and, more so, in the two screening strains (n=5; error bars to small to be visible; **: p<0.01, based on Dunnett’s test). (E) Aβ 1-40 and Aβ 1-42 expression was detected by immunoblot analysis of unboiled lysates using 6E10.
When expressed from a galactose-inducible (GAL1) promoter and a multi-copy plasmid, ssAβ 1-42 decreased cell growth (Fig. 1A). Using the same plasmid, Aβ 1-40 was less toxic, as were Pdi1 (an ER resident protein), BPTI (a small protein commonly used to study secretion), and even BPTIC51A (a variant that misfolds in the ER (21)) (Fig. 1A).
For genetic screens, strains with uniform stable ssAβ 1-42 expression were constructed by integrating tandem copies in the genome (Fig. S1B). We targeted a locus where insertions have no deleterious consequences and selected strains that grew slowly upon galactose induction, but with no major increase in lethality (Fig. S1C and Table S1). The peptide produced was of the expected size for processed Aβ (Fig. S1D), verified by mass spectrometry (Fig. S1E). Localization to secretory compartments was confirmed by immunofluorescence (Fig. 1B).
When lysates were not subjected to boiling, which disrupts oligomeric species, we detected Aβ oligomers on Bis-Tris gels (Fig. S1D). These forms reacted more strongly with the antibody NAB61, which preferentially recognizes toxic Aβ oligomers in AD patients (Fig. 1C) (10). These species disappeared upon boiling in lithium dodecyl sulfate (LDS) buffer. Assaying lysates by size-exclusion chromatography with a monoclonal IgM anti-Aβ antibody specific for Aβ oligomers detected a broad range of oligomeric species (Fig. 1D and Fig. S1F) (22). Eliminating preparation artifacts, these were not seen when purified monomeric Aβ peptide was added to control cultures prior to lysis (Fig. 1D). In strains that produced the same levels of Aβ 1-40 and 1-42 monomer after boiling (Fig. S1G), oligomers were much more abundant for Aβ 1-42 prior to boiling (Fig. 1E). Thus, oligomeric Aβ forms contribute to toxicity in yeast, as in neurons.
Screen for genetic modifiers of Aβ toxicity
We transformed a screening strain with an overexpression library of 5532 full length open reading frames (ORFs) (~90% of yeast ORFs) under control of the same promoter used for ssAβ 1-42 (Fig. S2A). Individual transformants were arrayed in media that prevented induction of either ssAβ 1-42 or the library constructs, then plated (four replicates each) onto several types of inducing media, chosen to support different levels of mitochondrial respiration (Fig. S2B; (23), Table S2). Intermediate levels of Aβ toxicity enabled the identification of enhancers and suppressors in the same screen (Fig. S1B; (23), Table S2). Genes that decreased or increased growth (Fig. S2B) were retested in an independently derived screening strain. Secondary screens eliminated genes that simply altered expression of Aβ from the GAL1 promoter (Fig. S3) or growth in the absence of Aβ.
We identified 23 suppressors and 17 enhancers (Table S2). Only a few modifiers were strongly affected by the state of respiration (Table S2). The screen hits comprised a wide range of cellular functions. Numerous hits had sequence similarity to human genes, and twelve had very clear human homologs (determined by HomoloGene or by analogous functionality [SLA1 – SH3KBP1] (24)) (Table 1). We focused further analysis on these.
Table 1.
Genes that modify Aβ toxicity. Yeast genes with clear nematode and human homologs were identified in an unbiased screen of 5,532 single copy plasmids carrying yeast ORFs under the control of the GAL1 promoter in three independent screens in three inducing medias (23).
| Yeast Aβ Suppressors |
Cellular Function |
C. elegans Homolog |
Human Homolog |
Connection of human homologs to AD Risk |
|---|---|---|---|---|
| YAP1802 | Endocytosis | unc-11* | PICALM | Validated risk factor1,2 |
| INP52 | Endocytosis | unc-26* | SYNJ1 | Interacts with validated risk factor BIN1 (27) |
| SLA1 | Endocytosis | Y44E3A.4* | SH3KBP1 | Interacts with validated risk factor CD2AP (28) |
| RTS1 | Phosphatase regulation |
pptr-2* | PPP2R5C | |
| ADE12 | Adenylosuccinate synthesis |
C37H5.6b* | ADSSL1 | Potential risk factor, this study3 |
| CRM1 | Nuclear protein export |
xpo-1* | XP01 | Potential risk factor, this study2 |
| GRR1 | Ubiquitination | C02F5.7 | FBXL2 | |
| VPS9 | Vesicle transport | rabx-5 | RABGEF1 | Potential risk factor, this study3 |
|
Yeast Aβ Enhancers |
||||
| PBS2 | Osmotic stress response |
mkk-4* | MAP2K4 | Activated by Aβ oligomers in cortical neurons (36) |
| KEM1 | RNA processing | xrn-1 | XRN1 | |
| MVP1 | Vacuolar sorting | lst-4 | SNX8 | |
| PMT2 | Mannosylation | - | POMT2 |
Three of these twelve genes had functions related to clathrin-mediated endocytosis (YAP1802, INP52 & SLA1; P=3.89e-4) and seven were functionally associated with the cytoskeleton (YAP1802, INP52, SLA1, CRM1, GRR1, KEM1 & RTS1; P=6.06e-8). None were identified in our previous screen for modifiers of α-syn toxicity (5, 7), establishing their specificity for the type of toxicity caused by Aβ 1-42.
Modifiers of Aβ toxicity are associated with AD susceptibility
Several human homologs of our yeast hits had connections to human AD risk factors, particularly those involved in clathrin-mediated endocytosis (Table 1). The human homolog of yeast YAP1802, PICALM, is one of the most highly confirmed risk factors for sporadic AD (25, 26). Another AD risk factor, BIN1, is involved in synaptic vesicle endocytosis and is believed to interact with synaptojanin, the human homolog of yeast INP52 (27). The functional homolog of yeast SLA1, SH3KBP1 (28), directly interacts with the risk factor CD2AP (29, 30). CD2AP links endocytosis to cytoskeletal dynamics and our other major class of screen hits.
To assess the potential clinical relevance of other screen hits with highly conserved human homologs we examined association with AD susceptibility using data from a published family-based GWAS (31, 32). Using a family-based association test, we discovered a suggestive association of XPO1 (CRM1 homolog, rs6545886, P=0.003) with AD susceptibility ((23), Table S3).
Next, we leveraged genotyping with extensive clinical and pathological data from two large epidemiological studies of aging, cognition, and AD (33, 34) ((23), Table S4–7). Using a quantitative summary measure of global AD pathologic burden available in these cohorts, counting both amyloid plaques and NFTs, we found that two additional loci identified by our yeast screen, ADSSL1 (ADE12 homolog, rs1128880, P=0.001) and RABGEF1 (VPS9 homolog, rs17566701, P=0.002) showed evidence of association with AD neuropathology (Table S7). Both loci also harbored suggestive association signals with episodic memory decline (Table S6). Thus, our yeast screen connects multiple human AD risk factors, and suggested risk factors, to Aβ toxicity.
C. elegans model of Aβ toxicity
To directly test our modifiers for effects on Aβ toxicity in neurons, we created a transgenic C. elegans model that expressed Aβ 1-42 in glutamatergic neurons, a neuronal subtype particularly vulnerable in AD. (A previous model expressed Aβ in body-wall muscles (35).) We used the eat-4 promoter, which regulates the BNPI glutamate transporter and, again, targeted Aβ to the secretory pathway (35). The Aβ transgene was integrated into chromosomal DNA to ensure the same Aβ 1-42 expression levels in all animals.
C. elegans has highly stereotyped cell lineages. Wild-type worms invariably have five glutamatergic neurons in their tails, which we visualized with eat-4-regulated GFP. Crossing worms expressing ssAβ to worms carrying this reporter resulted in the loss of GFP-marked glutamatergic neurons in an age-related manner: at day three only 48% of worms had five intact glutamatergic neurons, and at day seven only 25% did (Fig. 2A).
Figure 2.
Hits from the yeast screen modify the toxicity of Aβ in C. elegans glutamatergic neurons in the same direction as in yeast. (A) A new animal model of Aβ toxicity. Aβ 1-42, carrying a signal sequence targeting it to the secretory compartment, was expressed in glutamatergic neurons that also expressed GFP. Neuronal death increased from 50% at day 3 to 75% at day 7, establishing that this worm model exhibits age-dependent neurodegeneration. Control genes mCherry and LacZ had no effect on Aβ-induced neurodegeneration. Measurements are relative to the number of neurons found in WT worms expressing only GFP. Bar patterns indicate distinct functional categories (crosshatches for endocytic genes). The genetic modifiers tested influenced Aβ toxicity significantly (p < 0.05, Student’s t-test). XPO1 was derived from human cDNA. For each worm (w) or human (h) gene, three transgenic lines were established and 90 worms examined for each. (B) Representative examples of worms scored in Fig. 2A at the third day of development. Arrowheads indicate neuronal cell bodies, marked by transgenic expression of GFP.
To test the effects of our screen hits (Table 1) we established three independent transgenic worm lines for each gene, again expressing the protein from the eat-4 promoter. Unrelated control proteins mCherry and LacZ had no effect on Aβ toxicity. All three C. elegans homologs of the genes involved in clathrin-mediated endocytosis, unc-11, unc-26, and Y44E3A.4, increased the percentage of worms with five intact glutamatergic neurons (Fig. 2).
Finally, we tested four hits, three suppressors and one enhancer, involved in a diverse array of pathways. The yeast RTS1 gene encodes a phosphatase regulatory subunit that controls several stress-response pathways. The ADE12 gene product catalyzes the first step in the synthesis of adenosine monophosphate from inosine 5-monophosphate. The C. elegans homologs of each gene suppressed the Aβ-induced loss of glutamatergic neurons (Fig. 2A). We were unable to clone the worm homologs of GRR1, VPS9, or CRM1. However, CRM1 encodes a highly conserved nuclear export receptor. Expression of CRM1’s human homologue, XPO1, protected nematode glutamatergic neurons from Aβ (Fig. 2A). PBS2/MAP2K4, a MAP kinase involved in stress responses, increased neuronal loss (Fig. 2).
The effect of each gene was statistically significant (p < 0.05) for both the modest neuronal toxicity evinced at three days and the more severe toxicity at seven days. Importantly, the modifiers did not alter the levels of Aβ mRNA, which we tested by semi-quantitative RT-PCR (Fig. S5). Thus, every gene we tested in C. elegans glutamatergic neurons modified Aβ toxicity in the same direction (suppression vs. enhancement) as they did in yeast.
PICALM suppresses the toxicity of soluble Aβ oligomers with rat cortical neurons
PICALM is one of the most highly validated AD risk factors and its efficacy in our yeast and nematode models strongly suggests it alters Aβ toxicity. Modeling this in cultured mammalian neurons is not trivial, because any Aβ peptide expressed in the secretory pathway would simply diffuse away from the cell. Exogenously applied, preformed oligomeric Aβ species are often employed to model toxicity (13, 36), but their relevance is highly debated. We reasoned that a positive result for the highly validated AD risk factor PICALM might not only validate this assay but confirm the role of PICALM in Aβ detoxification.
We analyzed cortical neurons, a neuronal population particularly relevant to AD. Embryos from female rats with timed pregnancies were harvested at 18 days of gestation. Cortices dissected from these embryos were dissociated, plated and cultured for up to 21 days (23). The production of toxic Aβ oligomers is notoriously variable. We prepared oligomers according to several published methods, characterized them biochemically, and tested them for producing consistent levels of toxicity in our cortical neuronal preparations (Fig. 3, Fig. S6 and Fig. S7). The loss of toxicity when the same samples were allowed to form Aβ fibers (Fig. S6A and S6C) confirmed the importance of the oligomeric species (Fig. S6D).
Figure 3.
PICALM protects cultured rat cortical neurons from exogenously applied Aβ oligomers. Cortical neuron cultures prepared from rat embryos at embryonic day 18 were cultured for 5 days, transduced, cultured for 13 days, and then incubated for 20 hours with 750nM of soluble Aβ oligomers prepared from synthetic peptide (23). Infection with a PICALM lentiviral construct diminished toxicity in a dose-dependent, statistically significant manner. Cell viability was assessed by both ATP content (A) and MAP2 positive cell counting (B and C). Data are representative of three independent experiments and shown as mean +/− SEM ( *: p<0.05; **: p<0.01, based on Dunnett’s test for (A) and two-tail t-test for (B)).
Next, we infected neurons with lentiviruses engineered to express GFP or PICALM. When Aβ oligomers were added to these neurons, GFP had no effect but PICALM partially rescued the cells from Aβ induced cell death in a dose-dependent manner (Fig. 3). Rescue was significant whether assayed by cellular ATP levels (Fig. 3A) or by counting the number of MAP2 positive neurons (Fig. 3B and Fig. 3C). As previously described for midbrain neurons (7), we found that RAB1 protected cortical neurons from α-syn toxicity when this protein was expressed intracellularly by viral transfection. However, RAB1 was ineffective against Aβ oligomers, confirming the specificity of PICALM for the type of toxicity caused by Aβ oligomers (Fig. S7).
Effect of Aβ on endocytosis and trafficking
The role of PICALM in AD is unknown, but it has been postulated to affect disease by modifying APP trafficking (25). However, our experiments in yeast, nematode and rat neurons clearly establish PICALM as a modifier of Aβ toxicity itself. To investigate the mechanism for PICALM and the other modifiers that affect clathrin-mediated endocytosis, we returned to yeast.
One possibility is that clathrin-mediated endocytosis modulates Aβ toxicity simply by shunting toxic Aβ species to the lysosomal/vacuolar system for degradation. However, immunoblotting of yeast lysates indicated these modifiers had little effect on Aβ levels (Fig. S4). Alternatively, if Aβ specifically perturbs endocytic homeostasis, up-regulation of this pathway might ameliorate the defect.
To determine if Aβ altered clathrin distributions, we used a strain in which endogenous clathrin light chain (Clc1) was tagged with GFP, a fully functional fusion. A control strain exhibited the expected distribution of Clc1-GFP foci (37). Aβ perturbed clathrin localization, increasing both the number and brightness of foci, but decreasing their average size (Fig. 4A).
Figure 4.
Aβ causes defects in endocytosis and receptor protein trafficking. (A) In a yeast strain with GFP-tagged clathrin light chain (Clc1-GFP), ssAβ 1-42 caused an increase in Clc1-GFP foci. (B) A halo assay, which measures secretion of the α-factor pheromone, was used to assess secretion. In the screening strain ssAβ 1-42 diminished growth but did not reduce secretion compared to the vector control. In contrast, a strain expressing α-syn, which is known to impede ER-Golgi trafficking, exhibits a strong defect in secretion. (C) Aβ 1-42 caused a defect in the normal trafficking of Ste3 to the vacuole. A Ste3-YFP fusion is normally trafficked to the vacuole. Aβ-expressing cells showed accumulation of Ste3-YFP in cytoplasmic foci. Co-expression of YAP1802, INP52, and SLA1 partially restored Ste3-YFP trafficking to the vacuole. A constitutively-expressed Ste3-YFP gene was integrated in the genome of the control and screening strain. Scale bars are 5um. All images are on the same scale.
Such a pattern might indicate a defect in clathrin-mediated secretion as well as in endocytosis. To test effects of Aβ on secretion, we used a halo assay for secretion of the α-factor mating pheromone. As a control, we also tested the effects of α-syn expression, which produces a strong defect in secretion (7) (Fig. 4B). Unlike α-syn, Aβ did not inhibit secretion (Fig. 4B).
To assess the impact of Aβ on clathrin-mediated endocytosis specifically, we examined the well-characterized substrate Ste3, a member of the highly conserved G-protein coupled receptor family. Ste3 is targeted to the plasma membrane. In the absence of its ligand, the yeast mating-factor, it is constitutively endocytosed and degraded in the vacuole (38). As expected, a Ste3-YFP fusion was primarily localized to the vacuole in a control strain (Fig. 4C). In Aβ-expressing strains, endocytic trafficking of Ste3-YFP was profoundly perturbed and the protein was localized to numerous foci ((23), Fig. 4C). Aβ expression resulted in a reduction of vacuolar organelle size without a disruption in morphology, consistent with reduced delivery of cargo to this organelle. Finally, we tested the effects of the three Aβ toxicity suppressors that function in endocytic trafficking, YAP1802, INP52, and SLA1. Each partially reversed the defect in Ste3-YFP trafficking (Fig. 4C).
Conclusions and Perspectives
Our yeast model allowed us to conduct an unbiased screen of an entire genome for modifiers of Aβ toxicity. The emergence of three different genes involved in the process of clathrin-mediated endocytosis from nearly 6,000 tested ORFs confirms that the Aβ peptide in our model is trafficking through the secretory compartments as expected. More importantly, the ability of endocytic genes to rescue Aβ toxicity, together with the effects of Aβ on clathrin localization and the trafficking of a G protein-coupled receptor, establish that within these highly diverse organisms clathrin-mediated endocytosis is a critical point of vulnerability to Aβ.
Aβ oligomers have been reported to increase endocytosis in cultured cells (39), and human-induced neuronal cells derived from the fibroblasts of AD patients exhibit defects in endocytosis (40). Mechanistically, in our Aβ-expressing cells, the increased number of clathrin foci, the internalized foci of Ste3, and the effects of genetic modifiers on vacuolar localization all suggest that Aβ affects this pathway by interfering with the ability of endocytosed transmembrane receptors to reach their proper destinations.
PICALM, as well as two genes whose protein products (BIN1 and CD2AP) interact with hits from our screen are AD risk factors. Given the diversity of pathologies however, their connection to Aβ toxicity was unknown. Our work in yeast, nematodes, and rat cortical neurons clearly places these factors within the Aβ cascade, linking Aβ to the genetics of sporadic AD.
Neurons are particularly vulnerable to perturbations in the homeostasis of endocytosis, because they must constantly recycle both neurotransmitters and their receptors (41). Aβ interacts with, and alters signaling by, a variety of neuronal receptors (42). We propose that the conformational flexibility of these oligomers allows them to interact rather promiscuously with conformationally flexible unliganded receptors, which, in turn, disrupts endocytic homeostasis.
Our yeast screen also identified seven conserved genes functionally associated with the cytoskeleton. Because yeasts do not express tau, our findings may indicate that the connection between Aβ toxicity and the cytoskeleton is more deeply rooted than tau alone, probably involving clathrin-mediated endocytosis. In analyzing human GWAS data we also uncovered suggestive associations between AD and three other genes, XPO1, ADSSL1, and RABGEF1, and confirmed their Aβ relationships in yeast and nematode.
The treatments available for AD are few and their efficacy limited. Determining how best to rescue neuronal function in the context of the whole brain is a problem of staggering proportions. Our yeast model provides a tool for identifying genetic leads, investigating their mechanisms of action, and screening for genetic and small molecule modifiers of this devastating and etiologically complex disease.
Supplementary Material
Acknowledgments
We thank L. Chibnik, B. Keenan and D. Ng for helpful discussions. D. Wittrup, V. Lee, M.Vidal and C. Link for materials, J. Corneveaux, M. Huentelman, and other Translational Genomics investigators for assistance with the human study cohorts and participants in the Religious Orders Study and the Rush Memory and Aging Project. This work was supported by an HHMI Collaborative Innovation Award, NRSA fellowship F32 NS067782-02, the Cure Alzheimer’s Fund, NIH grants K08AG034290, P30AG10161, R01AG15819, and R01AG17917, the Kempe foundation and Alzheimerfonden.
Footnotes
Supplemental data are available in the Supplemental Online Material.
Supplemental Online Material
Materials and Methods
Figs. S1 to S7
Tables S1 to S7
References
REFERENCES
- 1.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker's yeast? Nat Rev Neurosci. 2010;11:436. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 2.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thayanidhi N, et al. Alpha-synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Mol Biol Cell. 2010;21:1850. doi: 10.1091/mbc.E09-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winslow AR, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190:1023. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gitler AD, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet. 2009;41:308. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su LJ, et al. Compounds from an unbiased chemical screen reverse both ER-to- Golgi trafficking defects and mitochondrial dysfunction in Parkinson's disease models. Dis Model Mech. 2009;3:194. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 9.Knobloch M, Konietzko U, Krebs DC, Nitsch RM. Intracellular Abeta and cognitive deficits precede beta-amyloid deposition in transgenic arcAbeta mice. Neurobiol Aging. 2007;28:1297. doi: 10.1016/j.neurobiolaging.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Lee EB, et al. Targeting amyloid-beta peptide (Abeta) oligomers by passive immunization with a conformation-selective monoclonal antibody improves learning and memory in Abeta precursor protein (APP) transgenic mice. J Biol Chem. 2006;281:4292. doi: 10.1074/jbc.M511018200. [DOI] [PubMed] [Google Scholar]
- 11.Shankar GM, et al. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh DM, Tseng BP, Rydel RE, Podlisny MB, Selkoe DJ. The oligomerization of amyloid beta-protein begins intracellularly in cells derived from human brain. Biochemistry. 2000;39:10831. doi: 10.1021/bi001048s. [DOI] [PubMed] [Google Scholar]
- 13.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 14.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 15.LaFerla FM. Pathways linking Abeta and tau pathologies. Biochem Soc Trans. 2010;38:993. doi: 10.1042/BST0380993. [DOI] [PubMed] [Google Scholar]
- 16.Ittner LM, et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell. 2010;142:387. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 17.Roberson ED, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 18.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Pelham HR. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- 21.Kowalski JM, Parekh RH, Wittrup KD. Secretion efficiency in Saccharomyces cerevisiae of bovine pancreatic trypsin inhibitor mutants lacking disulfide bonds is correlated with thermodynamic stability. Biochemistry. 1998;37:1264. doi: 10.1021/bi9722397. [DOI] [PubMed] [Google Scholar]
- 22.Lindhagen-Persson M, Brannstrom K, Vestling M, Steinitz M, Olofsson A. Amyloid-beta oligomer specificity mediated by the IgM isotype--implications for a specific protective mechanism exerted by endogenous auto-antibodies. PLoS One. 2010;5:e13928. doi: 10.1371/journal.pone.0013928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Please refer to Supplemental Online Material.
- 24.Stamenova SD, Dunn R, Adler AS, Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J Biol Chem. 2004;279:16017. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- 25.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert JC, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 27.Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272:16700. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 28.Gaidos G, Soni S, Oswald DJ, Toselli PA, Kirsch KH. Structure and function analysis of the CMS/CIN85 protein family identifies actin-bundling properties and heterotypic-complex formation. J Cell Sci. 2007;120:2366. doi: 10.1242/jcs.004333. [DOI] [PubMed] [Google Scholar]
- 29.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naj AC, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertram L, et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacker D, et al. ApoE-4 and age at onset of Alzheimer's disease: the NIMH genetics initiative. Neurology. 1997;48:139. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 33.Bennett DA, De Jager PL, Leurgans SE, Schneider JA. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009;72:1495. doi: 10.1212/WNL.0b013e3181a2e87d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulman JM, et al. Intermediate phenotypes identify divergent pathways to Alzheimer’s disease. PLoS ONE. 2010;5:e1124. doi: 10.1371/journal.pone.0011244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bozyczko-Coyne D, et al. CEP-1347/KT-7515, an inhibitor of SAPK/JNK pathway activation, promotes survival and blocks multiple events associated with Abeta-induced cortical neuron apoptosis. J Neurochem. 2001;77:849. doi: 10.1046/j.1471-4159.2001.00294.x. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y, Carroll S, Kaksonen MY, Toshima J, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldonado-Baez L, et al. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol Biol Cell. 2008;19:2936. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minano-Molina AJ, et al. Soluble Oligomers of Amyloid-{beta} Peptide Disrupt Membrane Trafficking of {alpha}-Amino-3-hydroxy-5-methylisoxazole-4-propionic Acid Receptor Contributing to Early Synapse Dysfunction. J Biol Chem. 2011;286:27311. doi: 10.1074/jbc.M111.227504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiang L, et al. Directed conversion of Alzheimer's disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Jung N, Haucke V. Clathrin-mediated endocytosis at synapses. Traffic. 2007;8:1129. doi: 10.1111/j.1600-0854.2007.00595.x. [DOI] [PubMed] [Google Scholar]
- 42.Verdier Y, Zarandi M, Penke B. Amyloid beta-peptide interactions with neuronal and glial cell plasma membrane: binding sites and implications for Alzheimer's disease. J Pept Sci. 2004;10:229. doi: 10.1002/psc.573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.