Abstract
We have developed a method for studying cellular adhesion by using a custom-designed microfluidic device with parallel non-connected tapered channels. The design enables investigation of cellular responses to a large range of shear stress (ratio of 25) with a single input flow-rate. For each shear stress, a large number of cells are analyzed (500–1500 cells), providing statistically relevant data within a single experiment. Besides adhesion strength measurements, the microsystem presented in this paper enables in-depth analysis of cell detachment kinetics by real-time videomicroscopy. It offers the possibility to analyze adhesion-associated processes, such as migration or cell shape change, within the same experiment. To show the versatility of our device, we examined quantitatively cell adhesion by analyzing kinetics, adhesive strength and migration behaviour or cell shape modifications of the unicellular model cell organism Dictyostelium discoideum at 21 °C and of the human breast cancer cell line MDA-MB-231 at 37 °C. For both cell types, we found that the threshold stresses, which are necessary to detach the cells, follow lognormal distributions, and that the detachment process follows first order kinetics. In addition, for particular conditions’ cells are found to exhibit similar adhesion threshold stresses, but very different detachment kinetics, revealing the importance of dynamics analysis to fully describe cell adhesion. With its rapid implementation and potential for parallel sample processing, such microsystem offers a highly controllable platform for exploring cell adhesion characteristics in a large set of environmental conditions and cell types, and could have wide applications across cell biology, tissue engineering, and cell screening.
INTRODUCTION
Cell adhesion is a key process in many physiological and pathological situations. It is known to trigger cell growth1 and cell migration.2 During tumor progression, cancer cells are able to tune their adhesion in order to escape from primary tumor site and colonize other organs.3
Many cells are also sensitive to external forces, in a process generally called mechanotransduction.4 In blood circulation, shear stress is the most common mechanical stimulation sensed and transduced by the cells. Shear stress has been shown to influence adhesion of immune system cells (e.g., leukocytes5 and neutrophils6), and gene expression in endothelial cells,7, 8 which alters adhesion on vessel walls of metastasizing cancer cells.9
Cell adhesion has been thoroughly investigated for mammalian cells, and many proteins and signaling pathways are known.10 On the other hand, mechanical aspects of cell adhesion are still under intensive theoretical and experimental considerations.11
Cell adhesion strength is usually estimated using controllable external forces that enable gradual cell detachment.12 This can be performed using centrifugal forces13, 14, 15 or other macroscale’ set-ups such as radial flow-chambers,16 spinning disks,17 and rectangular parallel-plate flow chambers.18, 19 To get inside into cell detachment dynamics for a given shear-stress, early experiments have combined parallel-plate flow chamber with time-lapse videomicroscopy.6 More recently, microfluidic-based adhesion chambers have been developed with tunable ranges of applied hydrodynamic forces, enabling real-time analysis of cell detachment kinetics,20, 21, 22 including cancer cell experiments.23, 24 Microfluidic dimensions require only small amounts of cells and reagents, and allow laminar flow, high throughput and the possibility of parallelization. Most of these devices, however, have a rather biological approach and, therefore, are satisfied with yielding the same results that could be achieved similarly by macroscopic approaches, that is, counting the fraction of cells that detaches when exposed to a specific shear stress.12 Little effort has been made so far to resolve the time dependence of this fraction, i.e., the detachment kinetics, which have been the object of mathematical and numerical modeling in several biophysical papers.25, 26, 27, 28
We aim at filling that gap with a simple microfluidic device consisting of four independent channels enabling investigation of cell detachment in a large shear stress range (more than 1 order of magnitude), with a single input flow rate. Microfluidic set-ups enable time-lapse investigation not only on detachment kinetics but also on cell motion under shear stress. We show how this can be used to enhance the understanding of cell adhesion. Both kinetics of cell detachment and cell motion are investigated using two different cell models with different adhesion strengths: the amoeba Dictyostelium discoideum and the metastatic human breast cancer cell line MDA-MB-231.
D. discoideum is a well-investigated model cell organism. In particular, it is a model of choice for the study of amoeboid behaviour, not only because the key signaling pathways implicated in cell movement are conserved from the amoeba to higher eukaryotes but also because this haploid unicellular eukaryote has the advantage of being a genetically and biochemically tractable organism (its genome has been completely sequenced in 200529). This cell type is very motile, with largely non-specific and relatively low substrate adhesion.16 On the other hand, the MDA-MB-231 mammalian cell line is highly mobile, invasive, and tumorigenic in vivo.30 Thanks to the versatility of the microfluidic-device presented in this paper, it is possible to get results on adhesion strength for both cell types with only minor experimental modifications. In this work, results of detachment kinetics, adhesion strength and cell dynamics are provided for both cell types.
MATERIALS AND METHODS
Device fabrication
We used standard multilayer photolithography procedures31, 32 to create the microfluidic device with four parallel channels shown top-viewed in Fig. 1a.
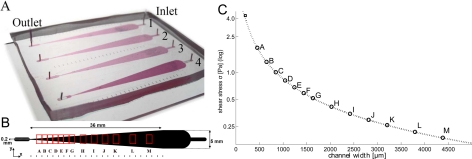
Figure 1.
Microfluidic device with four independent branches. (a) Photograph of the device, sealed on a 76 × 52 mm glass slide. The four channels are labeled with numbers. Inlet-outlet-distance is 50 mm. In- and outlet can be interchanged to avoid a high hydrodynamic pressure for large widths which occurs at the inlet site. (b) Photolithography mask (detail). The colored rectangles correspond to the visual field of different shear stress zones we used for our later discussed model experiment. (c) Shear stress (log-scale) related to the position in the tapered channel for a low flow rate Q = 5 ml/h. The corresponding letters are represented in (b). The square on the top left shows the highest possible shear stress that could be attained at this flow rate.
The photo mask pattern as shown in Fig. 1b was drawn with CleWin4 (wieweb software). The mask was transferred onto a glass plate as substrate. First, we applied one negative photoresist layer (Laminar Aqueous Resist E9220, thickness 50μm) and exposed it to UV light. This helps the further layers to stick properly. Then, 1, 2, or 3 layers of the same negative photoresist were put on the substrate and covered with the mask before exposing to UV light. This lead, after development with a carbonate solution (, heated to 67 °C), to profile heights of 50, 100, or 150 μm, respectively. We verified this height with a surface profiler (Veeco Dektak 150, contact stylus profilometry techniques).
The developed master was first silanized (Aldrich Octadecyltrichlorosilane, 90+%) to ease unpinning PolyDiMethylSiloxane (PDMS) afterwards, then covered with reticulating PDMS (10:1 ratio PDMS:reticulate, using Sylgard 184 Silicone Elastomer and the associated curing agent), and baked for 2 h at 75 °C. The reticulated PDMS was ablated from the master, pierced for in-/outlets with a Harris Micro-puncher 0.5 mm diameter, treated with O2-plasma (53 W, 2.5 min, 200 mTorr, distance from cathode about 5 cm) and irreversibly sealed to an equally treated clean glass plate (Scientific Glass Labs LTD, 76 × 52 mm, constituting the surface on which the cells adhere later). All operations so far had been done in a clean room to avoid contamination. Just after sealing, the channels were wetted with 0.22 μm-filtered ethanol to decrease surface tension and avoid bubble formation.
Channel coating
For Dictyostelium, the glass surface was used directly or further coated with (3-aminopropyl)triethoxysilane (APTES, Sigma-Aldrich). For APTES-coating, 1% APTES in 5 mM aqueous acetic acid was injected in the chamber. A stop-and-go flow of the solution was maintained for 20 min. The APTES-coated channels were then washed with distilled water before drying at 100 °C for 15 min. For MDA-MB-231, the glass surface inside the channel was coated with 20 μg/ml of collagen type IV for 30 min at 37 °C, and then blocked by rinsing the channels with a 0.4% Bovine Serum Albumin (BSA, Sigma Aldrich) solution for at least 30 min at 37 °C.
The channels were then thoroughly rinsed with fresh cell medium (HL5 for Dictyostelium and CO2-independent medium for MDA-MB-231) and used directly.
Flow creation
Both inlet and outlet of the four channels of our device were connected to flexible tubes (PORTEX Fine Bore Polythene Tubing 0.28 mm inner diameter), the inlet being connected to a switchable T-connectic, allowing easy cleaning and exchanging of hand- and pump-driven syringes. The tightly closed system allowed a precise measurement of fluid flow by collecting and weighing the liquid that had passed by.
For precise flow control, we used a syringe pump (PHD ULTRA infuse/withdraw, Harvard Apparatus) with up to four BRAUN Omnifix 20 ml Luer Lock syringes for Dictyostelium and up to two BRAUN 60 ml Luer Lock syringes for MDA-MB-231. Of major importances were tight syringes with a rubber piston which shows no stick-and-slip behaviour, in order to assure a steady-going flow. With 20 ml syringes, experiments of up to 3 h with a flow rate of 6–7 ml/h could be easily managed.
Strain and culture, cell injection
D. discoideum DH1 was grown in HL5 medium (formedium, UK) in plastic culture dishes (Falcon) at 21 °C. Vegetative cells were harvested at a density of 1 · 106 cells/ml, suspended in fresh medium and injected into the microchannel via the channel outlet using a micro-pipette. A careful injection of about 24 μl into the outlet with the inlet being kept open was sufficient to obtain cell densities of (4 ± 1) · 104 cells/cm2. The injected volume exceeds the volume of one channel (14 μl) to assure the coverage of those parts of the channel which are far from the injection point.
Human metastatic breast cancer cell line MDA-MB-231 was cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) medium supplemented with 10% Fetal Bovine Serum (FBS, PanBiotech) and 1% penicillin/streptomycin at 37 °C and 5% CO2 in culture petri dish (Falcon), as previously described by Vidal et al.33 We preliminarily verified survival and proliferation for MDA-MB-231 in microfluidic systems, which gave normal behavior for t > 72 h, with non-detaching shear stress (0.04 Pa) being applied. We also verified that cell shape, movements and rate of cell division were similar in petri dishes and in microfluidic channels (data not shown). Prior to each experiment, cells were detached using trypsin solution (0.25% w/v). The activity of trypsin was stopped with 10% FBS-containing medium, and the suspension was centrifuged (660 g, 5 min). The supernatant was aspirated and the cells were resuspended in medium to a density of 1 · 106 cells/ml. Injection followed the protocol for Dictyostelium, the cell densities were one order of magnitude below those for Dictyostelium.
Settling time at the surface was 30 min for Dictyostelium and 2 h for MDA-MB-231.
After the experiment, we checked that the cells are able to divide and migrate normally after having been subjected to an experiment with high shear-stress (data not shown).
Microscopy
The connected device was installed in a home-made temperature-controlled chamber, which enabled adhesion analysis in physiological temperature conditions (21 °C for Dictyostelium and 37 °C for MDA-MB231), and observed with an inverted light microscope (Nikon TE2000) equipped with a 4 × objective (field of view 2.2 × 1.7 mm, at a resolution of 1.61 μm/pixel), a cooled CCD camera (CoolSNAP HQ2 Monochrome, Roper Scientific) and a motorized xy-stage (Marzhauser). A low resolution 4 × objective was chosen to get high statistics in single experiments. Images were captured automatically with a commercial program (NIS Elements AR) along a path scanning the different positions (and therefore the different applied stresses), as visualized schematically in Fig. 1b by colored rectangles. One image contained typically 500–1500 cells for Dictyostelium and 40–120 cells for MDA-MB-231. A scanning path with 52 positions (4 channels, 13 positions each) took about 45 s, which determined the temporal resolution of detachment kinetics recording. A lower number of channels and of positions per channel enabled much higher time resolution. Higher spatial resolution (20×) but with lower statistics could be accomplished without change of the settings as well.
It turned out that setting out of focus the lenses delivered an optimal performance in practice for Dictyostelium: even if the precise cell shapes were not visible any longer, the cells were easier to distinguish from device impurities, thus facilitating subsequent cell counting. For MDA-MB-231, which has thinner cell bodies than Dictyostelium when adhering, phase contrast illumination was used. See supplementary material for typical images obtained for both cell types.34
Image processing
To count the number of cells for each image and each timestep, the recorded movies were analyzed with the free, platform-independent program imagej using a custom-written batch algorithm based on the Find maxima routine which gave the number of cells (and the cell positions, if tracking was desired) for each image. In the supplementary material, we give an example with identified cells.34
The data obtained were further analyzed with a single matlab program which calculated the cell density (in cells/cm2), the cell detachment in dependence of time and the parameters which characterize a cell population and its detachment kinetics (see Sec. 3). We thereby used the free matlab fitting toolbox EzyFit.
Within the same program, error calculus was done, effecting error propagation for stress incertitudes and analysis of the statistical error due to finite number of cells, both by means of straightforward resampling methods, described in the supplementary material.34 The matlab-code with a working data sample is available upon request.
RESULTS AND DISCUSSION
Device design and rationale
Several microfluidic cell detachment devices use parallely connected channels.21, 22 We tested such approaches, but a general problem with complex interconnected channel systems is, that a defect in one of the channels changes all hydrodynamical resistances and impairs the local flow rate estimates. Parallel channel systems bring along also other disadvantages, such as difficulties to dispose of air bubbles or problems with seeding the cells uniformly in the parallel branches. All these drawbacks could be avoided or downsized by our device which uses simple tapered channels without interconnections, corners and acute angles, as shown in Fig. 1.
The use of large glass slides (52 × 76 mm) enables to have 4 independent tapered channels in parallel. This offers the possibility of exploring different cell types and/or environmental conditions (coating, medium) in a single experiment. This can be pivotal for cell biology experiments where results from different conditions should be compared as much as possible from the same experimental day.
Tapered shapes of the channels enable the exploration of a wide range of shear stresses along the channel direction x. Our tapered device which is shown in Fig. 1 is characterized by a linearly decreasing width w given as (wmax = 5 mm and wmin = 200 μm, channel length L = 36 mm). As the tapering slope is small (dw/dx ≈ 0.13), the classic wall shear stress expression12 (at the bottom of the channel where the cells adhere) for a rectangular tube of width (h is the channel height) can be locally applied
| (1) |
where η is the viscosity and Q is the flow rate. In our case, is not fulfilled for all channel positions, and a 3D analysis of the velocity field is necessary, details are shown in the supplementary material.34
In a single experiment, the maximal range of shear stress is given by the ratio σmax/σmin = wmin/wmax = 25. This is large compared to microfluidic devices of similar design, for instance, σmax/σmin = 8 in Gutierrez and Groisman.22
With h varying between 50, 100, and 150 μm, and Q up to 50 ml/h, wall shear stresses up to 100 Pa can be realized; however, the device is designed to work best at 0.1–5 Pa, which covers the physiological range.12 The two cell types which we will discuss here cover this range. To increase the shear stress range for one experiment, different flows can be applied for parallel channels by using syringes with different inner diameters.
The large dimensions lead us to revisit microfluidic assumptions, notably laminarity. The Reynolds number can be calculated using the average flow velocity U = Q/(wh) and the hydraulic equivalent diameter for rectangular channels Dh = 2hw/(w + h)
| (2) |
Laminar flow can be assumed for Re < 2000.35 For this work, we have Re < 5 for experiments with Dictyostelium and Re < 20 for MDA-MB-231 experiments, thus indicating laminar flow. This could be supported by optical flow observations via fluorescent beads which showed linear trajectories (data not shown).
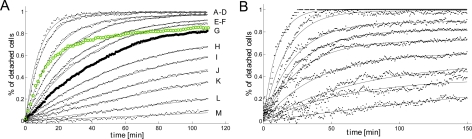
After cell injection and scanning path selection of the motorized stage (Fig. 1b), the experiment starts when both the syringe pump infusing at a constant flow rate Q and timelapse videomicroscopy are switched on (t = 0). The image treatment described in Sec. 2 results in the typical cell detachment curves displayed in Fig. 2. Each curve represents the proportion of detached cells as a function of time for a given position along the channel. The initial cell number decreases exponentially, before reaching an asymptotic regime where almost no cell detachment is detectable. Due to a limited recording time, the asymptotic regime is not fully reached for Dictyostelium in fresh medium (Fig. 2a) at low stress ranges but can be observed for data with MDA-MB-231 (Fig. 2b) and Dictyostelium in highly conditioned medium (HCM) (Fig. 2a, open circles). The meaning of these asymptotic states will be discussed later on in the framework of a threshold stress model. For each experiment, channel height and/or flow rate are adjusted in order to cover the whole range of detachment from complete detachment up to almost zero detachment (100 μm and 5 ml/h, respectively, for Dictyostelium cells; 50 μm and 17 ml/h, respectively, for MDA-MB-231 cancer cells).
Figure 2.
Detachment curves with first order kinetic fits. (a) Cell detachment curves for D. Discoideum DH1 in fresh medium, fitted with first order kinetics. The letters on the right show the channel position (cf. Fig. 1) which is used for the corresponding detachment curve. Higher shear stresses correspond to greater detachment. The detachment curve for 0.50 Pa (letter G) is highlighted by larger dots, the detachment curve for DH1 in HCM at the same shear stress is drawn in green open circles. This shows that the final detachment is very similar for both conditions, whereas the kinetics are much faster for HCM. (b) Detachment curves for MDA-MB-231 human breast cancer cells. The more significant fluctuations are due to a lower cell concentration and image treatment.
Mathematical description of adhesion and detachment
For describing the adhesive behavior of these two cell lines, we mostly follow Décavé et al.;16 similar ideas were also used by Cheung et al.24 Three major assumptions are made: (1) There is a threshold shear stress σt for each cell, which, if exceeded, leads sooner or later to the detachment of the respective cell. (2) This threshold stress is lognormally distributed over a given population. (3) The detachment dynamics follow first order kinetics.
The lognormal threshold stress distribution f(σt) is specified by its median σ1/2 (the stress at which half the cell population detaches) and the dimensionless distribution width
| (3) |
The cumulative distribution (i.e., the integral of f(σt)) gives the proportion of cells which will be detached by a given stress σ; we call this proportion the detachment efficiency e(σ),
| (4) |
The first order kinetics presuming threshold stresses—dn/dt = +kNc where Nc is the number of cells at a given time for cells with σt < σ and dn/dt = 0 for σt > σ—lead to a time-dependent expression for the percentage of detached cells16
| (5) |
The detachment efficiency, which stands for the percentage of detached cells for t = ∞, links the kinetics with the threshold stress distribution Eq. 4.
Adhesion thresholds for amoeboid and breast cancer cells follow lognormal distribution
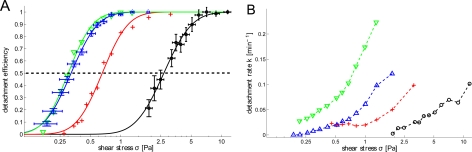
The experimentally obtained detachment curves shown in Fig. 2 are very well fitted with first order kinetics for each cell line, leading to a stress dependent detachment rate k (Fig. 3b). The detachment efficiency which corresponds to the asymptotic state value in Fig. 2 is plotted as a function of σ in Fig. 3a. The experimental values for each cell line are fitted by the lognormal cumulative distribution function.
Figure 3.
Detachment efficiency and detachment rate for different experiments. (a) The cumulative lognormal threshold-stress distribution function (cdf), fitted with Eq. 4 for four datasets, listed from left to right: Dictyostelium on glass substrate in highly conditioned medium (green, downward pointing triangles) and in fresh medium (blue, upward pointing triangles), in fresh medium on Aptes-coated substrate (red, crosses), and MDA-MB-231 on collagen-coated substrate (black, circles). Every datapoint for the Dictyostelium data in fresh medium corresponds to one position in Fig. 1. Error bars are shown for one condition each for Dictyostelium and MDA-MB-231 to give an idea of magnitude. σ1/2 can be read off at the point where the dashed line intersects the fits. (b) Detachment rates k, obtained by first order kinetics fits. The color/symbol code is the same as in (a). Stress errors are the same as for (a), fit errors for k are of the size of the symbols. Data points without significant detachment (Aptes for σ < 0.5 Pa) could not be fitted properly and were left out. Data points are connected as a guide for the eye.
The occurrence of lognormal distributions for biological quantities is well-known and also successfully applied for adhesive strength distributions,16, 24 but not well-understood. There is not a single standard generative model which produces lognormal populations: several possible mechanisms can lead to lognormality. Yet, the key idea is the accumulation of multiplicative (and not additive, which leads to normal distributions) random effects. For cellular objects, these multiplicative random effects may occur by asymmetrical cell division or external random effects, whose impact depends on the actual state of the cell. We refer to Koch36 for a biology-oriented introduction to generative models for lognormal distributions. Our data (Fig. 3a) fit very well to the lognormal cumulative distribution function and, therefore, give support to the idea of lognormal distributed threshold stresses for single cells, independently of the cell type.
Quantitative comparison of detachment parameters
For a quantitative discussion of adhesion parameters and to assess the precision and the robustness of our device, we investigate two main possible error sources (see supplementary material for more details34):
-
1.
Statistical and counting errors δN; a limited number of cells creates a statistical error when data are described by a given distribution law as given in Eq. 3.
-
2.
Physical incertitudes and device approximations of the wall shear stress δσ: flow velocity field and thus shear stress values may be modified by PDMS deformation due to internal pressure; temperature-dependency of viscosity can affect the shear stress as well; also channel width and thus shear stress are not constant over a given field of view.
As far as the second error source is concerned, δσ mostly depends on the channel height error ( and is almost independent of x). However, a proper measurement of h(x) by confocal microscopy was performed for high shear stress conditions (MDA-MB-231 cancer cell experiment); in a first approximation, deformation increases linearly with channel width (cf. again the supplementary information cited above, where we refer in detail to Gervais et al.37 for a theoretical framework). To estimate the error of the fit parameters σ1/2 and in Eq. 4 due to the limited experimental sample, i.e., the finite cell number, we used a numerical resampling method. Combining the primary error sources as root of the squares, we are able to give a total error estimation. For best conditions, for Dictyostelium, it is about 6% of the value for σ1/2 and 13% of the value for . The experiments with MDA-MB-231 cancer cells were all carried out at lower cell densities, resulting in larger statistical errors (cf. y-errorbars in Fig. 3a), but only slightly larger errors on fitted parameters (see Table TABLE I.).
TABLE I. Left:
Reproductibility among channels. Adhesion strength distribution parameters for three parallel channels with same conditions (D. discoideum DH1 cells in fresh medium on glass substrate at Q = 10 ml/h). Right: The results for adhesion on glass in fresh medium (FM), on glass in HCM, and in FM on an aptes-coated surface for Dictyostelium at Q = 5 ml/h, and on collagen-coated surfaces for MDA-MB-231.
| DH11 | DH12 | DH13 | DH1gl (FM) | DH1gl (HCM) | DH1AP(FM) | MDA | |
|---|---|---|---|---|---|---|---|
| σ1/2 (Pa) | 0.30 ± 0.02 | 0.31 ± 0.02 | 0.30 ± 0.02 | 0.32 ± 0.02 | 0.32 ± 0.02 | 0.65 ± 0.03 | 2.7 ± 0.2 |
| 0.47 ± 0.08 | 0.54 ± 0.08 | 0.54 ± 0.08 | 0.49 ± 0.08 | 0.48 ± 0.08 | 0.52 ± 0.07 | 0.51 ± 0.08 |
The experiments with Dictyostelium, carried out at Q = 5 ml/h, were compared to experiments which were carried out at Q = 10 ml/h (Fig. S3). Similar σ1/2 and (Table TABLE I.) as well as detachment efficiencies were found; further detail for this point is shown in the supplementary information.34 In other words, varying flow rates and channel widths in such a way, that shear stress is the same, yields similar detachment rates, as expected by Eq. 4. Within this test experiment, reproductibility among different parallel channels was verified with good accordance of adhesion strength from one channel to another, see Table TABLE I., left hand side.
The reproductibility among different channels allows to fully use the independent channels of our device in parallel to compare simultaneously the cells from the same culture in different environments. In Fig. 3, three different conditions for Dictyostelium are shown. Décavé et al.16 already made detachment experiments with Dictyostelium, which may serve, although made with different mediums, as a first point of reference to what follows. For our experiment, Dictyostelium in fresh HL5 medium on glass substrate yields σ1/2 = 0.32 Pa. Coating the glass substrate with Aptes increases adhesion strength significantly to σ1/2 = 0.65 Pa; this increase has already been reported by Décavé et al.16 For comparison, Fig. 3 also contains data for an MDA-MB-231 experiment on collagen substrate. Here, we found a characteristic adhesion strength σ1/2 ≈ 2.7 Pa, which shows that our device works for cell types of very different threshold shear stresses. The same order of magnitude was found by Cheung et al.24 for MDA-MB-231 -N (N-cadherin expressing MDA-MB-231) for antibody-coated surfaces (σ1/2 ≈ 2.1 Pa for step function accelerating and 5 min remaining PBS flow). This demonstrates that MDA-MB-231 cells exhibit similar adhesion strength for different families of adhesive proteins (cadherin-antibody for Cheung et al.24 versus integrin-collagen in our case).
Another possibility besides surface treatment to modify detachment of Dictyostelium is modifying the medium. HCM is obtained from Dictyostelium cell cultures; the medium therefore is enriched with various biomolecules that are naturally produced by Dictyostelium in cell culture. Some of these molecules can dock to extracellular receptors, which impedes non-attached receptors to interact with the substrate. We discussed this recently in the context of cell migration, where we observed lower motility and more rounded cells for Dictyostelium in HCM.38 For adhesion, we found no difference for σ1/2 for fresh medium and HCM, see Fig. 3a. By contrast, detachment kinetics were much faster in HCM (Figs. 3b, 2a, highlighted curves). These findings suggest that attachment and re-attachment of receptors, which is reduced by HCM as described above, play an important role in the detachment process, but does not significantly determine the threshold stress value for our experiments with Dictyostelium, i.e., the decision if a cell is detached at a given stress or not. This example illustrates the need for having acces to detachment kinetics. It enabled to highlight differences in cell detachment which are not revealed by σ1/2 values.
In Décavé et al.,16 experiments with Dictyostelium were carried out in buffer (whereas we used HL5 medium); buffer media is of simple constitution, the number of parameters which may influence an experiment is reduced. On the other hand, a buffer is not a natural environment, leading to complications such as osmotic shocks and starvation. We found that adhesion is weaker for Dictyostelium in HL5 compared to values given by Décavé et al.16 for experiments using buffer. The difference is about one order of magnitude. This indicates that adhesive contacts are strengthened in buffer medium. Due to lower applied shear stresses, detachment in HL5 is much slower (Figs. 2a, 3b) than in buffer. In buffer, Décavé et al.16 found typical detachment times of ≈5 min, whereas we got characteristic times of 1 h (Fig. 2a). As shown in Fig. 3b, kinetics are generally faster with increasing shear stresses. We will discuss a theoretical model for k(σ) in the context of cell motion/migration below.
We would like to stress here that kinetics could be obtained by a single experiment, observed at more than 200 time-steps. In contrast, Décavé et al.16 did one experiment for one experimental data point. We hold this for a major improvement of our device: evidently, it reduces the effort; second, it can be checked if an asymptotic state is attained in a single experiment. Generally, it is often preferable to acquire biological data for one issue at the same time for reasons of comparability. This allows to distinguish variations due to biological heterogeneity of a sample and those due to other changes of experimental parameters.
Cell tracking, migration, detachment mode
In addition to kinetics resolution and the possibility of parallel experiments, our device is also adapted to measurements of adhesion and cell tracking simultaneously. This allows to enhance the comprehension of detachment kinetics by analyzing cell shape modifications and migrative behaviour.
Dictyostelium, as stated above, is detached on a typical timescale of up to 1 h for an applied hydrodynamic stress close to σ1/2 in fresh medium. The detachment for Dictyostelium is described by Garrivier et al.25 as a peeling process which begins with the onset of the experiment. In this model, the microscopic peeling velocity vp, which is a function of some microscopic parameters, is connected with the detachment rate k and the peeling time τ using the cell dimension lc: . Our tracking analysis, however, shows that cells are able to migrate normally during up to 1 h before detachment. The trajectories show diffusive behaviour for low shear stresses and only partially directed flow-driven movement for high stresses (Figs. 4b, 4c, 4d). The diffusion coefficients in the tapered channel are similar to those obtained by measurements in petri dishes, which we carried out recently.38 The motility of Dictyostelium is therefore not disabled by shear stresses, and the peeling process does not start with flow onset, but at a stochastically determined moment. This is clearly visible on the cell of Fig. 4a (enhanced) submitted to a very high applied shear stress (2.6 Pa ≈ 4σ1/2). It first moves by extending several forward but also lateral pseudopods (see arrows in Fig. 4a (enhanced)) but finally stops migration, rounds up and is detached. It is therefore not possible to link microscopic and macroscopic kinetics within the framework of Garrivier et al.25 for Dictyostelium in natural-like environment (HL5 medium). This conclusion would not have been possible without the possibility to track the motion of cells.
Figure 4.
Dictyostelium cells motility during a detachment experiment on APTES coated substrate. (a) Detachment of a single Dictyostelium cell at 20× magnitude, at 2.6 Pa: despite the very high stress, the cell extends also lateral pseudopods (arrows) and migrates. Finally, the cell rounds and detaches. The arrow in the last frame indicates the flow direction. The migrative behavior comes out better in a time-lapse movie. (b) (c) Centered trajectories of at least 150 cells submitted to a shear stress of 0.25 Pa (b) and 0.70 Pa (c), respectively, over 52 min. Cell migration is almost random at low shear stresses, whereas it is strongly biased in flow direction for higher shear stresses. Flow direction is given by arrows. (d) Mean directionality of cell movement as a function of applied shear stress, indicating to which extent migration is aligned with flow direction. Directionality is defined as the angle between the flow direction and the cell movement direction over a 52 min interval. Therefore, cos(θ) = 1 indicates fully biased cell movement in flow direction, cos(θ) = 0 indicates random direction of migration. The chosen positions (b) and (c) are represented by black arrows, σ1/2 = 0.65 Pa by the red arrow (enhanced online) .
For MDA-MB-231, a very different detachment processes could be observed (Fig. 5a (enhanced) and Fig. 5d (enhanced)). At applied stresses around σ1/2, no significant migration could be observed. The detachment process starts just after shear flow onset. The process can be subdivided in several phases. They are most clearly visible in a time-lapse movie, Fig. 5d (enhanced). First, the cells, which are randomly oriented on the surface, align with the flow. The alignment is mainly a fast and discrete process, often caused by the rupture of anchor points (5D (enhanced)). In the next phase, the cell body is dragged by the flow with nearly constant velocity (center of mass movement, Fig. 5b), whereas the anchor points are more strongly bound. They are still spread out and detach with a rupture (cell shape evolution, Fig. 5c). The interplay between the cell body and the anchor points cannot be described by a simple peeling model without focal contacts.39 In the last phase, the cell adopts a waterdrop shape, with no more outlying anchor points (last frame of Fig. 5a (enhanced)). The cell slowly continues to be dragged in the direction of the shear flow, and finally detaches from the substrate.
Figure 5.
Detachment mode of MDA-MB-231 cancer cells. (a) Example of cell alignment in response to the imposed shear stress (see the time-lapse movie online for the different phases of detachment). (b) Single cell drop formation at 4×. The black arrow indicates flow direction. (c) The cell (its center of mass) moves after a short acceleration phase with constant velocity in flow direction. Leaps (marked by dotted lines) occur, when adhesion points break off. (d) Cell deformability is analysed by computing the solidity parameter s = A/Ach, which uses the cell area A and its convex hull Ach (shown in the 5th frame). A sudden increase of s corresponds to a rupture of an anchor point (see dashed lines) (enhanced online) .
Example of cell alignment in response to the imposed shear stress (enhanced online: time-lapse movie showing the different phases of detachment).
To highlight the anchor point ruptures, cell deformability is analyzed by computing the solidity parameter s = A/Ach, where A is the cell area and Ach the area of its convex hull, i.e., the percentage of the area spanned by the cell’s corner points which is occupied by the cell. A sudden augmentation of s corresponds to a rupture of an outlying anchor point, which reduces the convex hull, at almost constant cell area. Fig. 5c shows clearly two anchor point ruptures.
Such cell alignment, elongation and drop formation occurred above a shear stress threshold which is comparable to σ1/2 = 2.7 Pa. Similar shear stress dependent shape elongation has already been reported for breast cancer lines HBL100 submitted to centrifugal forces.14 This study evidenced a coordinated work between microtubule (helping elongation), actin (fighting against elongation) and focal adhesion, but it did not enable real-time analysis. More recent results using a microfluidic set-up24 reported that cell deformation is not only dependent on shear stress but also on shear stress acceleration: deformation was only observed for low acceleration conditions. This is in accordance with the results presented here, where a cell in a given position experienced one single shear stress for up to 150 min, with zero acceleration. The effect of acceleration on cell shape and cytoskeletal organisation could be easily further investigated with our set-up. However, for in-depth quantitative analysis of this process, higher magnification (10× or 20×, to the detriment of statistics) is required for automatic cell contour detection and is beyond the scope of this paper.
CONCLUSION
We have designed and tested a simple 4-channel tapered microfluidic device for cell adhesion strength measurements based on the analysis of cellular response to varying hydrodynamic shear stresses. Compared to other recent microfluidic shear stress designs, our device offers a larger shear stress range and a better and very precise detachment kinetics measurement. This allowed us to confirm that first order kinetics and lognormal distributions for the proportion of detached cells are very general laws describing well either amoeboid Dictyostelium or mesenchymal breast cancer cells. Another advantage of our device is the possibility to track cells (single cells or few hundreds as well) and therefore to better understand adhesion through migration analysis. In addition, visualization during experiments gives feedback over possible problems and allows to show, e.g., the mode of detachment, which can bring forward cell detachment modeling. Because each channel is fully independent of the others, the effect of several environmental parameters (substrate coating, buffers, drugs, etc.) on adhesion and migration can be tested simultaneously. Since this paper has its main focus on presenting the design and the potential it offers, we leave more systematic studies of environmental parameters to future publications. Besides, influence of shear stress on other parameters of interest can be easily measured without device alterations. For instance, influence of shear stress on cell division for Dictyostelium was an issue that occurred while testing our device. We think that the device presented in this paper is a versatile tool that could be used to analyze not only detachment strength and kinetics for different cell types in varying environmental conditions but also putative shear stress dependent cell behaviour such as cell motion.
ACKNOWLEDGMENTS
The LPMCN research team belongs to the CNRS consortium CellTiss and ELyTLab (Lyon-Tohoku) international laboratory. This work was supported by CNRS PEPS grant, Région Rhône-Alpes cible grant, by a grant from the Canceropole Lyon Auvergne Rhône-Alpes (CLARA-Axis 6) and by ANR ONCOSCREEN. NanoLyon clean room facilities were used in this work to fabricate microfluidic structures. We thank M. Faivre for her help in photolithography and microfluidics, and F. Letourneur for providing Dictyostelium discoideum cell lines. We thank P. Cosson and F. Bruckert for helpful discussions and preliminary cell detachment experiments.
References
- Huang S. and Ingber D. E., Nat. Cell Biol. 1, E131 (1999). 10.1038/13043 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Choi C. K., and Horwitz A. R., J. Cell Sci. 122, 1473 (2009). 10.1242/jcs.052894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Curr. Opin. Cell Biol. 16, 14 (2004). 10.1016/j.ceb.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P., and Bershadsky A. D., Nat. Rev. Mol. Cell Biol. 10, 21 (2009). 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Fukuda S., Yasu T., Predescu D. N., and Schmid-Schönbein G. W., Circ. Res. 86, E13 (2000). 10.1161/01.RES.86.1.e13 [DOI] [PubMed] [Google Scholar]
- Lawrence M. B., Smith C. W., Eskin S. G., and McIntire L. V., Blood 75, 227 (1990), http://bloodjournal.hematologylibrary.org/content/75/1/227. [PubMed] [Google Scholar]
- Resnick N., Yahav H., Shay-Salit A., Shushy M., Schubert S., Zilberman L. C. M., and Wofovitz E., Prog. Biophys. Mol. Biol. 81, 177 (2003). 10.1016/S0079-6107(02)00052-4 [DOI] [PubMed] [Google Scholar]
- Davies P. F., Spaan J. A., and Krams R., Ann. Biomed. Eng. 33, 1714 (2005). 10.1007/s10439-005-8774-0 [DOI] [PubMed] [Google Scholar]
- Gomes N., Legrand C., and Fauvel-Lafve F., Clin. Exp. Metastasis 22, 215 (2005). 10.1007/s10585-005-7359-5 [DOI] [PubMed] [Google Scholar]
- DiMilla P. A., Stone J. A., Quinn J. A., Albelda S. M., and Lauffenburger D. A., J. Cell Biol. 122, 729 (1993). 10.1083/jcb.122.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., J. Biomech. 33, 23 (2000). 10.1016/S0021-9290(99)00163-3 [DOI] [PubMed] [Google Scholar]
- Young E. W. K. and Simmons C. A., Lab Chip 10, 143 (2010). 10.1039/b913390a [DOI] [PubMed] [Google Scholar]
- Jay P. Y., Pham P. A., Wong S. A., and Elson E. L., J. Cell Sci. 108(Pt 1), 387 (1995), http://jcs.biologists.org/content/108/1/387. [DOI] [PubMed] [Google Scholar]
- Thoumine O. and Ott A., Cytoskeleton 35, 269 (1996). [DOI] [PubMed] [Google Scholar]
- Koo L. Y., Irvine D. J., Mayes A. M., Lauffenburger D. A., and Griffith L. G., J. Cell Sci. 115, 1423 (2002), http://jcs.biologists.org/content/115/7/1423. [DOI] [PubMed] [Google Scholar]
- Décavé E., Garrivier D., Bréchet Y., Fourcade B., and Bruckert F., Biophys. J. 82, 2383 (2002). 10.1016/S0006-3495(02)75583-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A. J., Ducheyne P., and Boettiger D., Biomaterials 18, 1091 (1997). 10.1016/S0142-9612(97)00042-2 [DOI] [PubMed] [Google Scholar]
- Kapur R. and Rudolph A. S., Exp. Cell Res. 244, 275 (1998). 10.1006/excr.1998.4156 [DOI] [PubMed] [Google Scholar]
- Boura C., Muller S., Vautier D., Dumas D., Schaaf P., Claude Voegel J., Franois Stoltz J., and Menu P., Biomaterials 26, 4568 (2005). 10.1016/j.biomaterials.2004.11.036 [DOI] [PubMed] [Google Scholar]
- Young E. W. K., Wheeler A. R., and Simmons C. A., Lab Chip 7, 1759 (2007). 10.1039/b712486d [DOI] [PubMed] [Google Scholar]
- Lu H., Koo L. Y., Wang W. M., Lauffenburger D. A., Griffith L. G., and Jensen K. F., Anal. Chem. 76, 5257 (2004). 10.1021/ac049837t [DOI] [PubMed] [Google Scholar]
- Gutierrez E. and Groisman A., Anal. Chem. 79, 2249 (2007). 10.1021/ac061703n [DOI] [PubMed] [Google Scholar]
- Kwon K. W., Choi S. S., Lee S. H., Kim B., Lee S. N., Park M. C., Kim P., Hwang S. Y., and Suh K. Y., Lab Chip 7, 1461 (2007). 10.1039/b710054j [DOI] [PubMed] [Google Scholar]
- Cheung L. S. L., Zheng X., Stopa A., Baygents J. C., Guzman R., Schroeder J. A., Heimark R. L., and Zohar Y., Lab Chip 9, 1721 (2009). 10.1039/b822172c [DOI] [PubMed] [Google Scholar]
- Garrivier D., Décavé E., Bréchet Y., Bruckert F., and Fourcade B., Eur. Phys. J. E 8, 79 (2002). 10.1140/epje/i2002-10010-8 [DOI] [PubMed] [Google Scholar]
- Cozens-Roberts C., Lauffenburger D. A., and Quinn J. A., Biophys. J. 58, 841 (1990). 10.1016/S0006-3495(90)82430-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M. D., Dembo M., and Hammer D. A., Biophys. J. 67, 2522 (1994). 10.1016/S0006-3495(94)80742-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. A. and Calderwood D. A., Science 316, 1148 (2007). 10.1126/science.1137592 [DOI] [PubMed] [Google Scholar]
- Eichinger L., Pachebat J. A., Glöckner G., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q. et al. , Nature 435, 43 (2005). 10.1038/nature03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajchowski D. A., Bartholdi M. F., Gong Y., Webster L., Liu H.-l., Munishkin A., Beauheim C., Harvey S., Ethier S. P., and Johnson P. H., Gene Ther. 61, 5168 (2001). [PubMed] [Google Scholar]
- Zhao X.-M., Xia Y., and Whitesides G. M., J. Mater. Chem. 7, 1069 (1997). 10.1039/a700145b [DOI] [Google Scholar]
- Stephan K., Pittet P., Renaud L., Kleimann P., Morin P., Ouaini N., and Ferrigno R., J. Micromech. Microeng. 17, N69 (2007). 10.1088/0960-1317/17/10/N01 [DOI] [Google Scholar]
- Vidal L. J.-P., Perry J. K., Vouyovitch C. M., Pandey V., Brunet-Dunand S. E., Mertani H. C., Liu D.-X., and Lobie P. E., Mol. Cancer Res. 8, 444 (2010). 10.1158/1541-7786.MCR-09-0368 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3673802E-BIOMGB-6-002201 for more details on these points.
- Batchelor G. K., An Introduction to Fluid Dynamics (Cambridge University Press, United Kingdom, 2002). [Google Scholar]
- Koch A. L., J. Theor. Biol. 12, 276 (1966). 10.1016/0022-5193(66)90119-6 [DOI] [PubMed] [Google Scholar]
- Gervais T., El-Ali J., Günther A., and Jensen K. F., Lab Chip 6, 500 (2006). 10.1039/b513524a [DOI] [PubMed] [Google Scholar]
- Golé L., Rivière C., Hayakawa Y., and Rieu J.-P., PLoS ONE 6(11), e26901 (2011) 10.1371/journal.pone.0026901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M. D. and Hammer D. A., Biophys. J. 64, 936 (1993). 10.1016/S0006-3495(93)81456-5 [DOI] [PMC free article] [PubMed] [Google Scholar]