Abstract
Background
Primary tumor recurrence commonly occurs after surgical resection of lung squamous cell carcinoma (SCC). Little is known about the genes driving SCC recurrence.
Methods
We used array comparative genomic hybridization (aCGH) to identify genes affected by copy number alterations that may be involved in SCC recurrence. Training and test sets of resected primary lung SCC were assembled. aCGH was used to determine genomic copy number in a training set of 62 primary lung SCCs (28 with recurrence and 34 with no evidence of recurrence) and the altered copy number of candidate genes was confirmed by quantitative PCR (qPCR). An independent test set of 72 primary lung SCCs (20 with recurrence and 52 with no evidence of recurrence) was used for biological validation. mRNA expression of candidate genes was studied using qRT-PCR. Candidate gene promoter methylation was evaluated using methylation microarrays and Sequenom EpiTYPER analysis.
Results
18q22.3 loss was identified by aCGH as being significantly associated with recurrence (p = 0.038). Seven genes within 18q22.3 had aCGH copy number loss associated with recurrence but only SOCS6 copy number was both technically replicated by qPCR and biologically validated in the test set. SOCS6 copy number loss correlated with reduced mRNA expression in the study samples and in the samples with copy number loss, there was a trend for increased methylation, albeit non-significant. Overall survival was significantly poorer in patients with SOCS6 loss compared to patients without SOCS6 loss in both the training (30 vs. 43 months, p = 0.023) and test set (27 vs. 43 months, p = 0.010).
Conclusion
Reduced copy number and mRNA expression of SOCS6 are associated with disease recurrence in primary lung SCC and may be useful prognostic biomarkers.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide, accounting for greater than one million deaths annually [1]. Non-small cell lung cancer (NSCLC) accounts for 80% of all lung cancer diagnoses. Conventionally NSCLC has been divided into three subtypes: adenocarcinoma (AC), squamous cell carcinoma (SCC) and large cell carcinoma (LC), with AC and SCC accounting for 85% of NSCLC cases [2]. The treatments for NSCLC have been generic and largely ineffective resulting in a five-year survival of 15% [3]. Early diagnosis followed by surgical resection remains the most effective treatment strategy [4]. However, even in stage I patients undergoing surgical resection, recurrence of the primary tumor occurs in 30–35% of cases [2]. Molecular alterations are likely to be involved in driving disease recurrence, but the specific genes involved remain to be elucidated.
DNA copy number alterations are ubiquitous to almost all human malignancies [5]. The identification of tumor-specific DNA copy number alterations can assist in the discovery of oncogenes or tumor suppressor genes which are typically located within genomic regions of amplifications or loss respectively [5]. Array-comparative genomic hybridization (aCGH) has been used to investigate copy number alterations in several malignancies, including lung SCC [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]. Karyotyping and conventional CGH characterized lung SCCs, as having frequent copy number gains in 1p, 3q, 5p, 7q and 8q and copy number loss in 3p, 5q, 8p, 9p, and 14q [13], [17], [18], [19], [20], [21], [22]. High-resolution copy number characterization of these regions resulted in the discovery of the driver oncogenes SOX2 [14] and FGFR1 [23]. To our knowledge, the few studies that have specifically evaluated genomic differences unique to lung SCC recurrence and/or metastases have used low-resolution platforms on relatively small sample sizes [18], [24]. Consequently, while these studies have identified genomic regions with copy number alterations associated with recurrence and/or metastases, they have not been able to identify the driving gene/s associated with recurrence.
In this study we analyzed lung SCC tumors using a whole-genome aCGH microarray platform to identify genomic copy number alterations specific to tumors, which developed early recurrence of primary tumor post-resection. To identify recurrence specific genes within the candidate genomic regions, we used an independent method of copy number determination (quantitative PCR) and confirmed the findings in an independent set of SCC tumors. Finally, to assess whether candidate gene/s copy number alterations have prognostic value, we analyzed the survival data of training and test set subjects.
Materials and Methods
Subjects and Tumor Samples
The training set consisted of sixty-two tumor samples, which were collected from patients with histologically proven primary lung SCC. The tumor samples were obtained from patients who underwent curative-intent surgical resection at The Prince Charles Hospital between 1990 and 2004. Formalin fixed paraffin embedded tissue samples of normal lung and tumor tissue, adjacent to the frozen tumor sample, was used for hematoxylin and eosin examination and only those tumor samples that contained at least 50% tumor cells and all surgical bronchial margins were free of disease were used as training set samples and underwent aCGH experiments. The subjects were fitted to one of our two disease recurrence outcome criteria: non-recurrence, clinically disease-free for at least 36 months following surgery; or recurrent disease, unambiguous clinical, imaging, or histopathologic evidence of local or distant recurrence of the original primary lung cancer in a local or distant metastatic site occurring between 3 and 18 months post-resection. The threshold of 36 months for non-recurrence cases was selected since most patients develop disease recurrence within this period of time and to allow for comparison with other similarly designed studies. In addition, an independent test set, consisting of seventy-two tumor samples that were collected and stored in The Prince Charles Hospital Lung Tissue Bank were utilized for validation purposes. Inclusion and exclusion criteria for the test set were identical to that used for the training set.
Ethics Statement
Ethics approval was granted from TPCH Human Research Ethics Committee (HREC/09/QPCH/17) and The University of Queensland (Project number: 2009000727) and all subjects provided written informed consent prior to inclusion in the study.
Nucleic Acid Extraction
Tumor and paired normal lung tissue collected from surgical resection specimens, as previously described [25], were snap frozen and processed for genomic DNA and total RNA extraction. A total of 300–500 mg of frozen tissue was used for genomic DNA extraction using a modified salt-precipitation method [26]. High molecular weight genomic DNA was purified using a Blood and Cell Culture mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Purified genomic DNA was quantitated with a NanoDrop spectrophotometry system (NanoDrop Technologies, Wilmington, DE, USA) using 2 µL of DNA. Total RNA was extracted as described previously [25].
Array Comparative Genomic Hybridization
aCGH experiments were performed on Human Genome Microarray 44B (Agilent Technologies Inc.) microarrays, a high-resolution 60-mer oligonucleotide-based microarray that contains 42,920 probes sourced from the NCBI human genome reference sequence. These probes represent 24,983 genes. Microarray images were analyzed using Feature Extraction Software, version 8.0 (Agilent Technologies, Inc.) and assessed for relative data quality in CGH Analytics, version 3.4.27 (Agilent Technologies, Inc.). For each aCGH microarray, a tumor sample (test) was compared with a commercially available Female Genomic DNA (Promega, Madison, USA). A triangular smoothing algorithm with a moving average window of 2 Mb was applied to log ratio aCGH data. aCGH microarray experiments were designed in compliance with the MIAME guidelines (http://www.mged.org/Workgroups/MIAME/miame.html). The aCGH raw data (unchanged), metadata and clinical information of the subjects in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) public repository (http://www.ncbi.nlm.nih.gov/geo) and can be accessed through the accession number GSE32058. Normalized aCGH data was analyzed using Genomic Identification of Significant Target in Cancer (GISTIC), a bioinformatics method that identifies genomic regions most likely to contain oncogenes and tumor suppressor genes [27]. Segmented regions were estimated with Circular Binary Segmentation (CBS) using the R package “DNACopy” (http://www.r-project.org/). Copy number variation data from the Human Genome Build 35 (hg17) was obtained from the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation). GISTIC scores for locus (G score) were obtained as the product of frequency and mean amplitude of amplifications or deletions. Only amplifications exceeding a log2 copy number ratio of 0.848 for amplifications or below 0.737 for deletions were included, accounting for 2.8 copies per cell and 1.6 copies per cell in samples respectively. G scores were compared against a null model to determine a false discovery rate (q value) and peaks with q values below 0.05 were considered.
Real-time Quantitative PCR and Quantitative Reverse Transcription PCR
To validate copy number alterations detected by aCGH, real-time quantitative PCR (qPCR) assays were used. Pre-designed QuantiTect primers (Qiagen) were used to measure candidate gene copy number by qPCR, details of which are provided in Table S3. Information pertaining to the location of the primers was obtained from Qiagen (http://www.qiagen.com/geneglobe). β-actin was used as the reference locus for qPCR. Normal human pooled genomic DNA was used as reference DNA (Promega, Madison, USA). All qPCR experiments were performed using the Rotor-Gene 6000™ (Qiagen, Hilden, Germany). Each assay was performed in triplicate in 10 µL reactions containing 5 µL QuantiFast SYBR Green PCR Master Mix (2×; Qiagen); 2 ul QuantiTect Primer assay (10×; Qiagen); and 10 ng of genomic DNA. PCR product amplification was performed according to the following conditions: 1 cycle at 95°C for 10 minutes, 40 cycles, of 95°C for 10 seconds and 60°C for 30 seconds.
To evaluate the mRNA expression levels of candidate genes, we used qRT-PCR. mRNA levels of the candidate gene were compared to those of housekeeper genes. The geometric mean of the relative gene expression of BAT1, SEPT2 and 18s were used as the comparator reference for RT-PCR. Relative gene expression was calculated using the Pfaffl method [28]. qRT-PCR was carried out using QuantiTect primer sets and 30 ng of cDNA as template. All assays were performed in triplicate in 10 µL reactions containing 5 µL QuantiFast SYBR Green PCR Master Mix (2×; Qiagen) and 2 ul QuantiTect Primer assay (10×; Qiagen).
Methylation Analysis
Microarray
Methylation microarray data (manuscript in preparation) was available for 49 tumor samples from the training set. Genomic DNA was bisulphite converted using an EZ DNA Methylation Kit (Zymo Research, CA, USA) and hybridized to Illumina Infinium Methylation 27 V1.0 microarrays. The β-value was determined as the ratio of methylated fluorescence signal to the combined signal of the methylated and unmethylated alleles, giving a value between 0 and 1.
MassARRAY ® EpiTYPER
We employed the MassARRAY® EpiTYPER analysis (Sequenom), for the detection and quantitation of DNA methylation of the promoter regions of SOCS6. The EpiTYPER Sequenom Mass Array service was provided by Sequenom, Inc. (Brisbane, Australia) and for each sample, 1.5 µg of DNA in a volume of 30 µL was sent to the service. EpiTYPER is a validated approach in providing a highly quantitative view of CpG dinucleotide methylation, with up to single nucleotide resolution, using the technique of MALDI TOF (Matrix-assisted laser desorption/ionization Time of Flight) mass spectrometry. Primers for SOCS6 were designed using the Epidesigner software (primers available on request). The SOCS6 promoter was divided into one or more amplicons, within a region comprising 2500 bp upstream of the transcription start site and the regions analyzed corresponded to annotated CpG islands identified using the UCSC Genome Browser (http://genome.ucsc.edu). Genomic DNA was bisulfite treated using EZ-96 DNA methylation kits (Zymo Research, CA, USA), followed by PCR amplification using primers directed to the promoter regions of SOCS6. Amplicons were then subjected to the EpiTYPER assays, the products analyzed by mass spectrometry and methylation ratios obtained using EpiTYPER v1.0.5 software (SEQUENOM). The relative amount of methylation (% methylation) was determined by comparing the signal intensities between the mass signals of methylated and non-methylated template.
Statistical Analysis
All statistical analyses were performed using SPSS (Version 17, SPSS Inc., Chicago, USA). Fisher's exact test was applied to assess the relationship between copy number as a bivariate categorical variable (above or below the log base 2 threshold) and clinico-pathological characteristics. The Mann-Whitney U test was used to measure differences in candidate gene copy number between recurrence and non-recurrence samples. The censored five-year overall survival after surgical resection was estimated using the Kaplan-Meier method and survival differences were analyzed using the log-rank test.
Results
aCGH Profile of Lung SCC Tumors and Recurrence Phenotype
In the training and test set subjects, there were no significant differences in clinical or pathological characteristics between those with disease recurrence and those without (Table 1).
Table 1. Subject Characteristics.
| Training set | Test set | |||||
| Recurrence SCC, n (%) | Non-Recurrence SCC, n (%) | p-value* | Recurrence SCC, n(%) | Non-Recurrence SCC, n(%) | p-value* | |
| Subjects, n | 28 | 34 | 20 | 52 | ||
| Age, years | ||||||
| Median (range) | 67 (39–81) | 69 (44–84) | 69 (39–82) | 69 (44–91) | ||
| Gender, n(%) | ||||||
| Males | 21 (75) | 25 (73) | 1.0 | 14 (70) | 43 (83) | 0.33 |
| Females | 7 (25) | 9 (27) | 6 (30) | 9 (17) | ||
| Smoking history, n(%) | ||||||
| Never | 3 (11) | 1 (3) | 0.32 | 2 (10) | 4 (7) | 0.67 |
| Ever smokers | 25 (89) | 33 (97) | 18 (90) | 48 (93) | ||
| Pack years, median (range) | 35 (0–100) | 38 (0–135) | 39 (0–75) | 35 (0–243) | ||
| Stage, n(%) | ||||||
| I–II | 23 (82) | 30 (88) | 0.72 | 14 (70) | 44 (85) | 0.19 |
| III–IV | 5 (18) | 4 (12) | 6 (30) | 8 (15) | ||
| Tumor differentiation, n(%) | ||||||
| Well to moderate | 14 (50) | 18 (53) | 1.0 | 9 (45) | 26 (50) | 0.80 |
| Poor | 14 (50) | 16 (47) | 11 (55) | 26 (50) | ||
| Tumor invasion, n(%) | ||||||
| Lymphatic | 8 (29) | 4 (12) | 0.12 | 2 (10) | 5 (10) | 1.00 |
| Vascular | 11 (39) | 8 (24) | 0.27 | 10 (50) | 15 (29) | 0.10 |
| Perineural | 3 (11) | 0 (0) | 0.09 | 2 (10) | 4 (8) | 0.67 |
| Pleural | 6 (21) | 8 (24) | 1.00 | 5 (33) | 16 (31) | 0.78 |
*p-value determined using Fisher's exact test.
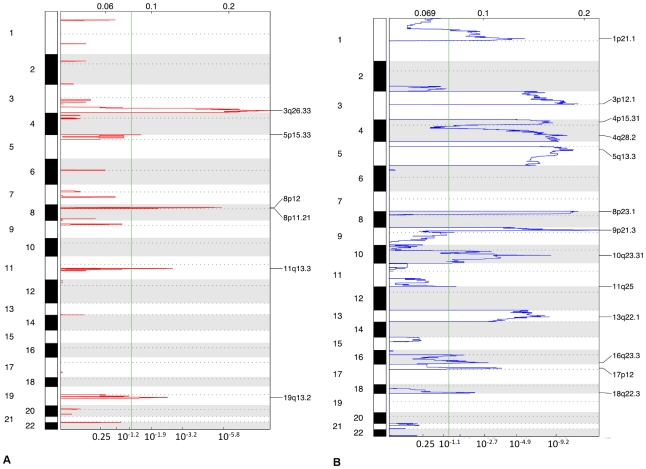
We initially assayed copy number alterations in all 62 tumors of the training set. GISTIC identified nineteen genomic regions (thirteen deletions and six amplifications) with a false discovery rate <0.05 (Figure 1) (Table 2). The frequency of copy number alteration at each of these regions was compared between disease recurrence groups. Only 18q22.3 was significantly different, showing more frequent loss in recurrence compared with non-recurrence tumors (54% vs. 24%, p = 0.038) (Table 3). When the tumors were stratified by recurrence phenotype and analyzed separately, GISTIC identified loss of 18q22.3 only in tumors, which recurred, while loss in 1p, 3p, 4q, 5q, 8p, 9p, 10q, 13q, 16q, 17p were common to both phenotypes. Conversely, amplifications in 5p15.33, 8q24.21, 9p21.1 and 19q13.2 were unique to non-recurrence tumors while amplifications in 3q and 8p occurred in both phenotypes (Table S1).
Figure 1. Genomic copy number alterations in lung squamous cell carcinomas.
Plots of high-level amplifications (a) and deletions (b) in 62 lung SCCs from GISTIC analysis of aCGH data. X-axis shows the G score (top) and false discovery rate (q value; bottom) with a green line demarcating a false discovery rate of 0.05. Labels on the right denote the peaks of the most significantly altered regions.
Table 2. GISTIC Identified Chromosomal Regions with Copy Number Alterations in Training Set SCC Tumors.
| Cytoband | Broad or Focal | Region Limits | Frequency of deletions in test set samples, n (%) | FDR values | GISTIC genes within region | Cancer-associated genes |
| Regions with Copy Number Losses | ||||||
| 9p21.3 | Both | 21733411–21957547 | 33 (53) | 2.30×10−18 | 4 | CDKN2A, CDKN2B |
| 8p23.1 | Broad | 3313385–12711819 | 40 (65) | 1.04×10−13 | 67 | ANGPT2 |
| 3p12.1 | Broad | 84414538–85858393 | 38 (61) | 1.04×10−13 | 6 | POU1F1 |
| 5q13.3 | Broad | 76066702–76539988 | 38 (61) | 7.97×10−13 | 76 | PIK3R1, HTR1A |
| 4q28.2 | Broad | 130278523–134426567 | 37 (60) | 1.06×10−11 | 12 | PLK4 |
| 10q23.31 | Broad | 89540134–89615643 | 25 (40) | 2.67×10−09 | 4 | PTEN |
| 13q22.1 | Broad | 72396304–72490843 | 25 (40) | 4.32×10−09 | 7 | |
| 1p21.1 | Broad | 101664084–102994090 | 30 (48) | 2.41×10−06 | 8 | COL11A1 |
| 17p12 | Broad | 12394069–12459756 | 24 (39) | 1.50×10−04 | 17 | |
| 4p15.31 | Broad | 20391110–20848036 | 35 (56) | 4.09×10−03 | 59 | BST1, SLIT2, PI4K2B |
| 16q23.3 | Broad | 80626349–81449546 | 23 (37) | 1.03×10−03 | 16 | |
| 18q22.3 | Broad | 64017241–75522477 | 24 (39) | 4.96×10−03 | 18 | SOCS6, CD226, RTTN, MBP |
| 11q25 | Broad | 132926079–134452384 | 17 (27) | 2.83×10−02 | 10 | ACAD8 |
| Regions with Copy Number Gains | ||||||
| 3q26.33 | Focal | 183473858–183941579 | 15 (24) | 1.38×10−13 | 9 | SOX2, TP73L |
| 8p12 | Focal | 38703882–38796309 | 8 (13) | 9.01×10−09 | 2 | FGFR1 |
| 19q13.2 | Focal | 43840068–44336812 | 4 (6) | 2.73×10−04 | 66 | AKT2, ECH1 |
| 11q13.3 | Focal | 68821296–70110392 | 5 (8) | 3.06×10−02 | 13 | CCND1, PPFIA1 |
| 5p15.33 | Focal | 1–1510601 | 6 (10) | 1.46×10−02 | 21 | TERT |
| 8p11.21 | Focal | 41969702–42416269 | 2 (3) | 4.19×10−02 | 630 | TACC1 |
Table 3. Frequency of Copy Number Alterations of GISTIC Identified Chromosomal Regions in Recurrence and Non-recurrence SCC Training Set Samples.
| Cytoband | Loss in SCC Recurrence, n (%) | Loss in SCC Non-Recurrence, n (%) | p-value* |
| Regions with Copy Number Losses | |||
| 18q22.3 | 15 (54) | 8 (24) | 0.038 |
| 4q28.2 | 14 (50) | 23 (68) | 0.198 |
| 1p21.1 | 11 (39) | 19 (56) | 0.213 |
| 10q23.31 | 9 (32) | 16 (47) | 0.301 |
| 11q25 | 6 (21) | 11 (32) | 0.400 |
| 8p23.1 | 20 (71) | 20 (59) | 0.425 |
| 3p12.1 | 19 (68) | 19 (56) | 0.434 |
| 5q13.3 | 17 (61) | 21 (62) | 0.604 |
| 13q22.1 | 10 (36) | 15 (44) | 0.606 |
| 9p21.3 | 16 (57) | 17 (50) | 0.617 |
| 17p12 | 10 (36) | 14 (41) | 0.795 |
| 4p15.31 | 15 (54) | 20 (59) | 0.798 |
| 16q23.3 | 10 (36) | 13 (38) | 1.000 |
| Regions with Copy Number Gains | |||
| 3q26.33 | 4 (14) | 11 (32) | 0.139 |
| 19q13.2 | 1 (4) | 3 (9) | 0.620 |
| 5p15.33 | 2 (7) | 4 (12) | 0.681 |
| 8p12 | 4 (14) | 4 (12) | 1.000 |
| 11q13.3 | 2 (7) | 3(9) | 1.000 |
| 8p11.21 | 1 (4) | 1 (3) | 1.000 |
*p-value determined using Fisher's exact test.
Candidate Genes within Recurrence-Associated Copy Number Loss at 18q22.3
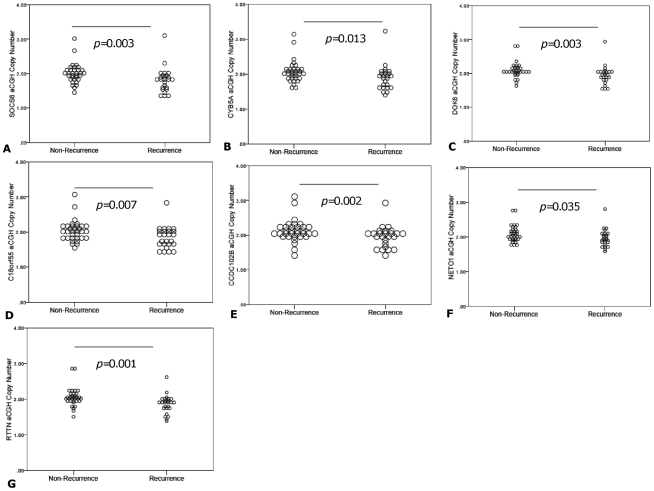
The GISTIC algorithm identified a region of loss within 18q22.3 from 64017241 to 75522477 (Table 2). Within 18q22.3 were 18 candidate genes (Table S2), which were further studied to determine whether they were recurrence-specific. Seven of these (CYB5A, SOCS6, DOK6, C18orf55, CCDC102B, NETO1 and RTTN) had lower copy number in primary tumors of patients that recurred (Figure 2). Table S2 lists the oligonucleotide probe ID and location for all probes within the 18q22.3 region of interest.
Figure 2. aCGH copy number of genes within 18q22.3 demonstrating preferential loss in SCC recurrence.
The Y-axis represents the derived DNA copy number from aCGH log2 normalized data and the X-axis represents the recurrence phenotype. Mann-Whitney U test to was used to assess for any differences in copy number between recurrence phenotypes and p values<0.05 were deemed significant.
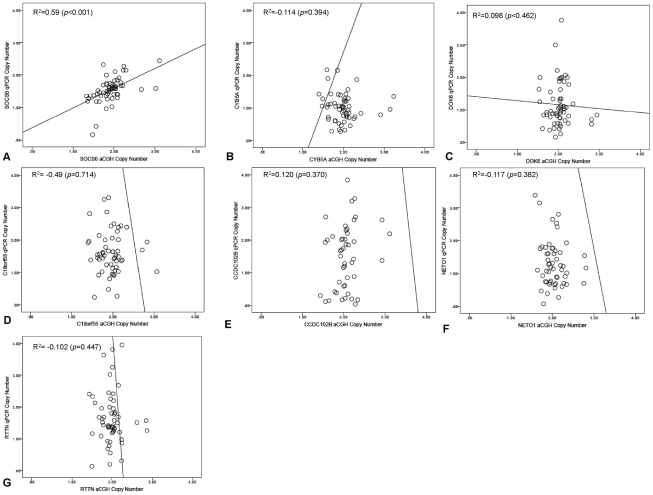
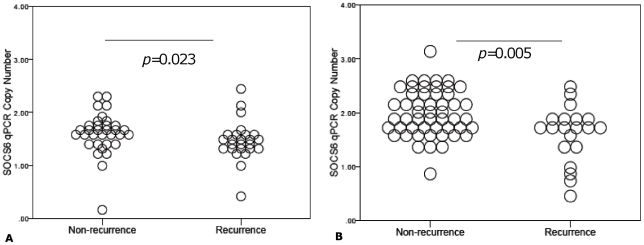
aCGH based copy number assessment of the seven candidate genes was technically validated by qPCR (Figure 3). A high degree of concordance between aCGH and qPCR data was observed for SOCS6 (R2 = 0.59, p<0.001), but CYB5A, DOK6, C18orf55, CCDC102B, NETO1 and RTTN showed Pearson coefficients of <0.5 (Figure 3). Given these results, we also tested the association between qPCR-determined copy number in the training and test set tumors. qPCR confirmed significantly lower SOCS6 copy number in the group with tumor recurrence in both training (p = 0.023) (Figure 4A) and test (p = 0.005) (Figure 4B) sets.
Figure 3. Quantitative PCR (qPCR) validation of array CGH identified candidate genes preferentially lost in SCC recurrence.
The Y-axis represents the derived DNA copy number from qPCR normalized to house-keeper genes, b-actin while the X-axis represents the DNA copy number derived from aCGH. Pearson's correlation coefficient was used to assess for any relationship between the copy number derived from the methods and p values<0.05 were deemed significant. The aCGH copy number of onlySOCS6 (a) was validated by qPCR.
Figure 4. Dot plot of qPCR-derived SOCS6 copy number (y-axis) is compared to the recurrence phenotype (x-axis) in the training (n = 62) and test set (n = 72) subjects.
Figure 4A and 4B represent training set and test set subjects respectively. Mann-Whitney U test to was used to assess for any differences in copy number between recurrence phenotypes and p values<0.05 were deemed significant.
To confirm that the SOCS6 copy number alterations were somatically acquired, SOCS6 copy number was determined by qPCR in the paired normal lung of the training set tumors. The median copy number of SOCS6 in normal lung was higher than in tumor samples (p<0.001) (Figure S1A), and the number of SOCS6 copies in normal lung did not differ between patients with recurrence and non-recurrence tumors (p = 0.321) (Figure S1B).
SOCS6 mRNA Expression in SCC Tumors
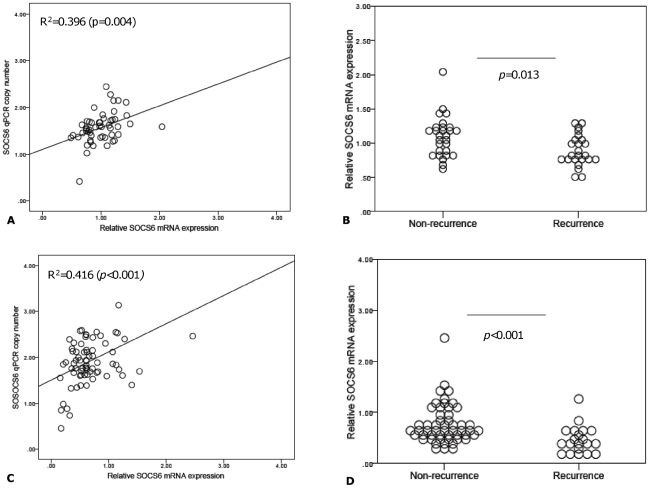
To determine if SOCS6 DNA copy number was an important regulator of mRNA expression, qRT-PCR was performed in the training and test set tumor cDNA samples. There was a modest correlation between SOCS6 copy number and mRNA expression in the training set (r2 = 0.396, p = 0.004) (Figure 5A) and test set (r2 = 0.416, p<0.001) (Figure 5C). SOCS6 mRNA expression was significantly lower in recurrence samples in the training (p = 0.013, Figure 5B) and test sets (p = <0.001, Figure 5D).
Figure 5. Relationship between SOCS6 mRNA expression, copy number and recurrence phenotype.
Figure 5A and 5C are scatter plots of qRT-PCR derived SOCS6 mRNA expression (x-axis) and qPCR-derived SOCS6 copy number (y-axis) in training set (n = 62) (a) and test set (n = 72) (c) respectively. Figure 5B and 5D represent SOCS6 mRNA levels (y-axis) ad recurrence phenotype (x-axis) in the training and test sets respectively. Pearson's correlation coefficient was used to determine any association between SOCS6 qPCR derived copy number and mRNA levels. Mann-Whitney U test to was used to assess for any differences in copy number between recurrence phenotypes and p values<0.05 were deemed significant.
SOCS6 Methylation Status in SCC tumors
An alternative mechanism responsible for reduced mRNA expression, in addition to gene copy number loss, is promoter hypermethylation. We therefore evaluated the methylation status of SOCS6 in the training set of SCC tumor samples using two independent techniques: Illumina Infinium Methylation microarrays and MassARRAY® EpiTYPER analysis. Methylation microarray analysis of 30 non-recurrence and 19 recurrence tumors from the training set found the methylation index (β) was lower in tumors with SOCS6 loss (i.e. <2 copies/cell) (n = 29) than in tumors with normal copy number (i.e. ≥2 copies/cell) (n = 20), but the difference was not statistically significant (0.047±0.047 vs. 0.063±0.041, p = 0.211). There was no correlation between SOCS6 methylation and mRNA expression either in samples with <2 SOCS6 copies/cell (r2 = −0.065, p = 0.748) or in samples with ≥2 SOCS6 copies/cell (r2 = 0.061, p = 0.810).
Quantitation of the degree of SOCS6 methylation using mass spectrometry of amplification products with the Sequenom EpiTYPER assay was performed in 62 SCC tumor samples from the training set. Five primer sets were designed covering 95 CpG dinucleotides within the promoter region upstream from the transcription site. Thirty-two amplicons failed to provide any analyzable data and another eight had unreliable results and were excluded. The average methylation for the 62 samples across all 55 CpG sites was 9%. There was no difference in the levels of methylation between recurrence and non-recurrence samples (8.9% vs. 9%, p = 0.966) and there was no difference between methylation levels according to recurrence phenotype compared to reference DNA (recurrence 8.9% vs. 12%, p = 0.189; non-recurrence 9% vs. 12%, p = 0.137). As observed in the methylation microarray data, there was a trend for negative correlation between SOCS6 methylation and mRNA expression in the samples with <2 SOCS6 copies/cell (R2 = −0.377, p = 0.092) while this was not the case in samples with ≥2 SOCS6 copies/cell (R2 = 0.232, p = 0.210).
SOCS6 Loss and Decreased mRNA Expression was Associated with Poor Survival in SCC tumors
Training and test subjects were deemed to have ‘SOCS6 loss’ if their tumors had less than 1.45 copies/cell, as measured by qPCR. The qPCR threshold was derived from the mean copy number minus two standard deviations of the training set tumors with ≥2 copies/cell (aCGH-derived) (n = 26) (SOCS6 mean copy number ± standard deviation = 2.03±0.29). This approach has been previously used to determine qPCR thresholds for aCGH gene copy number alterations [29].
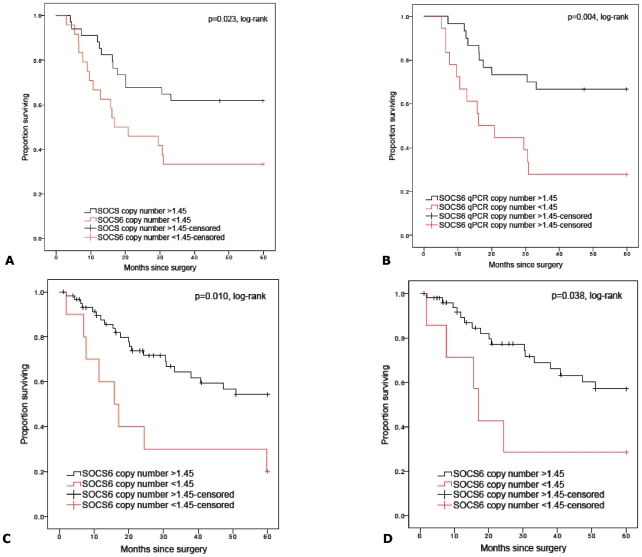
Evaluation of overall survival at five years post surgery in the training and test set subjects demonstrated that those with ‘SOCS6 loss’ had significantly worse survival compared to subjects without ‘SOCS6 loss’ (training set: 30 months vs. 43 months, Log-rank p = 0.023; test set: 27 months vs. 43 months, p = 0.010) (Figure 6A and 6C, respectively). Stratification by TNM stage found ‘SOCS6 loss’ was associated with worse survival in early stage (stage I–II) tumors (training set: 28 months vs. 46 months, p = 0.004; test set: 27 months vs. 45 months, p = 0.038) (Figure 6B and 6D, respectively) but not in the smaller cohort with advanced stage disease (stage III–IV) (training set: 35 months vs. 22 months, p = 0.518 and test set: 26 months vs. 34 months, p = 0.293, Figure S2A and S2B respectively). ‘SOCS6 loss’ was also associated with shorter recurrence free survival in both the training (28 months vs. 46 months, Log-rank p = 0.007) and test set subjects (28 months vs. 46 months, Log-rank p = 0.019) (Figure S3A and S3B, respectively).
Figure 6. Kaplan-Meier curves of overall survival and SOCS6 qPCR-derived copy number in study subjects with follow-up duration of 5 years after surgical resection in the training set (n = 62) and test set (n = 72).
Figure 6A and 6B represent overall survival in all training set and TNM early stage subjects while 6C and 6D represent all test set and TNM early stage test set subjects. Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
Stratification of samples into low SOCS6 mRNA expression (≤0.85 relative mRNA expression) compared to normal SOCS6 mRNA expression (>0.85 relative mRNA expression), was associated with worse overall survival (training set: 25 months vs. 44 months, p = 0.002; test set: 31 months vs. 48 months, p = 0.004) (Figure S4A and S4C, respectively). In the subjects with early stage tumors, decreased SOCS6 mRNA expression was associated with worse overall survival (training set: 23 months vs. 46 months, p<0.001; test set: 35 months vs. 48 months, p = 0.033) (Figure S4B and S4D, respectively). However, this was not demonstrable in the small cohort with advanced stage tumors (training set: 28 months vs. 29 months, p = 0.973; test set: 12 months vs. 37 months, p = 0.101, Figure S2C and S2D respectively).
Discussion
Recent improvements in clinical outcome in NSCLC has been achieved by using biologically targeted therapies, underpinning the importance of recognizing the molecular heterogeneity of lung cancer [30]. The tumor-node-metastasis (TNM) staging system based on tumor size, nodal involvement and the presence of distant metastases is the current standard for predicting prognosis in NSCLC patients [31], [32]. However, TNM stage cannot fully encompass the heterogeneous biology of NSCLC tumors. In primary lung SCC tumors with well-annotated recurrent disease follow-up, we have tried to identify novel gene/s that may be involved in the pathophysiology of tumor recurrence after surgical resection. In this study, we found that loss of 18q22.3 occurred more often in primary tumors that recurred than those that did not. Located within the 18q22.3 region is SOCS6, whose copy number was significantly lower in recurrence tumors compared to the non-recurrence tumors. We also found that DNA copy number loss and that promoter methylation may not regulate SOCS6 mRNA expression. Importantly, loss of SOCS6 copy number and reduced mRNA expression had prognostic significance.
The loss of 18q11-23 is a well-recognized marker of poor prognosis in many solid organ malignancies including esophageal squamous cell cancer [33], head and neck squamous cell cancer [34] and colorectal cancer [35]. Recent evidence in gastric cancer suggests that SOCS6 maybe the candidate gene for the 18q22 copy number alteration and the loss of SOCS6 appears to be a critical genetic alteration in the development of certain subtypes of gastric cancer [36]. The suppressor of cytokine signaling (SOCS) family comprises eight members SOCS (1–7) and CISH. The SOCS family of proteins negatively regulate the cytokine-induced Janus family tyrosine kinase/signal transducers and activators of transcription signaling pathway, thereby inhibiting the cellular growth and proliferation of tumor cells [37]. Unlike other SOCS family members, SOCS6 does not interact with JAK2 but has a direct effect on the insulin receptor (IR) and KIT signaling pathways [38], [39]. The deregulation of both insulin and KIT-signaling are known to play an important role in the proliferation of several malignancies [40], [41]. Consequently, tumor cells with loss of SOCS6 may have increased activation of insulin and KIT-signaling resulting in uncontrolled growth. In gastric cancer, SOCS6 loss in conjunction with promoter hypermethylation results in transcriptional silencing [36], [42]. Here we show that SOCS6 mRNA expression is positively correlated with DNA copy number. In our study we found a trend for increased methylation with reduced mRNA expression in the samples with SOCS6 copy number loss. Further study is warranted in order to better understand the regulatory mechanisms involved in SOCS6 transcription regulation in lung SCC.
We have shown that the loss of SOCS6 copy number and corresponding decreases in mRNA expression are related to significantly shorter overall survival, particularly in subjects with early stage SCC tumors. This is clinically important since a prognostic marker for early stage SCCs is definitely needed for the improvement of patients' outcome. Such a prognostic marker may allow clinicians to select the most efficacious adjuvant therapy with consequent improvements in survival. Therefore, if our findings are confirmed in a prospective study, SOCS6 copy number and/or mRNA expression can be used as a molecular marker for prediction of prognosis in patients with early stage lung SCC. Additionally we have demonstrated that reliable screening for SOCS6 copy number loss can be performed using the rapid and simple method of qPCR. The small cohort size of subjects with advanced stage SCC, limits our ability to make definitive conclusions about the role of SOCS6 copy number and mRNA expression as a prognostic biomarker in this cohort of subjects.
Copy number analysis can be useful to identify ‘driver’ (causative of disease) oncogenes or tumor suppressor genes [43]. Causal focal regions of gain (harboring oncogenes) and loss (harboring tumor suppressor genes) in lung SCCs are now being elucidated through high-resolution copy number analyses [14], [18], [23]. These include amplifications in 1p34.2, 2p15, 3q11.2, 3q26.33, 3q29, 4q13.1, 5p15.1, 7p11.2, 8p12, 8q24.21, 9p13.3, 11q13.2, 12p12.1, 14q21.1, 19q12, 19q13.2, 20p12.3 and 22q12.2 and deletions in 3p12.3, 3q24.2, 3q12.1, 4p15.31, 4q32.1, 5p14.2, 5q31.1, 7p11.2, 8p21.3, 9p21.3, 14q21, 17p11.2 and 18q22.1 [14], [18], [23]. In our analysis on a platform of >40,000 elements we found gain in 3q26.33 and loss in 9p21.3 as the most significant alterations in lung SCC, demonstrating the consistency of results from independent studies using high-resolution aCGH platforms. Copy number analysis has also been used to identify genomic alterations associated with metastatic behavior of primary lung SCC [18], [24]. Yan et al. used CGH to analyze 21 non-metastatic and 18 metastatic lung SCC tumors and found that when taking advanced stage into consideration, gains on 2p, 20p and losses on 2q, 4q and 18q were associated with metastases [24]. The aCGH study of Boelens et al, demonstrated that gains of 7q36, 8p12 and 10q22 were specific to SCC tumors with lymph node metastasis, while gain of 8q22-q24 and loss of 8p23 and 13q21 were specific to SCC tumors that developed distant metastasis within three years of surgical resection [18]. While Boelens et al. used aCGH, they did not find an association of 18q22.3 loss with tumor recurrence as we report here, which may reflect differences in the study population, sample size (34 versus 62), and aCGH platforms (6000-element bacterial artificial chromosome-based array versus 44,000-element oligonucleotide array).
Data generated from analysis of high-throughput methods such as aCGH needs to be validated by an alternative method such as qPCR [44]. Inadvertent false discovery due to the large number of probes on micoroarray platforms is the major reason for the need to have biological validation and technical replication. In our study, we noted differences in the aCGH-based and qPCR copy number data. A similar lack of correlation has been noted by others [10]. There are a number of potential reasons for this ranging from false discovery to technical limitations, such as differences between the microarray platform and the qPCR-based copy number assays. For some genes it may relate to limitations of the microarray platform. The Agilent 44B aCGH platform is a platform with 60-mer oligonucleotide probes with resolution of 35–75 kb including coding and noncoding sequences. Low representation of aCGH probes in some regions could prevent accurate quantification of specific genes. Another possible explanation of the poor correlation is the possibility of small SOCS6 intra-genic copy number variations. Microdeletions within a gene may be missed due to the resolution of this Agilent 44B aCGH platform which has an average resolution of approximately 35–75 kb, but will be less likely by newer higher resolution, platforms, such as the 1 M Agilent array [45], [46]. Other studies have used qPCR assays similar to those used in our study to validate aCGH findings [47]. These qPCR assays are designed to span the coding sequence of the candidate gene, while the aCGH probes span both coding and non-coding sequences. Despite this, aCGH and SNP platforms with lower resolution have been able to provide novel insights into disease biology in solid organ malignancies, such as ovarian cancer (50K SNP arrays) [10] and male breast cancer (44B Agilent aCGH arrays) [48].
The tissue samples in this study were macrodissected not microdissected. Microdissection enriches for tumor cells and increases the ability to detect tumor-specific copy number changes. The admixture of normal cells, infiltrating blood and lymphoid cells in our samples may have influenced detection of copy number alterations despite the selection of tumor samples with at least 50% tumor cell content. Despite this limitation, our data showed that loss of SOCS6 copy number is associated with poor prognosis. This suggests that even despite the presence of non-tumor cells, the detection of SOCS6 copy number alteration may have potential as a prognostic biomarker. In this study, when we applied the GISTIC algorithm to SCC recurrent and non-recurrent tumors separately, we identified several genomic alterations unique to non-recurrent tumors (5p15.33, 8q24.21, 9p21.1 and 19q13.2). While amplifications in these regions are typically associated with worse clinical outcome in other solid organ malignancies [49], [50], further study is warranted to understand the biological significance, if any, of genomic alterations unique to non-recurrent tumors in lung SCC.
In our study we used an array approach (Illumina BeadsArray Technology) to screen for promoter methylation of SOCS6. We then used a quantitative approach (MassARRAY® EpiTYPER analysis) to validate the methylation microarray findings. There are now several approaches available for the study of DNA methylation, some suited for studying single-locus methylation [51] and others for genome-wide approaches [52]. The Illumina BeadArray technology is appropriate technology for genome-wide methylation analysis and the results have been validated against methylation-sensitive PCR, a single gene locus methylation detection method, with high analytical sensitivity [53]. On the other hand, EpiTYPER is a highly accurate quantitative method for that has been validated [54] and used for the evaluation of methylation status in several malignancies including non-small cell lung cancer [55]. Both methods demonstrated that the SOCS6 gene promoter is not hypermethylated in our samples. However, in the tumors with low copy number of SOCS6, there was a trend for increasing methylation, albeit non-significant. This raises the possibility that low copy number and promoter methylation of SOCS6 may be responsible for reduced mRNA expression. However this will require confirmation in a larger study cohort.
In conclusion, we showed that SOCS6 located in the genomic locus 18q22.3, has reduced gene copy number and reduced mRNA expression in primary lung SCCs that recur early after surgical resection. Since SOCS family proteins are known to inhibit a potentially important growth-signaling pathway, a tumor suppressor function in lung SCC is a possibility requiring further study to elucidate mechanisms underlying disease recurrence.
Supporting Information
qPCR-derived SOCS6 copy number in normal lung of training set (n = 62). Figure S1A compares the qPCR-derived SOCS6 copy number (y-axis) and paired normal lung and tumor tissue (x-axis). Figure S1B compares qPCR-derived SOCS6 copy number (y-axis) and in training set normal lung recurrence phenotype of the tumor (x-axis) (non-recurrence = 34 and recurrence = 28). Mann-Whitney U test to was used to assess for any differences in copy number and p values<0.05 were deemed significant.
(TIF)
Kaplan-Meier curves of overall survival in advanced stage training set (n = 9) and test set (n = 14) study subjects with follow-up duration of 5 years after surgical resection. Figure S4A and S4B represent qPCR-derived SOCS6 copy number while Figure S4C and S4D represent SOCS6 mRNA expression and overall survival in TNM advanced stage training set and test set subjects. Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
(TIF)
Kaplan-Meier curves of qPCR-derived SOCS6 copy number and recurrence-free survival in study subjects with follow-up duration of 5 years after surgical resection. Figure S2A represents training set subjects (n = 62) and Figure S2B represents test set subjects (n = 72). Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
(TIF)
Kaplan-Meier curves of overall survival and relative SOCS6 mRNA expression in training set (n = 62) and test set (n = 72) study subjects with follow-up duration of 5 years after surgical resection. Figure S3A and S3B represent overall survival in all training set and TNM early stage subjects while Figure S3C and S3D represent all test set and TNM early stage test set subjects. Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
(TIF)
GISTIC Identified Chromosomal Regions with Copy Number Alterations in Non-recurrence and Recurrence SCC Tumors.
(DOC)
RefSeq Genes Contained Within the GISTIC Identified Cytoband 18q22.3.
(DOC)
aCGH Probes and QuantiTect (Qiagen) Primers used for qPCR.
(DOC)
Acknowledgments
The authors wish to thank the patients of TPCH for their participation in this study, TPCH thoracic surgeons (Dr. Kevin Matar and Dr. Morgan Windsor) and UQ Thoracic Research Nurses (Linda Passmore, Elizabeth McCaul and Deborah Courtney), Professor Alexander Dobrovic (Peter MacCallum Cancer Centre) for reviewing the manuscript and Laboratory staff for their assistance with specimen collection and preparation (Maria Martins, Maxine Tan, Kylie Parsonson, Rebecca Stockwell, Morgan Davidson, Jane Tjahjadi, Brielle Parris, Jennifer Campbell, John De Laat, Annalese Semmler, Samuel Kim, Jeremy Barr, Nick Chiang, Glenn Rabnott, Ainsley Tunnicliffe, Alex Ruddy, Ting Phila and Jessie Kelly).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by NH&MRC and Queensland Smart State project grants; UQ Postgraduate Medical Scholarship and NH&MRC Postgraduate Medical Scholarship (to KBS), NH&MRC Practitioner Fellowship (KMF), NH&MRC Career Development Award (IAY), NH&MRC Biomedical Scholarship (SMS, CMW), Cancer Council Queensland Senior Research Fellowship (KMF), Queensland Clinical Research Fellowship (KMF, IAY) and The Prince Charles Hospital Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer Statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. The Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 3.Spiro SG, Silvestri GA. One Hundred Years of Lung Cancer. Am J Respir Crit Care Med. 2005;172:523–529. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- 4.Scott WJ, Howington J, Feigenberg S, Movsas B, Pisters K. Treatment of Non-small Cell Lung Cancer Stage I and Stage II. Chest. 2007;132:234S–242S. doi: 10.1378/chest.07-1378. [DOI] [PubMed] [Google Scholar]
- 5.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nature Genetics. 2003;33:238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 6.Kang JU, Koo SH, Kwon KC, Park JW, Kim JM. Identification of novel candidate target genes, including EPHB3, MASP1 and SST at 3q26.2-q29 in squamous cell carcinoma of the lung. BMC Cancer. 2009;9:237. doi: 10.1186/1471-2407-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonsson G, Staaf J, Vallon-Christersson J, Ringner M, Holm K, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Research. 2010;12:R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco D, Tonon G, Huang Y, Zhang Y, Sinha R, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Sung JS, Park KH, Kim YH. Genomic alterations of chromosome region 11p as predictive marker by array comparative genomic hybridization in lung adenocarcinoma patients. Cancer Genetics and Cytogenetics. 2010;198:27–34. doi: 10.1016/j.cancergencyto.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, et al. Integrated Genome-Wide DNA Copy Number and Expression Analysis Identifies Distinct Mechanisms of Primary Chemoresistance in Ovarian Carcinomas. Clin Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Järvinen A-K, Autio R, Kilpinen S, Saarela M, Leivo I, et al. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes, Chromosomes and Cancer. 2008;47:500–509. doi: 10.1002/gcc.20551. [DOI] [PubMed] [Google Scholar]
- 12.Pei J, Balsara B, Li W, Litwin S, Gabrielson E, et al. Genomic imbalances in human lung adenocarcinomas and squamous cell carcinomas. Genes, Chromosomes and Cancer. 2001;31:282–287. doi: 10.1002/gcc.1145. [DOI] [PubMed] [Google Scholar]
- 13.Choi Y-W, Choi JS, Zheng LT, Lim YJ, Yoon HK, et al. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the lung. Lung Cancer. 2007;55:43–51. doi: 10.1016/j.lungcan.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir BA, Woo MS, Getz G, Perner S, Ding L, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newnham G, Conron M, McLachlan SA, Dobrovic A, Do H, et al. Integrated mutation, copy number and expression profiling in resectable non-small cell lung cancer. BMC Cancer. 2011;11:93. doi: 10.1186/1471-2407-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balsara BR, Testa JR. Chromosomal imbalances in human lung cancer. Oncogene. 2002;21:6877–6883. doi: 10.1038/sj.onc.1205836. [DOI] [PubMed] [Google Scholar]
- 18.Boelens MC, Kok K, van der Vlies P, van der Vries G, Sietsma H, et al. Genomic aberrations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer. 2009;66:372–378. doi: 10.1016/j.lungcan.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Petersen S, Aninat-Meyer M, Schluns K, Gellert K, Dietel M, et al. Chromosomal alterations in the clonal evolution to the metastatic stage of squamous cell carcinomas of the lung. British Journal of Cancer. 2000;82:65–73. doi: 10.1054/bjoc.1999.0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sy SM, Wong N, Lee T-W, Tse G, Mok TS-K, et al. Distinct patterns of genetic alterations in adenocarcinoma and squamous cell carcinoma. Eur J Cancer. 2004;40:1082–1094. doi: 10.1016/j.ejca.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Luk C, Tsao MS, Bayani J, Shepherd F, Squire JA. Molecular cytogenetic analysis of non-small cell lung carcinoma by spectral karyotyping and comparative genomic hybridization. Cancer Genetics and Cytogenetics. 2001;125:87–99. doi: 10.1016/s0165-4608(00)00363-0. [DOI] [PubMed] [Google Scholar]
- 22.Pei J, Balsara BR, Li W, Litwin S, Gabrielson E, et al. Genomic imbalances in human lung adenocarcinomas and squamous cell carcinomas. Genes, Chromosomes and Cancer. 2001;31:282–287. doi: 10.1002/gcc.1145. [DOI] [PubMed] [Google Scholar]
- 23.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, et al. Frequent and Focal FGFR1 Amplification Associates with Therapeutically Tractable FGFR1 Dependency in Squamous Cell Lung Cancer. Science Translational Medicine. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan W, Song L, Liang Q, Fang Y. Progression Analysis of Lung Squamous Cell Carcinomas by Comparative Genomic Hybridization. Tumor Biology. 2005;26:158–164. doi: 10.1159/000086488. [DOI] [PubMed] [Google Scholar]
- 25.Larsen JE, Pavey SJ, Passmore LH, Bowman R, Clarke BE, et al. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28:760–766. doi: 10.1093/carcin/bgl207. [DOI] [PubMed] [Google Scholar]
- 26.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proceedings of the National Academy of Sciences. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwakawa R, Kohno T, Kato M, Shiraishi K, Tsuta K, et al. MYC Amplification as a Prognostic Marker of Early-Stage Lung Adenocarcinoma Identified by Whole Genome Copy Number Analysis. Clinical Cancer Research. 2011;17:1481–1489. doi: 10.1158/1078-0432.CCR-10-2484. [DOI] [PubMed] [Google Scholar]
- 30.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, et al. Non-small-cell lung cancer. The Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 31.Chansky K, Sculier J-P, Crowley JJ, Giroux D, Meerbeeck JV, et al. The International Association for the Study of Lung Cancer Staging Project Prognostic Factors and Pathologic TNM Stage in Surgically Managed Non-small Cell Lung Cancer. Journal of Thoracic Oncology. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 32.Detterbeck FC, Boffa DJ, Tanoue LT. The New Lung Cancer Staging System. Chest. 2009;136:260–271. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 33.Ando T, Ishiguro H, Kimura M, Mitsui A, Mori Y, et al. Frequent loss of the long arm of chromosome 18 in esophageal squamous cell carcinoma. Oncology Reports. 2007;17:1005–1011. [PubMed] [Google Scholar]
- 34.Takebayashi S, Hickson A, Ogawa T, Jung K-Y, Mineta H, et al. Loss of chromosome arm 18q with tumor progression in head and neck squamous cancer. Genes, Chromosomes and Cancer. 2004;41:145–154. doi: 10.1002/gcc.20066. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka T, Watanabe T, Kitayama J, Kanazawa T, Kazama Y, et al. Chromosome 18q Deletion as a Novel Molecular Predictor for Colorectal Cancer With Simultaneous Hepatic Metastasis. Diagn Mol Pathol. 2009;18:219–225. doi: 10.1097/PDM.0b013e3181910f17. [DOI] [PubMed] [Google Scholar]
- 36.Lai R-H, Hsiao Y-W, Wang M-J, Lin H-Y, Wu C-W, et al. SOCS6, down-regulated in gastric cancer, inhibits cell proliferation and colony formation. Cancer Letters. 2010;288:75–85. doi: 10.1016/j.canlet.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Larsen L, Ropke C. Suppressors of cytokine signalling: SOCS. APMIS. 2002;110:833–844. doi: 10.1034/j.1600-0463.2002.1101201.x. [DOI] [PubMed] [Google Scholar]
- 38.Bayle J, Letard Sb, Frank R, Dubreuil P, De Sepulveda P. Suppressor of Cytokine Signaling 6 Associates with KIT and Regulates KIT Receptor Signaling. Journal of Biological Chemistry. 2004;279:12249–12259. doi: 10.1074/jbc.M313381200. [DOI] [PubMed] [Google Scholar]
- 39.Krebs DL, Uren RT, Metcalf D, Rakar S, Zhang J-G, et al. SOCS-6 Binds to Insulin Receptor Substrate 4, and Mice Lacking the SOCS-6 Gene Exhibit Mild Growth Retardation. Mol Cell Biol. 2002;22:4567–4578. doi: 10.1128/MCB.22.13.4567-4578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyd DB. Insulin and Cancer. Integrative Cancer Therapies. 2003;2:315–329. doi: 10.1177/1534735403259152. [DOI] [PubMed] [Google Scholar]
- 41.Hassan HT. c-Kit expression in human normal and malignant stem cells prognostic and therapeutic implications. Leukemia Research. 2009;33:5–10. doi: 10.1016/j.leukres.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Lai R-H, Wang M-J, Yang S-H, Chen J-Y. Genomic organization and functional characterization of the promoter for the human suppressor of cytokine signaling 6 gene. Gene. 2009;448:64–73. doi: 10.1016/j.gene.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 43.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan DSP, Lambros MBK, Natrajan R, Reis-Filho JS. Getting it right: designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Lab Invest. 2007;87:737–754. doi: 10.1038/labinvest.3700593. [DOI] [PubMed] [Google Scholar]
- 45.Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Human Mutation. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Przybytkowski E, Ferrario C, Basik M. The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome. BMC Medical Genomics. 2011;4:16. doi: 10.1186/1755-8794-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charchar FJ, Kaiser M, Bingham AJ, Fotinatos N, Ahmady F, et al. Whole Genome Survey of Copy Number Variation in the Spontaneously Hypertensive Rat. Hypertension. 2010;55:1231–1238. doi: 10.1161/HYPERTENSIONAHA.109.141663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tommasi S, Mangia A, Iannelli G, Chiarappa P, Rossi E, et al. Gene copy number variation in male breast cancer by aCGH. Analytical Cellular Pathology. 2010;33:113–119. doi: 10.3233/ACP-CLO-2010-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueno T, Tangoku A, Yoshino S, Abe T, Toshimitsu H, et al. Gain of 5p15 Detected by Comparative Genomic Hybridization as an Independent Marker of Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Clinical Cancer Research. 2002;8:526–533. [PubMed] [Google Scholar]
- 50.Yamamoto S, Tsuda H, Honda K, Onozato K, Takano M, et al. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod Pathol. 2009;22:499–507. doi: 10.1038/modpathol.2008.234. [DOI] [PubMed] [Google Scholar]
- 51.Kristensen LS, Hansen LL. PCR-Based Methods for Detecting Single-Locus DNA Methylation Biomarkers in Cancer Diagnostics, Prognostics, and Response to Treatment. Clin Chem. 2009;55:1471–1483. doi: 10.1373/clinchem.2008.121962. [DOI] [PubMed] [Google Scholar]
- 52.Kondoa Y, Issa J-PJ. DNA methylation profiling in cancer. 2010. Expert Reviews in Molecular Medicine Vol. 12. [DOI] [PubMed]
- 53.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Research. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ehrich M, Field JK, Liloglou T, Xinarianos G, Oeth P, et al. Cytosine Methylation Profiles as a Molecular Marker in Non-Small Cell Lung Cancer. Cancer Research. 2006;66:10911–10918. doi: 10.1158/0008-5472.CAN-06-0400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qPCR-derived SOCS6 copy number in normal lung of training set (n = 62). Figure S1A compares the qPCR-derived SOCS6 copy number (y-axis) and paired normal lung and tumor tissue (x-axis). Figure S1B compares qPCR-derived SOCS6 copy number (y-axis) and in training set normal lung recurrence phenotype of the tumor (x-axis) (non-recurrence = 34 and recurrence = 28). Mann-Whitney U test to was used to assess for any differences in copy number and p values<0.05 were deemed significant.
(TIF)
Kaplan-Meier curves of overall survival in advanced stage training set (n = 9) and test set (n = 14) study subjects with follow-up duration of 5 years after surgical resection. Figure S4A and S4B represent qPCR-derived SOCS6 copy number while Figure S4C and S4D represent SOCS6 mRNA expression and overall survival in TNM advanced stage training set and test set subjects. Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
(TIF)
Kaplan-Meier curves of qPCR-derived SOCS6 copy number and recurrence-free survival in study subjects with follow-up duration of 5 years after surgical resection. Figure S2A represents training set subjects (n = 62) and Figure S2B represents test set subjects (n = 72). Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
(TIF)
Kaplan-Meier curves of overall survival and relative SOCS6 mRNA expression in training set (n = 62) and test set (n = 72) study subjects with follow-up duration of 5 years after surgical resection. Figure S3A and S3B represent overall survival in all training set and TNM early stage subjects while Figure S3C and S3D represent all test set and TNM early stage test set subjects. Censored values (+) indicate the last known follow-up time for those subjects still alive after surgical resection.
(TIF)
GISTIC Identified Chromosomal Regions with Copy Number Alterations in Non-recurrence and Recurrence SCC Tumors.
(DOC)
RefSeq Genes Contained Within the GISTIC Identified Cytoband 18q22.3.
(DOC)
aCGH Probes and QuantiTect (Qiagen) Primers used for qPCR.
(DOC)