Abstract
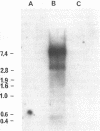
The nucleotide sequence of an infectious clone of rice tungro bacilliform virus (RTBV) DNA has been determined. The circular genome has 8002 bp and one strand contains four open reading frames (ORFs). One ORF is potentially capable of encoding a protein of 24 kD (P24) and has no initiation (ATG) codon. The other three ORFs potentially encode proteins of 12 kD, 194 kD and 46 kD (P12, P194, P46) respectively. The functions of P24, P12 and P46 are unknown. Comparative analyses with retroviruses and Commelina yellow mottle virus suggest that the 194 kD putative product is a polyprotein that is proteolytically cleaved to yield the virion coat protein, a protease and replicase (reverse transcriptase and RNase H) characteristic of retroelements. The DNA sequence reveals other features which strongly support our belief that RTBV is a pararetrovirus. These include sequences at the mapped positions of two discontinuities in the virion DNA which are complementary to tRNA metinit and purine-rich, and may be the priming sites for minus- and plus-strand DNA synthesis respectively. As the positions of likely transcriptional signals suggest, a full-length viral transcript is observed by northern analysis. The predicted folding of the 645 bp 5'-region of this RNA resembles that of caulimoviruses. Comparisons with other reverse transcribing elements are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonneville J. M., Sanfaçon H., Fütterer J., Hohn T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989 Dec 22;59(6):1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- Bouchez D., Tokuhisa J. G., Llewellyn D. J., Dennis E. S., Ellis J. G. The ocs-element is a component of the promoters of several T-DNA and plant viral genes. EMBO J. 1989 Dec 20;8(13):4197–4204. doi: 10.1002/j.1460-2075.1989.tb08605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaday J., Guillemaut P., Weil J. H. The nucleotide sequences of the initiator transfer RNAs from bean cytoplasm and chloroplasts. Nucleic Acids Res. 1980 Mar 11;8(5):999–1008. doi: 10.1093/nar/8.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S. N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986 Jan 24;14(2):623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S. N., Turner D., Mulder G. A small DNA molecule containing covalently-linked ribonucleotides originates from the large intergenic region of the cauliflower mosaic virus genome. Nucleic Acids Res. 1983 Jan 25;11(2):251–264. doi: 10.1093/nar/11.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Gordon K., Bonneville J. M., Sanfaçon H., Pisan B., Penswick J., Hohn T. The leading sequence of caulimovirus large RNA can be folded into a large stem-loop structure. Nucleic Acids Res. 1988 Sep 12;16(17):8377–8390. doi: 10.1093/nar/16.17.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Gordon K., Sanfaçon H., Bonneville J. M., Hohn T. Positive and negative control of translation by the leader sequence of cauliflower mosaic virus pregenomic 35S RNA. EMBO J. 1990 Jun;9(6):1697–1707. doi: 10.1002/j.1460-2075.1990.tb08293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh H. P., Ghosh K., Simsek M., RajBhandary U. L. Nucleotide sequence of wheat germ cytoplasmic initiator methionine transfer ribonucleic acid. Nucleic Acids Res. 1982 May 25;10(10):3241–3247. doi: 10.1093/nar/10.10.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Wu F. C., Scholthof H. B., Shepherd R. J. Gene VI of figwort mosaic virus (caulimovirus group) functions in posttranscriptional expression of genes on the full-length RNA transcript. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9203–9207. doi: 10.1073/pnas.86.23.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M. A., Spielmann A., Caboche M. Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature. 1989 Jan 26;337(6205):376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- Hasegawa A., Verver J., Shimada A., Saito M., Goldbach R., Van Kammen A., Miki K., Kameya-Iwaki M., Hibi T. The complete sequence of soybean chlorotic mottle virus DNA and the identification of a novel promoter. Nucleic Acids Res. 1989 Dec 11;17(23):9993–10013. doi: 10.1093/nar/17.23.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R., Covey S. N., Maule A. J. Structure and replication of caulimovirus genomes. J Cell Sci Suppl. 1987;7:213–229. doi: 10.1242/jcs.1987.supplement_7.16. [DOI] [PubMed] [Google Scholar]
- Hull R., Sadler J., Longstaff M. The sequence of carnation etched ring virus DNA: comparison with cauliflower mosaic virus and retroviruses. EMBO J. 1986 Dec 1;5(12):3083–3090. doi: 10.1002/j.1460-2075.1986.tb04614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Meshi T., Motoyoshi F., Takamatsu N., Okada Y. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 1986 Nov 11;14(21):8291–8305. doi: 10.1093/nar/14.21.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. C., Gough K., Dasgupta I., Rao B. L., Cliffe J., Qu R., Shen P., Kaniewska M., Blakebrough M., Davies J. W. Rice tungro disease is caused by an RNA and a DNA virus. J Gen Virol. 1991 Apr;72(Pt 4):757–761. doi: 10.1099/0022-1317-72-4-757. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver C. J., Ingle J. The molecular integrity of chloroplast ribosomal ribonucleic acid. Biochem J. 1971 Jun;123(2):235–243. doi: 10.1042/bj1230235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. L., Ellis T. H., Turner L., Hellens R. P., Cleary W. G. A copia-like element in Pisum demonstrates the uses of dispersed repeated sequences in genetic analysis. Plant Mol Biol. 1990 Nov;15(5):707–722. doi: 10.1007/BF00016121. [DOI] [PubMed] [Google Scholar]
- Mares M., Meloun B., Pavlik M., Kostka V., Baudys M. Primary structure of cathepsin D inhibitor from potatoes and its structure relationship to soybean trypsin inhibitor family. FEBS Lett. 1989 Jul 17;251(1-2):94–98. doi: 10.1016/0014-5793(89)81435-8. [DOI] [PubMed] [Google Scholar]
- Mason W. S., Taylor J. M., Hull R. Retroid virus genome replication. Adv Virus Res. 1987;32:35–96. doi: 10.1016/s0065-3527(08)60474-1. [DOI] [PubMed] [Google Scholar]
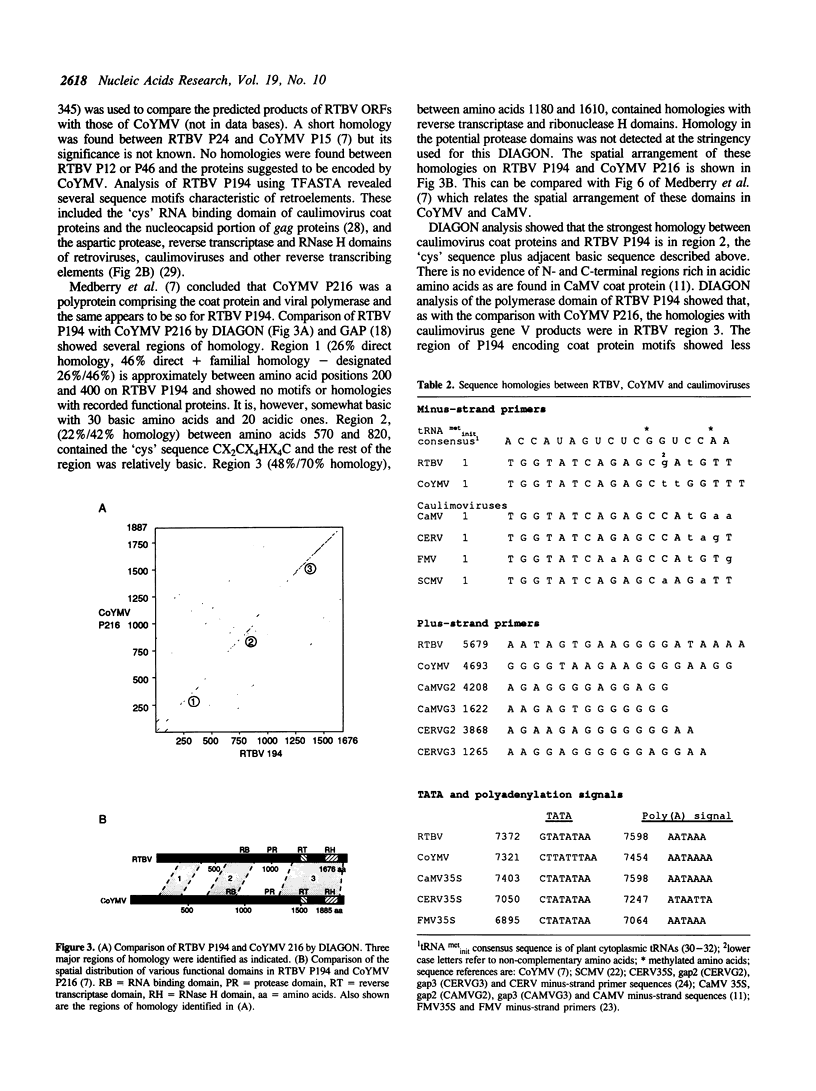
- Medberry S. L., Lockhart B. E., Olszewski N. E. Properties of Commelina yellow mottle virus's complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic Acids Res. 1990 Sep 25;18(18):5505–5513. doi: 10.1093/nar/18.18.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penswick J., Hübler R., Hohn T. A viable mutation in cauliflower mosaic virus, a retroviruslike plant virus, separates its capsid protein and polymerase genes. J Virol. 1988 Apr;62(4):1460–1463. doi: 10.1128/jvi.62.4.1460-1463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Richins R. D., Scholthof H. B., Shepherd R. J. Sequence of figwort mosaic virus DNA (caulimovirus group). Nucleic Acids Res. 1987 Oct 26;15(20):8451–8466. doi: 10.1093/nar/15.20.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sibbald P. R., Argos P. Scrutineer: a computer program that flexibly seeks and describes motifs and profiles in protein sequence databases. Comput Appl Biosci. 1990 Jul;6(3):279–288. doi: 10.1093/bioinformatics/6.3.279. [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Kalitsis P., Joseph J. L., Sentry J. W. Plant retrotransposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5015–5019. doi: 10.1073/pnas.86.13.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Meissner F., Moll J., Vorderwülbecke T. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1987;15 (Suppl):r53–188. doi: 10.1093/nar/15.suppl.r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcriptases. Retrons in bacteria. Nature. 1989 May 25;339(6222):254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Volovitch M., Drugeon C., Yot P. Studies on the single-stranded discontinuities of the cauliflower mosaic virus genome. Nucleic Acids Res. 1978 Aug;5(8):2913–2925. doi: 10.1093/nar/5.8.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. F., Ausubel F. M. A copia-like transposable element family in Arabidopsis thaliana. Nature. 1988 Nov 17;336(6196):242–244. doi: 10.1038/336242a0. [DOI] [PubMed] [Google Scholar]