Abstract
Microbial metagenomes are DNA samples of the most abundant, and therefore most successful organisms at the sampling time and location for a given cell size range. The study of microbial communities via their DNA content has revolutionized our understanding of microbial ecology and evolution. Iron availability is a critical resource that limits microbial communities' growth in many oceanic areas. Here, we built a database of 2319 sequences, corresponding to 140 gene families of iron metabolism with a large phylogenetic spread, to explore the microbial strategies of iron acquisition in the ocean's bacterial community. We estimate iron metabolism strategies from metagenome gene content and investigate whether their prevalence varies with dissolved iron concentrations obtained from a biogeochemical model. We show significant quantitative and qualitative variations in iron metabolism pathways, with a higher proportion of iron metabolism genes in low iron environments. We found a striking difference between coastal and open ocean sites regarding Fe2+ versus Fe3+ uptake gene prevalence. We also show that non-specific siderophore uptake increases in low iron open ocean environments, suggesting bacteria may acquire iron from natural siderophore-like organic complexes. Despite the lack of knowledge of iron uptake mechanisms in most marine microorganisms, our approach provides insights into how the iron metabolic pathways of microbial communities may vary with seawater iron concentrations.
Introduction
Despite its high abundance in the Earth's crust, iron concentrations are very low in the ocean. This is due to the low solubility of iron in the oxic and slightly alkaline seawater of today's oceans, uptake by microorganisms and limited input from external sources. Consequently, the bioavailability of this element is very low in many oceanic regions [1]. Iron is essential for cell metabolism, especially for electron transport. Photosynthesis, respiration, and nitrogen fixation require high cellular concentrations of iron [2].
Microorganisms, particularly bacteria, have evolved several different iron uptake mechanisms [3], [4]. Inorganic iron can be acquired either in its reduced ferrous (Fe2+) [5], or oxidized ferric (Fe3+) state [6]. However, in well-oxygenated seawater, the free ion concentrations of both forms are low, and bacteria have developed alternative strategies to access the organically complexed pool. The synthesis and uptake of siderophores, strong chelators of ferric iron, is one example [7], but bacteria can also acquire iron from free heme (a prosthetic group of porphyrin containing an atom of iron) or heme-containing proteins using both direct uptake and hemophores [8]. Such strategies have been identified in diverse marine bacteria [9].
Many recent studies have highlighted the strategies used by diatoms and cyanobacteria to minimize their iron demand, such as reducing the expression of “expensive” iron genes in the photosystem I complex [10], [11] and nitrogenase [12], or else increasing flavodoxin production [13], [14]. Genomic analysis of Prochlorococcus clades isolated from iron-depleted oceanic regions showed that several genes encoding for iron-containing proteins were absent, thereby reducing the cellular iron quota [15].
The relation between genomic iron uptake strategies and oceanic environments has recently been investigated in cyanobacteria, the most abundant photosynthetic group of marine bacteria. Many Synechococcus genomes from strains isolated in the open ocean lack most known genes for iron stress, while genomes from strains isolated in coastal and upwelling areas contain many such genes, suggesting that maintaining multiple iron limitation compensation strategies is not a selective advantage in the open ocean [16]. Consistent with this, the light-harvesting gene isiA of Synechococcus has been proposed as a biomarker of HNLC regions [17]. However, heterotrophic bacteria also compete for iron at low concentrations, and can account for up to 50% of the total planktonic iron uptake [18]. Additionally, they may modify iron chemistry through the production of organic ligands and thereby regulating phytoplankton production [19]. Because metagenomes contain the gene content of the community of most abundant and therefore most successful microorganisms, they provide a complementary glimpse to organismal studies into the genetic basis of adaptation to the environment.
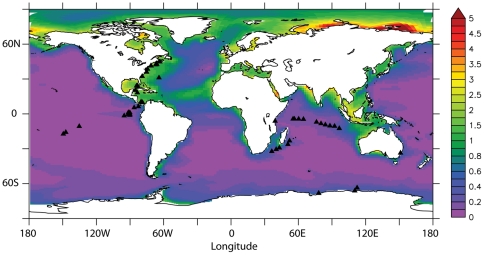
We set up a database of 2319 sequences, corresponding to 140 gene families and 10 different iron-related metabolic pathways, to explore the mechanisms involved in iron acquisition in the ocean. The caveats of using Blast Best Hits and evalue cutoffs have been discussed previously [20]. Here, we defined a stringent criterion to discard false positives: we only considered Reciprocal Best Hits with coverage and identity threshold empirically estimated from our database. We investigate the link between genomic iron metabolism strategies of microbial communities and iron concentrations among 54 worldwide distributed marine metagenomes (Figure 1). Iron concentrations at each location were taken from a biogeochemical model that incorporates information about the sources and cycling of iron in the ocean and were compared to independently acquired observed iron concentrations. We thus established the relationships between microbial communities' iron-related gene prevalence, taxonomic affiliation and iron concentrations in different marine habitats.
Figure 1. Map of annual average surface iron concentration (0–100 m) from the NEMO-PISCES model.
Metagenomic sample sites are represented by black triangles. Color scale stands for dissolved iron concentration (nM).
Methods
Iron Metabolic Pathway database
We selected genes involved in iron metabolisms from the literature in cases where the protein product had been characterized or where the function of the protein could be inferred by sequence analysis. In this way, we identified 140 genes specifically involved in iron metabolism. We assigned these genes to 10 iron-related metabolic pathways, summarized in Table 1. A maximal phylogenetic coverage for each gene was achieved by retrieving all available bacterial sequences using the NCBI search tool with the gene names as a query. All 9917 annotations and protein sequences were manually inspected to discard irrelevant or incomplete protein sequences. To discard redundant sequences from the same genus, we randomly selected one full-size sequence per gene per available genus, resulting in 1753 remaining sequences. Additionally, we screened the annotations of the protein sequences from the Moore Microbial Genome database (marine bacteria isolates) for genes involved in iron metabolism (http://www.moore.org/microgenome/) and retrieved 566 genes. After manual inspection of these sequences, 191 putative ABC iron transporters could not be assigned to any one of the previous pathways, and therefore constituted the unspecified iron transport category (TR). We thus obtained a dataset of 2319 sequences belonging to 11 phyla (Actinobacteria, alphaproteobacteria, Bacteroidetes, betaproteobacteria, Cyanobacteria, deltaproteobacteria, Deinococcus-thermus, Firmicutes, gammaproteobacteria, Spirochetes, epsilonproteobacteria) (Table S1). The 140 gene families with at least two sequences were aligned and processed to estimate an identity and coverage threshold within each gene family. These thresholds estimations are needed because many proteins involved in different pathways could share sequence similarities, like ABC transporters of different iron-related pathways. We found that 65% amino-acid identity over a minimum length of 100 amino acids (or 80% of query length coverage) corresponded to 96% of correct orthologous gene assignment and 100% of correct pathway assignment (Table S2). Our criteria are stringent and we therefore probably underestimated the number of matches, but this enables a robust analysis of the different proportion of each iron-related pathway between sites.
Table 1. Iron-related metabolic pathway database.
| Pathway | Abbreviation | Sequences | Genes in the database | Phylogenetic spread* | Genes with hits | Number of hits |
| Control (recA) | CN | 120 | 1 | 11 | 1 | 4396 |
| Flavodoxin switch [13] | FL | 181 | 1 | 11 | 1 | 46 |
| Fe2+ uptake [5] | F2 | 241 | 11 | 10 | 6 | 579 |
| Fe3+ uptake [6] | F3 | 104 | 8 | 9 | 9 | 586 |
| Heme uptake [8] | HE | 196 | 24 | 10 | 9 | 23 |
| Oxidative stress [56], [57] | OX | 168 | 4 | 10 | 4 | 789 |
| Regulation [58] | RG | 269 | 9 | 11 | 6 | 602 |
| Siderophore uptake | SU | 562 | 23 | 10 | 6 | 620 |
| Siderophore synthesis [7] | SS | 164 | 56 | 7 | 2 | 5 |
| Storage [48] | ST | 123 | 3 | 9 | 3 | 139 |
| Unspecified iron transport | TR | 191 | NA | 6 | 35 sequences | 3455 |
| TOTAL | 2319 | 140 | 47 | 11240 |
*number of most abundant phylogenetic groups represented (11 in total, see Methods).
Metagenome Data and Screen
We analyzed metagenomic data from Global Ocean Sampling sites for organisms collected within a 0.1–0.8 µm size range. We downloaded data for metagenomes containing at least 50 million base pairs (Mbp) from the CAMERA database [21]. Sargasso Sea sample GS000A was discarded from the analysis as it is suspected of contamination [22]. We thus collected 54 metagenomes corresponding to 10.5 billion nucleotides from 4 Habitat types: 22 open ocean, 24 coastal, 4 coral reef, and 4 marine-derived lake (Antarctic) sites (Figure 1, Table S3). We screened these datasets with our gene database using TBLASTN [23]. Non-redundant hits were numbered based on reciprocal best hits [24] with our identity and coverage thresholds (Table S4). We used the gene encoding the recombinase A, RecA, as a single copy control to estimate taxonomic diversity. All metagenomic sequences where searched against a database of 120 RecA protein sequences with representatives in all prokaryotic taxonomic groups. A taxonomic group was considered to be present in a metagenome when a sequence had a reciprocal best hit with a recA gene belonging to this taxonomic group (Tatusov et al 1997). In a preliminary analysis, we first checked that the proportion of our control gene, recA, relative to hits against our iron-related gene database, did not vary with metagenome size (Spearman Rho = 0.03 p = 0.79). We checked the congruence of our recA based taxonomic affiliation with the 16S rRNA based taxonomic affiliation obtained on 29 metagenomes [25]. We found a very good correlation between phylum prevalence for both genes (Spearman correlation coefficient of 0.86, p<10–16). However, the variance in the number of hits within phylum was significantly lower for the single copy recA gene, which is expected as a consequence of 16S rDNA copy number variations. Total recruited reads per metabolic pathways were obtained by summing up results for all genes from the same pathway. We used hits to recA as a proxy for the number of microbial genomes and inferred taxonomic diversity from reciprocal blast best hits. RecA is assumed to be in single copy in most genomes and belongs to the class of housekeeping genes that does not frequently undergo horizontal gene transfer [26].
Environmental Data
We extracted salinity, temperature, water depth and chlorophyll concentration for each sampling site from the CAMERA database [21]. We used the ocean general circulation and biogeochemistry model based NEMO-PISCES [27] to infer nitrate, phosphate and dissolved iron (dFe) concentrations. This resulted in 8 environmental variables per site (Table S3). Dissolved iron is an operational definition for the fraction of iron that passes through a 0.2 µm filter.
NEMO-PISCES simulates nanophytoplankton and diatoms, meso- and micro-zooplankton, small and large detritus, calcium carbonate, dissolved-inorganic-carbon, carbonate, dissolved-organic-carbon, oxygen, nitrate, phosphate, silicic acid, ammonium and dFe concentrations. The biotic iron demand varies between phytoplankton groups and as a function of dFe concentrations and light. dFe is removed via biotic uptake and particulate iron is remineralised back to dFe, with dFe scavenged as a function of the total particle load, with ligand complexation explicitly represented, assuming a uniform ligand concentration of 0.6 nM. For this study, we used a state-of-the-art version of NEMO-PISCES that includes aeolian, sedimentary, hydrothermal and fluvial dFe sources [27]. We verified the predictions of our model against the 2438 dFe observations (between 0 and 100 m) compiled in the database of Moore and Braucher [28]. We obtained a very good correlation (R2 = 0.56), with a bias of 0.09 nM between mean observed (0.32 nM) and modeled (0.23 nM) dFe (values compared at the same latitude, longitude, depth and month of sampling). Because our metagenomes all come from surface samples (<5 m) where dFe is expected to be more variable, we also checked the relationship between predicted and observed dFe for surface sites only (R2 = 0.54, Figure S1).
Statistical analysis
All analyses were performed with R [29]. We first normalized the number of hits for each gene per site to the metagenome size in kbp. We checked that the proportion of iron-related metabolic pathways was not correlated to metagenome size (Spearman Rho = 0.11 p = 0.47).
Multivariate comparisons were performed with numerical ecology tools from the ade4 package [30]. We used non-parametric tests; Kruskal-Wallis test to assess significance levels of differences between habitats and Spearman's rank correlation coefficient to assess the significance of the relationship between metabolic pathways or gene prevalences and iron concentration. We used Fisher's combined probability X2 = −2*Σln(p) (which follows a Chisquare distribution with 2n degrees of freedom) to test the overall significance of several independent p-values bearing upon the same null hypothesis [31].
We followed the biogeography approaches [32], [33] to test for correlations between taxonomic distribution, iron metabolism pathways and environmental variables. The three datasets were represented with rows as sites, and columns were either proportion of metabolic pathways, proportion of different taxonomic groups based on recA assignation or environmental variables. We then constructed a matrix of iron content differences as distances between pairs of sites. We computed the three site-site correlation matrices by computing the Spearman's correlation coefficients between each pair of sites from each data frames. These correlation coefficient matrices were transformed in relative rank matrices: pairs of sites with higher correlation coefficients received lower ranks, whereas the pairs of site having less correlation received higher ranks [33]. The rank matrices thus contain the between site correlation ranks for iron metabolism pathways, taxonomy or environmental variables. We then estimated the between matrices correlation using a Mantel test to assess whether closely related sites on the basis of metabolic pathways were also closer in terms of taxonomic distribution or/and environmental variables or/and iron concentration.
Iron Biomarker genes
The most abundant single genes of our database (i.e. 13 genes detected in at least 27/54 metagenomes) were tested individually for their distribution in habitats and their correlation with iron concentration. These genes were bfr (iron storage); exbB, fur (regulation); fbpC, futA (Fe3+ uptake); feoB, yfeA, yfeB and yfeC (Fe2+ uptake); fecA (siderophore uptake); isiA, sodA, sodB (protection against oxidative stress). We standardized the abundance of each gene against the number of single copy control recA gene per metagenome. We used Spearman's rank correlation in sites containing data for both iron-related pathway prevalence and dissolved iron concentrations.
Results
Iron-uptake gene prevalence depends on habitat types and is higher in low iron environments
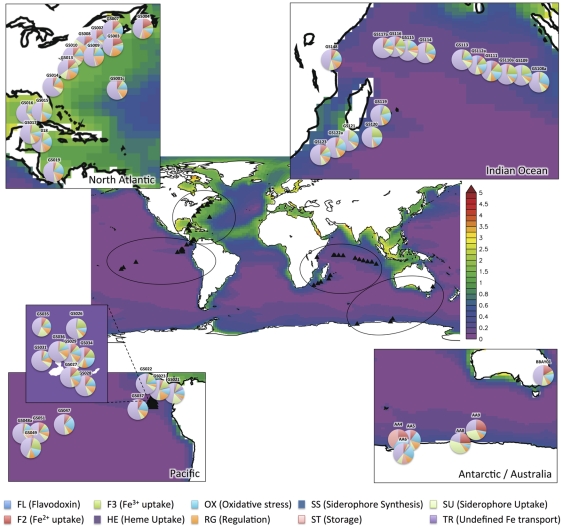
To screen marine metagenomes, we set up a database of 140 genes involved in 10 different pathways related to iron metabolism: flavodoxin switch, Fe2+ and Fe3+ uptake, heme uptake, response to oxidative stress, regulation, siderophore uptake, siderophore synthesis, storage, flavodoxin switch and unspecified iron transport (Table 1, Table S1). Microbial abundance and taxonomic diversity were inferred from the prevalence of the single copy recA gene. The 54 metagenomes larger than 50 Mbp have been obtained from 4 marine habitat types: coastal, open ocean, coral reef and marine-derived lakes (Figure S2, Table S3). First, we investigated whether the total proportion of iron related pathways varied between defined habitats. We found that the proportion of genes involved in iron metabolism pathways, relative to the control gene recA, is not equally distributed among habitat types (Kruskal-Wallis p = 0.002). Coastal sites contain a higher proportion of genes involved in iron metabolism, followed by coral reef, open ocean and marine-derived lake. This may reflect variations in the relative proportion of bacterial species, as compared to viral or picoeukaryotic communities, which have been shown to be present in 0.8 µm filtered metagenomes [34], [35]. Alternatively, this could reflect differences in iron-related gene copy numbers per genome between these habitats. Second, we investigated whether the proportion of iron-related pathways varied with predicted dissolved iron concentrations estimated from a biogeochemical model (Figure 2). Interestingly, the proportion of genes involved in iron metabolism pathways increases with decreasing iron concentration in open ocean and coastal habitats (Spearman Rho = −0.36, p = 0.014). This reflects a greater number of genes involved in iron uptake or metabolism in communities experiencing iron starvation. There is no significant correlation between the number of different iron-related pathways and iron concentrations. This suggests that the greater prevalence of iron-related genes in low iron environments is not the consequence of an increase in the number of different pathways, but the consequence of an increase in gene prevalence in several pathways.
Figure 2. Proportion of iron-related metabolic pathways for each site overlaid on iron concentrations.
Color scale stands for dissolved iron concentration (nM).
Given this global trend, we investigated the extent to which each pathway prevalence varies between habitat. The prevalence of three iron related pathways varies between habitats (Table 2): the iron uptake pathways of Fe2+ and Fe3+, and the regulation pathway. Fe2+ uptake (F2) is under-represented in open ocean sites (Kruskal-Wallis p = 0.023), whereas Fe3+ uptake (F3) is over-represented in the open-ocean (Kruskal-Wallis p = 0.005). Regulation (RG) is over-represented in coral reef sites (Kruskal-Wallis p = 0.004). Since the prevalence of each pathway normalized by the metagenome size can be considered as independent, we can combine the probabilities across pathways to test whether there is a global difference of pathway prevalence between habitat types. Consistent with the analysis of total iron-related gene prevalence, the prevalence of iron-related metabolic pathways differs significantly between habitats (p<10−5). We also found that taxonomic prevalence is significantly different between habitat type (Table 2) and this prompted us to investigate the relationship between iron concentration and pathway prevalence in each habitat separately.
Table 2. Relationship between iron pathway prevalence, habitat and iron concentration across sites.
| Abb | Habitat effect Kruskal-Wallis p-value | Iron concentration effect Spearman Rho | ||
| Coastal sites | Open ocean sites | |||
| Iron Pathway Prevalence | ||||
| Flavodoxin Switch | FL | 0.132 | 0.04 | −0.45 * |
| Fe2+ uptake | F2 | 0.023 | −0.33 | −0.26 |
| Fe3+ uptake | F3 | 0.005 | −0.23 | 0.06 |
| Heme uptake | HE | 0.474 | 0.04 | −0.57 ** |
| Oxidative stress | OX | 0.262 | −0.32 | −0.21 |
| Regulation | RG | 0.004 | −0.29 | 0.02 |
| Siderophore synthesis | SS | 0.324 | 0.35 | −0.26 |
| Storage | ST | 0.028 | 0.31 | 0.56 ** |
| Siderophore uptake | SU | 0.044 | −0.14 | −0.53 * |
| Unspecified iron transport | TR | 0.003 | −0.49 * | −0.31 |
| Fisher combined p-value | *** | * | *** | |
| Taxonomic prevalence | ||||
| Actinobacteria | AB | 0.951 | 0.34 | −0.38 |
| Bacteroidetes | BA | 0.521 | −0.19 | 0.20 |
| Cyanobacteria | CY | 9.10−5 | 0.11 | 0.13 |
| Deinococcus-Thermus | DT | 0.862 | −0.12 | 0.22 |
| Firmicutes | FI | 0.12 | 0.24 | 0.26 |
| Alpha-proteobacteria | aP | 0.010 | 0.20 | −0.06 |
| Beta-Proteobacteria | bP | 0.783 | 0.34 | 0.04 |
| Delta-Proteobacteria | dP | 0.630 | −0.17 | 0.51 * |
| Epsilon-Proteobacteria | eP | 0.025 | −0.30 | nd |
| Gamma-Proteobacteria | gP | 0.015 | 0.05 | −0.15 |
| Spirochetes | SP | 0.428 | 0.13 | −0.003 |
| Fisher combined p-value | *** | ns | ns | |
*: p<0.05,
**: p<0.01,
***: p<0.001,
ns: not significant.
Iron-related gene prevalence reveal biological adaptations to iron concentrations
To explore whether microbial communities have different strategies of iron metabolism as a consequence of iron availability, we investigated the statistical significance of the variation between the relative proportion of each of the 10 iron-related pathways and iron concentration for 23 coastal and 22 open ocean sites, where iron concentrations can be inferred from the biogeochemical model. While there is a globally significant correlation between pathway prevalence and iron concentrations (combined probabilities across pathways 0.038 and 0.006 for coastal and open ocean environments, respectively), most individually significant trends appear in the open ocean habitat (Table 2). Storage prevalence increases significantly with predicted iron concentration (Spearman Rho = 0.56 p = 0.007) (Table 2) whereas the siderophore uptake pathway decreases with increasing iron concentration in open ocean sites (Spearman Rho = −0.53 p = 0.011), suggesting that this is an iron starvation strategy in this habitat. Consistent with this idea, experimental evidence suggests that siderophore synthesis does not occur in iron-rich media for many species [36]. Although more rarely detected, heme uptake and flavodoxin switch show negative correlations with iron concentration in open ocean sites (Spearman Rho = −0.57, p = 0.006 and Spearman Rho = −0.45, p = 0.034, respectively).
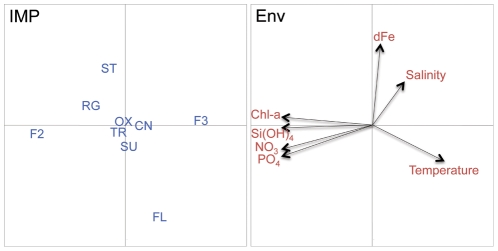
These correlations between different iron metabolism pathways and environmental variables are summarized in the canonical correlation analysis scattergram based on iron-related pathways proportion between sites (Figure 3). This analysis highlights two pairs of anti-correlated iron uptake strategies: Fe2+ uptake (F2) versus Fe3+ uptake (F3) (Spearman Rho = −0.47, p-value = 0.002) and storage (ST) versus flavodoxin switch (FL) (Rho −0.38, p = 0.015) and provides a further illustration of the positive correlation of storage (ST) with iron concentration. Nutrient concentrations (phosphate, silicate and nitrate) are correlated with each other and with Fe2+ uptake, which is typical of coastal sites, and negatively correlated with Fe3+ uptake, which is typical of open ocean sites. The concentration of these nutrients is not significantly correlated to iron concentrations.
Figure 3. Scatter diagram of Canonical Correspondence Analysis.
The iron-related metabolic pathways and environmental variables are projected as a result of CCA on 37 metagenomes. Left panel: position of iron-related metabolic pathways (IMP) on the canonical axes. Right panel: contribution of the environmental variables to the canonical space. (CN: control recA; F2: Fe2+ uptake; F3: Fe3+ uptake; FL: flavodoxin switch; OX: oxidative stress; RG: regulation; ST: storage; SU: siderophore uptake; TR: unspecified iron transport).
Taxonomic diversity and iron-related metabolic pathway prevalence covary between but not within habitats
In order to assess the relative importance of taxonomy in the observed correlation between dFe and iron metabolism pathway prevalence, we investigated the relationship between phylum prevalence and dissolved iron contration (dFe) (Table 2). Only one minor class (deltaproteobacteria) shows a significant relationship with dFe in open ocean. In a previous analysis of 16S rDNA prevalence [25], deltaproteobacteria were also poorly represented and mainly affiliated to the SAR324 clade. Overall, there is no significant relationship between phylum prevalence and dFe (Fisher exact test for coastal and open ocean sites). There is, however, a striking difference in taxonomic diversity between habitats.
To further test global correlations between iron-related metabolic pathway (IMP) prevalence, environmental variables and taxonomy, we followed classical biogeography analyses [32], [37] (see Methods). Essentially, we tested whether between site correlations, estimated from three different matrices (environmental variables, iron metabolism pathways prevalence and taxonomic diversity) showed a similar pattern. This enables to assess the strength of the possible covariations between environmental factors, taxonomic diversity, and functional diversity. The null hypothesis we test here is that there is no covariation, i.e. sites are correlated independently by any of the pair of matrices compared.
We performed these analyses on the 37 sites for which all 8 environmental variables were available: 20 open ocean, 15 coastal and 2 coral reef sites. We found a correlation between the prevalence of iron-related pathways and taxonomy between sites (Mantel test Rho = 0.19, p = 0.001). Environmental correlations were associated to iron metabolism pathways (Mantel test, Rho = 0.11 p = 0.008) and to taxonomy (Mantel test, Rho = 0.24, p = 0.001). Thus, we detected a significant correlation between pathway prevalence (functional diversity), taxonomy and environmental variables.
We then examined whether it was possible to detect a specific effect of iron concentration on iron metabolism pathway prevalence and taxonomy by taking iron content differences as distances between pairs of sites (Table 3). The taxonomy matrices and iron metabolism matrices were correlated (Mantel test, Rho = 0.38, p = 0.001) for the 45 sites for which dissolved iron concentration could be inferred (22 open ocean and 23 coastal sites). Iron concentration differences were weakly associated to iron-related pathway prevalences (Mantel test, Rho = 0.06, p = 0.043), whereas there was no correlation between iron concentration differences and taxonomy (Mantel test, Rho = 0.05, p = 0.092). Because taxonomy and pathway prevalence both vary with habitat type, the habitat type may explain part of the above correlations between sites. We therefore analyzed the dependency on iron concentrations in open ocean and coastal sites independently. In both habitats, taxonomy at the phylum level and iron-related metabolic pathways remain significantly correlated (Mantel test, open ocean Rho = 0.27, coastal Rho = 0.34, p = 0.001). Within open ocean sites, the correlations between iron concentration and iron metabolism pathways or taxonomy are no longer significant, probably because variation in iron concentration was too low within open ocean sites. However, in coastal sites, iron concentration and iron metabolism pathways remain significantly correlated (Mantel test, Rho = 0.14, p = 0.02). However, iron concentrations are no longer correlated to taxonomic diversity.
Table 3. Relationship between iron pathway prevalence, taxonomy and iron differences between pairs of sites.
| All sites | Coastal sites | Open Ocean sites | |
| Number of sites | 45 | 22 | 23 |
| IMP×dFe | 0.06* | 0.14* | ns |
| IMP×Taxo | 0.38** | 0.34** | 0.27** |
| Taxo×dFe | ns | ns | ns |
*: p<0.05,
**: p<0.01,
***: p<0.001,
ns: not significant.
Iron Biomarker Genes
To identify possible iron biomarker genes, we tested the relationship between the prevalence of the 13 most represented single genes of our database (i.e. detected in at least half of the 54 metagenomes screened) and iron concentration using Spearman's rank coefficient for sites where the gene was detected. Bfr (bacterioferritin) showed a strong positive correlation with predicted dissolved iron (Rho = 0.50, p = 0.007). Conversely, fecA (encoding the ferric dicitrate outer membrane transporter) was negatively correlated with iron concentration (Rho = −0.31, p = 0.03). In open ocean sites, this correlation was much stronger (Rho = −0.61, p = 0.002). We found no significant correlation between the prevalence of the cyanobacterial photosystem isiA gene and predicted iron concentrations. This gene is encoding the iron-stress chlorophyll-binding protein and had previously been suggested as an iron biomarker gene [17]. However, the prevalence of isiA is significantly higher in open ocean sites as opposed to coastal sites (Kruskal-Wallis p = 0.004).
Discussion
We show significant quantitative and qualitative variations in iron-related strategies with predicted iron concentrations, and a global trend of increasing proportions of iron uptake genes in low iron environments. The sign of observed correlations are consistent with results obtained from experimental studies: the iron storage pathway prevalence increases with simulated dissolved iron concentrations (Rho = 0.56), whereas siderophore uptake prevalence appears to be a low iron strategy (Rho = −0.53). A striking qualitative difference in iron uptake strategies between coastal and open ocean habitats is the negative correlation between Fe2+ uptake versus Fe3+ uptake. The bioavailability of iron in the ocean is linked to its chemical speciation, which is not well known. Most of dissolved iron is complexed by organic ligands (like siderophores); however, inorganic species also exist, though at much lower concentrations. The fraction of “free” iron, despite its extremely low steady state concentration, is thus an important resource for bacteria. This is consistent both with field data on iron speciation, which shows that unchelated iron can be an important source of iron to the phytoplankton in the sea [38], and with gene content analysis of marine cyanobacteria, suggesting that some strains are specialized in one ferric state uptake [39]. Fe2+ uptake prevalence is indeed significantly higher in coastal sites, whereas Fe3+ uptake is higher in the open ocean. Consistently, there is a significant relationship between the proportion of cyanobacteria and the habitat (Table 2). The prevalence of Fe2+ at a given region might result from elevated photoreduction of organically complexed Fe3+ (i.e. a greater source) which can be elevated in coastal sites [40] or reduced Fe2+ oxidation (i.e. reduced sink) due, for example, to low oxygen microenvironment arising from large particles or aggregates that are abundant in coastal waters [41]. We speculate that the striking differences between genomic prevalence of Fe2+ and Fe3+ uptake genes in open ocean versus coastal environments could reflect an as yet unreported bioavailability difference in these two oxidative states of iron. A better knowledge of the bioavailability of these inorganic forms will require precise determination of the supply rate in different environmental conditions.
From the 10 pathways identified from the literature, two pathways have very few sequence representatives in the metagenomes. The first rare pathway is heme uptake (9 genes of this pathway have at least one hit in 11 metagenomes), consistent with recent evidence that many free living marine bacteria lack orthologous genes of this pathway [9]. The second rare pathway is siderophore synthesis (detected in 3 of 54 sites), whereas siderophore uptake was present in all metagenomes screened. There are two kinds of hypotheses to explain this apparent paradox. First, genes involved in siderophore synthesis may be more species-specific than those involved in siderophore uptake, such that our similarity based approach cannot efficiently detect genes involved in siderophore synthesis. Siderophore synthesis is performed by nonribosomal peptide synthetases (NRPS) or NRPS-independent pathways, the latter pathway being much less well characterized [42]. Moreover, these biosynthesis pathways are very diverse and specific for each type of siderophore. In contrast, all siderophore receptor systems identified so far are composed of a specific membrane siderophore receptor and periplasmic binding proteins (Gram negative), and ABC-type transport proteins, showing many structural similarities [7]. Consistent with this, siderophore uptake is represented by fewer genes in our database (23), with an average of 15.6 sequences per gene and an average of 4 taxonomic groups per gene, whereas siderophore synthesis is represented by more genes (53) with an average of 3.8 sequences per gene and an average of 2.1 taxonomic groups per gene. Both differences in diversity are significant in terms of the average number of sequences (Wilcoxon test, p = 0.0004), and the number of taxonomic groups per gene (Wilcoxon test, p = 0.0003). The higher diversification of the siderophore synthesis gene family might thus explain the few number of hits observed. A second explanation could be that specific siderophore synthesis is not an evolutionary stable strategy [43] in the ocean, as it is too expensive and wasteful for marine microorganisms, and therefore those microorganisms producing siderophores are counter-selected. In contrast, the siderophore uptake strategy is advantageous, because “natural” siderophores, such as citrate, which is a metabolic byproduct, are present in the environment and can be taken up by siderophore uptake genes like the receptor fecA. Marine bacteria may thus take advantage of the presence of siderophore-iron complexes, which are not necessarily excreted by bacteria to the marine environment.
This “natural” siderophore uptake gene, fecA, is one of the two candidate genes for iron bioavailability we have identified, based on a significant correlation between their prevalence and predicted iron concentration. FecA is the outer membrane receptor component of ABC-transporter of dicitrate-type siderophores [44]. One of the main structural features of marine siderophores identified so far is that they contain predominantly α-hydroxy-carboxylic acids (like citric-acid) [45]. These are photoreactive siderophores such as petrobactin, ochrobactins, synechobactins, alterobactin, or dicitrate itself. In addition, in dilute environments like seawater where the synthesis of specific siderophores may be prohibitively wasteful for isolated cells, iron may be complexed to natural organic ligands [46]. Citrate is ubiquitous in nature and can complex Fe3+ in the form of ferric-dicitrate [47]. Our results suggest that uptake of ferric-dicitrate as a source of iron may be particularly important in open ocean waters. Since fecA is absent from cyanobacterial genomes, this gene is a good candidate for iron bioavailability for the heterotrophic bacterial community. This will have to be investigated experimentally in the ocean, e.g. with quantitative PCR.
The second candidate gene for iron bioavailability, bacterioferritin (bfr) is involved in iron storage inside the cell [48] and we found that its prevalence increases with predicted iron concentration. This positive relationship is consistent with a recent proteomic analysis in Acetinobacter that shows that it is upregulated in iron-rich culture conditions compared to iron-chelated media [49]. Moreover, expansion in the number of bfr copies in Synechococcus genome is associated with coastal environments [39].
All previous pan-oceanic metagenomic studies have evidenced a strong habitat effect [15], [17], [33], [37]. This is consistent with the large body of work from both geochemical and microbial biodiversity surveys. Coastal sites are nutrient-rich because of the proximity of land-based sources and account for approximately 30% of all marine biological productivity [50]. The open-ocean is typically more stable and generally poorer in nutrients with lower biomass levels, but with a remarkably high diversity.
Horizontal Gene Transfer (HGT), which is the exchange of genes between distantly related bacteria, is a major mechanism of genome evolution in prokaryotes [51], [52]. Not surprisingly, HGT has been found to be prevalent in marine bacterial genome evolution [53] and especially so for transporter genes [32], [54]. Therefore, one might expect that microbial communities' taxonomic and functional diversity are not strongly correlated. Consistent with this, previous studies have shown that there is a much stronger correlation between functional diversity, measured as membrane protein diversity, and environmental variables compared with that between taxonomic diversity, measured from 16S rDNA, and environmental variables for 29 metagenomes [33], [55]. Our results suggest that both iron-related metabolic pathway prevalence and taxonomy at the phylum level are correlated with dissolved iron concentrations, suggesting an important phylogenetic inertia between functional and taxonomic diversity (at the phylum level) on iron metabolism genes.
In conclusion, marine metagenomes enable us to investigate how growth limiting abiotic factors may shape the most abundant, and therefore most successful, genes in a community assemblage. Here, we show that different iron metabolism strategies, inferred from gene prevalence, vary with iron concentrations across marine environments, and that both habitat type and taxonomy are important factors to take into account at a global scale. Our analysis indicates that iron storage (especially bacterioferritin) and flavodoxin switch are the most prevalent iron response strategies, whereas siderophore uptake (especially the ferric-dicitrate receptor gene fecA) increases with iron depletion in the open ocean. The difference between Fe2+ and Fe3+ uptake between coastal and open ocean environments and the unexpected prevalence of dicitrate receptors shed new light on the bioavailability of iron for bacteria in the marine environment.
Supporting Information
Relationship between predicted and observed dFe for surface sites.
(TIF)
Proportion of iron-related metabolic pathways between habitats. Frequencies are relative to the number of control gene hits (recA) for each site.
(TIF)
Database of iron-related genes.
(XLS)
Identity between gene families in the database.
(XLS)
Metagenomes and associated environmental variables.
(XLS)
Number of RecA and iron metabolism genes for each metagenome.
(XLS)
Acknowledgments
We would like to thank Yves Desdevises for discussions on multivariate analysis. We also thank Nigel Grimsley, Hervé Moreau and Ingrid Obernosterer for insightful comments and support, and the Genomics of Phytoplankton group in Banyuls-sur-Mer for stimulating discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: ET was funded by the European network of excellence Marine Genomics Europe (GOGE-CT-505403) and the Agence Nationale de la Recherche (ANR-08-BLAN-0309). GP is funded by European Union FP7 program under grant agreement 254619. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boyd PW, Ellwood MJ. The biogeochemical cycle of iron in the ocean. Nat Geoscience. 2010;3:675–682. [Google Scholar]
- 2.Raven JA. The iron and molybdenum use efficiencies of plant growth with different energy, carbon and nitrogen sources. New Phytologist. 1988;109:279–287. [Google Scholar]
- 3.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 4.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 5.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo-transport of ferrous iron into bacteria. Biometals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 6.Katoh H, Hagino N, Grossman AR, Ogawa T. Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2001;183:2779–2784. doi: 10.1128/JB.183.9.2779-2784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong Y, Guo M. Bacterial heme-transport proteins and their heme-coordination modes. Arch Biochem Biophys. 2009;481:1–15. doi: 10.1016/j.abb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkinson BM, Roe KL, Barbeau KA. Heme uptake by Microscilla marina and evidence for heme uptake systems in the genomes of diverse marine bacteria. Appl Environ Microbiol. 2008;74:6263–6270. doi: 10.1128/AEM.00964-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey S, Melis A, Mackey KR, Cardol P, Finazzi G, et al. Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim Biophys Acta. 2008;1777:269–276. doi: 10.1016/j.bbabio.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Strzepek RF, Harrison PJ. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature. 2004;431:689–692. doi: 10.1038/nature02954. [DOI] [PubMed] [Google Scholar]
- 12.Kupper H, Setlik I, Seibert S, Prasil O, Setlikova E, et al. Iron limitation in the marine cyanobacterium Trichodesmium reveals new insights into regulation of photosynthesis and nitrogen fixation. New Phytol. 2008;179:784–798. doi: 10.1111/j.1469-8137.2008.02497.x. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan D, Folea IM, Jolley CC, Kouril R, Lubner CE, et al. A novel photosynthetic strategy for adaptation to low-iron aquatic environments. Biochemistry. 2011;50:686–692. doi: 10.1021/bi1009425. [DOI] [PubMed] [Google Scholar]
- 14.Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, et al. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc Natl Acad Sci USA. 2011;108:2184–2189. doi: 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA. 2010;107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivers AR, Jakuba RW, Webb EA. Iron stress genes in marine Synechococcus and the development of a flow cytometric iron stress assay. Environ Microbiol. 2009;11:382–396. doi: 10.1111/j.1462-2920.2008.01778.x. [DOI] [PubMed] [Google Scholar]
- 17.Bibby TS, Zhang Y, Chen M. Biogeography of photosynthetic light-harvesting genes in marine phytoplankton. PloS One. 2009;4:e4601. doi: 10.1371/journal.pone.0004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tortell PD, Maldonado MT, Granger J, Price NM. Marine bacteria and biogeochemical cycling of iron in the oceans. FEMS Microbiol Ecol. 1999;29:1–11. [Google Scholar]
- 19.Rue EL, Bruland KW. The role of organic complexation on ambient iron chemistry in the equatorial Pacific Ocean and the response of a mesoscale iron addition experiment. Limnol Oceanogr. 1997;42:901–910. [Google Scholar]
- 20.Temperton B, Gilbert JA, Quinn JP, McGrath JW. Novel analysis of oceanic surface water metagenomes suggests importance of polyphosphate metabolism in oligotrophic environments. 2011;6:e16499. doi: 10.1371/journal.pone.0016499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLong EF. Microbial community genomics in the ocean. Nat Rev Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 25.Biers EJ, Sun S, Howard EC. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl Environ Microbiol. 2009;75:2221–2229. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisen JA. The RecA protein as a model molecule for molecular systematic studies of bacteria: Comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tagliabue A, Bopp L, Dutay JC, Bowie AR, Chever F, et al. Hydrothermal contribution to the oceanic dissolved iron inventory. Nat Geosci. 2010;3:252–256. [Google Scholar]
- 28.Moore JK, Braucher O. Sedimentary and mineral dust sources of dissolved iron to the world ocean. Biogeosciences. 2008;5:631–656. [Google Scholar]
- 29.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 30.Thioulouse J, Dray S. Interactive multivariate data analysis in R with the ade4 and ade4TkGUI packages. J Stat Softw. 2007;22:1–14. [Google Scholar]
- 31.Fisher R. Statistical Methods for Research Workers. London: Oliver and Boyd; 1932. [Google Scholar]
- 32.Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 33.Patel PV, Gianoulis TA, Bjornson RD, Yip KY, Engelman DM, et al. Analysis of membrane proteins in metagenomics: networks of correlated environmental features and protein families. Genome Res. 2010;20:960–971. doi: 10.1101/gr.102814.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monier A, Claverie JM, Ogata H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008;9:R106. doi: 10.1186/gb-2008-9-7-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piganeau G, Desdevises Y, Derelle E, Moreau H. Picoeukaryotic sequences in the Sargasso sea metagenome. Genome Biol. 2008;9:R5. doi: 10.1186/gb-2008-9-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visca P, Leoni L, Wilson MJ, Lamont IL. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol. 2002;45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 37.Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci USA. 2009;106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morel FMM, Kustka AB, Shaked Y. The role of unchelated Fe in the iron nutrition of phytoplankton. Limnol Oceanogr. 2008;53:400–404. [Google Scholar]
- 39.Palenik B, Ren Q, Dupont CL, Myers GS, Heidelberg JF, et al. Genome sequence of Synechococcus CC9311: Insights into adaptation to a coastal environment. Proc Natl Acad Sci USA. 2006;103:13555–13559. doi: 10.1073/pnas.0602963103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagliabue A, Arrigo KR. Processes governing the supply of iron to phytoplankton in stratified seas. J Geophys Res Oceans. 2006;111:C6. [Google Scholar]
- 41.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 42.Gulick AM. Ironing out a new siderophore synthesis strategy. Nat Chem Biol. 2009;5:143–144. doi: 10.1038/nchembio0309-143. [DOI] [PubMed] [Google Scholar]
- 43.Maynard Smith J, Price GR. The Logic of Animal Conflict. Nature. 1973;246:15–18. [Google Scholar]
- 44.Härle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995;14:1430–1438. doi: 10.1002/j.1460-2075.1995.tb07129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler A. Marine siderophores and microbial iron mobilization. BioMetals. 2005;18:369–374. doi: 10.1007/s10534-005-3711-0. [DOI] [PubMed] [Google Scholar]
- 46.Völker C, Wolf-Gladrow DA. Physical limits on iron uptake mediated by siderophores or surface reductases. Mar Chem. 1999;65:227–244. [Google Scholar]
- 47.Pierre JL, Gautier-Luneau I. Iron and citric acid: a fuzzy chemistry of ubiquitous biological relevance. BioMetals. 2000;13:91–96. doi: 10.1023/a:1009225701332. [DOI] [PubMed] [Google Scholar]
- 48.Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBOJ. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nwugo CC, Gaddy JA, Zimbler DL, Actis LA. Deciphering the iron response in Acinetobacter baumannii: A proteomics approach. J Proteomics. 2010 doi: 10.1016/j.jprot.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longhurst AR. 2007. Ecological Geography of the Sea; Elsevier, editor: Academic Press.
- 51.Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- 52.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brochier-Armanet C, Deschamps P, Lopez-Garcia P, Zivanovic Y, Rodriguez-Valera F, et al. Complete-fosmid and fosmid-end sequences reveal frequent horizontal gene transfers in marine uncultured planktonic archaea. ISME J. 2011;5:1291–1302. doi: 10.1038/ismej.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol. 2011;7:473. doi: 10.1038/msb.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiancone E, Ceci P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 57.Cornelis P, Wei Q, Andrews SC, Vinckx T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics. 2011 doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 58.Escolar L, Pérez-Martín J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationship between predicted and observed dFe for surface sites.
(TIF)
Proportion of iron-related metabolic pathways between habitats. Frequencies are relative to the number of control gene hits (recA) for each site.
(TIF)
Database of iron-related genes.
(XLS)
Identity between gene families in the database.
(XLS)
Metagenomes and associated environmental variables.
(XLS)
Number of RecA and iron metabolism genes for each metagenome.
(XLS)