Abstract
Jak1/2 inhibition suppresses STAT3 phosphorylation that is characteristic of many cancers. Activated STAT3 promotes the transcription of factors that enhance tumor growth, survival, and angiogenesis. AZD1480 is a novel small molecule inhibitor of Jak1/2, which is a key mediator of STAT3 activation. This study examined the use of diffusion-weighted (DW) and dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) biomarkers in assessing early tumor response to AZD1480. Cediranib (AZD2171), a vascular endothelial growth factor signaling inhibitor, was used as a comparator. Thirty mice were injected with Calu-6 lung cancer cells and randomized into the three treatment groups: AZD1480, cediranib, and sham. DW-MRI and DCE-MRI protocols were performed at baseline and at days 3 and 5 after treatment. The percent change from baseline measurements for Ktrans, ADC, and ve were calculated and compared with hematoxylin and eosin (H&E), CD31, cParp, and Ki-67 histology data. Decreases in Ktrans of 29% (P < .05) and 53% (P < .05) were observed at days 3 and 5, respectively, for the cediranib group. No significant changes in Ktrans occurred for the AZD1480 group, but a significant increase in ADC was demonstrated at days 3 (63%, P < .05) and 5 (49%, P < .05). CD31 staining indicated diminished vasculature in the cediranib group, whereas significantly increased cParp staining for apoptotic activity and extracellular space by image analysis of H&E were present in the AZD1480 group. These imaging biomarker changes, and corresponding histopathology, support the use of ADC, but not Ktrans, as a pharmacodynamic biomarker of response to AZD1480 at these time points.
Introduction
Targeting signal transduction pathways has become a major strategy in the development of novel cancer therapies. Inhibition of these pathways can promote antiangiogenic, proapoptotic, and/or antiproliferative effects [1]. An example of this type of targeted therapy is a novel class of pharmaceuticals known as Jak inhibitors. The Janus family of kinases (Jak) is fundamental to mediating activation of signal transducers and activator of transcription (STAT) proteins. STAT proteins are responsible for mediating cytokine and growth factor responses. During the past two decades, it has been shown that persistent activation of STAT3 is found in a wide variety of human cancer cell lines [2] and that aberrant STAT3 activation is necessary for proliferation and survival of some cancer cell lines [3,4].
In addition to these tumor cell-autonomous effects, it has been reported that persistent activation of STAT3 directly targets vascular endothelial growth factor (VEGF) [5], one of the most potent angiogenesis-inducing signaling factors. STAT3 acts to direct transcriptional activation of VEGF, as well as having downstream effects that, when inhibited, may slow hypoxia-inducible factor 1 activation, which is another upregulator of VEGF [6].
A Jak1/2 small molecule inhibitor, AZD1480, has been shown to actively suppress the role of STAT3 in tumorigenesis. Hedvat et al. [7] demonstrated that AZD1480 significantly reduced STAT3 phosphorylation in multiple tumor xenograft models that exhibit aberrant, persistent STAT3 activation, and inhibited tumor xenograft growth.
In recent years, there has been considerable growth in the number and capabilities of noninvasive imaging biomarkers of tumor microenvironment and response to therapy. Diffusion-weighted magnetic resonance imaging (DW-MRI) provides the ADC, which has been shown to correlate with biophysical properties such as cell density, whereas dynamic contrast-enhanced MRI (DCE-MRI) provides Ktrans, a biomarker of tumor perfusion and microvascular permeability [8,9]. DCE-MRI also provides ve, a biomarker of the extravascular extracellular space, although changes in this biomarker can be difficult to interpret because the measurement is not always robust.
The microscopic thermally induced behavior of molecules moving in a random pattern is referred to as self-diffusion or Brownian motion [10]. In a system that is defined by small compartments, such as cells, that are separated by semipermeable barriers, such as cell membranes, the rate of self-diffusion is less than that of free diffusion and can be described by an apparent diffusion coefficient (ADC), which depends on the properties and spatial relationship between these barriers [11–13], which act to restrict the free diffusion of water molecules. Techniques such as a pulsed-gradient spin-echo (PGSE) sequence, which are sensitive to this diffusion and the effects of restrictions on water movements at spatial scales up to the order of several microns, have been designed; this spatial scale is on the order of the diameter of a cell and therefore can be used as an estimate of cellularity. In well-controlled situations, the variations in ADC have been shown to correlate inversely with tissue cellularity [14] and have been used to monitor treatment response in a variety of anticancer therapies, including both cytotoxic and antivascular/antiangiogenic drugs [15–17].
A second technique that has been shown to be sensitive to changes in physiological characteristics of tumor vasculature is DCE-MRI [8,18], which characterizes the pharmacokinetics of an injected contrast agent as it enters and exits a region of interest (ROI) or tissue of interest. By acquiring T1-weighted images of the MRI signal over time, the kinetics of the CA can be quantified, and using a model to describe its distribution, biologically relevant parameters can be extracted that represent changes in, for example, blood flow, permeability, and tissue volume fractions. These measurements have been used to diagnose cancer and monitor treatment [8].
Two important processes regulated by STAT3 are cellular proliferation and tumor angiogenesis, so we investigated the value of DW-MRI and DCE-MRI biomarkers for assessing the response to AZD1480, a novel Jak1/2 inhibitor, compared with a potent antiangiogenic agent, cediranib [19], at early treatment time points.
Materials and Methods
Tumor Model
Calu-6 human lung carcinoma cells were grown as described by Wedge et al. [20]. Cediranib [19] and AZD1480 (unpublished) have been shown to inhibit tumor growth in this cell line. Briefly, the cells were grown in medium containing 10% fetal calf serum and 2 mM l-glutamine in Eagle minimal essential medium with 1% sodium pyruvate (100 mM) and 1% nonessential amino acids (Life Technologies, Inc, Grand Island, NY). Cells growing at 80% confluence were harvested, and a single cell suspension containing 1 x 106 cells suspended in 100 µl of medium was injected subcutaneously on the flank. Tumors were grown for 15 to 20 days to reach a size of approximately 200 to 250 mm3 before imaging.
Animal Model
Thirty adult female fox nu/nu (8–10 weeks of age) were purchased from Charles River Laboratories (Raleigh, NC), housed in pathogen-free facilities with a 12-hour light/dark cycle (6 a.m. to 6 p.m.), and provided with rodent chow and tap water ad libitum. For DCE-MRI studies, a 26-gauge jugular catheter was surgically implanted to allow for delivery of the contrast agent. All animals were imaged at three time points: a baseline scan was acquired before any treatment and then two subsequent sessions at days 3 and 5 after the initiation of treatment. The mice were then humanely killed for histologic analysis of tumor tissues. During all imaging procedures, the mice were anesthetized using a 2%/98% isoflurane/oxygen mixture. Body temperature was maintained through a flow of warm air through the magnet bore, and temperature and respiratory rate were monitored throughout the entire experiment. All procedures in the experiment were reviewed and approved by our institutional animal care and use committee.
Treatment
The vehicle for AZD1480 was prepared using 0.1 ml of Tween-80 (Sigma-Aldrich, St Louis, MO), 0.5 g of methylcellulose powder, and 100 ml of distilled water stirred overnight. Vehicle was added to a dose of 50 mg/kg of AZD1480 such that the administered volume was 10 ml/kg, and pH was adjusted to 2.0. The vehicle for cediranib was prepared using 0.1 ml of Tween-80 added to 100 ml of distilled water. Vehicle was added to a dose of 6 mg/kg such that the administered volume was 10 ml/kg. All drugs and vehicles were stored at 4°C. Thirty mice were randomly assigned to one of the following three treatment groups once tumor volumes were approximately 200 mm3: 50 mg/kg AZD1480 per os daily for 6 days, 6 mg/kg, cediranib per os daily for 6 days, and vehicle control per os daily for 6 days.
Data Acquisition
Diffusion-Weighted MRI. Animals were imaged on a 9.4-T MR scanner (Varian, Palo Alto, CA) with a 38-mm quadrature coil (Varian) at all three time points. Because of technical issues, three animals had day 5 data and four animals had day 3 data acquired on a 7-T MR scanner (Varian) using identical protocols. A gradient echo scout sequence was used to locate the tumor tissue. Once the ROI was located, 15 slices were imaged (1 mm thick, interleaved) with diffusion weighting using a gated and navigated PGSE sequence. DW-MRI parameters were as follows: TR\TE\α = 2000 milliseconds\42 milliseconds\15°, acquisition matrix = 1282, field of view (FOV) = 35 mm2, and number of excitations (NEX) = 2, with Δ = 35.00 milliseconds and δ = 5.00 milliseconds, giving b values of 150.88, 500.2, and 800.22 mm2/s.
Dynamic Contrast-Enhanced MRI. After DW-MRI, precontrast T1 maps were obtained using an inversion recovery snapshot fast low-angle-shot gradient echo sequence with an adiabatic inversion pulse and nine inversion times (TIs) ranging from 20 to 10,000 milliseconds. Imaging parameters were as follows: TR\TE\α = 12100 milliseconds\3.44 milliseconds\15° and NEX = 4, FOV = 35 mm2, and matrix = 1282. The data were then fit to to obtain T1 maps for all 15 slices of data. The DCE-MRI protocol used a T1-weighted, gradient echo sequence to obtain 40 serial images for each of 15 axial planes in 18 minutes of imaging. The parameters were as follows: TR\TE\α = 100 milliseconds\ 2.83 milliseconds\25°, NEX = 2, with the same acquisition matrix and FOV as for the T1 map. A bolus of 0.1 mmol/kg gadopentetate (Magnevist; Bayer Healthcare Pharmaceuticals, Inc, Wayne, NJ) was delivered for 2.5 seconds with an automated Harvard pump (2.4 ml/min) through a jugular catheter beginning after the acquisition of the fifth dynamic image.
Data Analysis
Diffusion-Weighted MRI. For the PGSE sequence used, the signal (S) will change because of the diffusion and can be described as
| [1] |
where b is diffusion weighting imparted by magnetic field gradients and S0 is the signal without diffusion weighting. Changing the magnitude or duration of the diffusion gradients, thus changing the b values, yields multiple image sets with varying diffusion weighting. The data can then be exponentially fit to acquire ADC values on a voxel-by-voxel basis. The DW images acquired at three separate b values, 150.88, 500.2, and 800.22 mm2/s, were fit to to extract the ADC values at every ROI and voxel location.
Dynamic Contrast-Enhanced MRI. Data collected for the T1 map were fit using a nonlinear least squares method in Matlab 2008a (The MathWorks, Natick, MA). For each inversion pulse, eight lines of k space were acquired; these eight lines account for a single effective TI (TIeff), with the TIeff corresponding to the timing of the first collected line. A centric phase encode table was used to reconstruct the data, and no cardiac gating was used during the acquisition. Under the assumption that TR T1 and a sufficiently low flip angle, the signal at each acquisition can be described in the following equation:
| [2] |
where m1 is a free parameter to account for a partial inversion and S0 and S are the signal intensities at equilibrium at the effective inversion time (TIeff), respectively.
Once precontrast T1 maps were calculated, tumor signal curves were fit to a two-compartment model to extract pharmacokinetic parameters [21]. Briefly, if we assume that a homogeneous distribution of CA exists in both compartments and there exists no back flux to either compartment, then any of the previously mentioned cases can be generalized into a linear, first-order ordinary differential equation describing CA kinetics into the tissue
| [3] |
where Ct is the concentration in the tissue compartment, Cp is the concentration in the plasma compartment, whereas Ktrans is the transfer constant between Cp and Ct and ve is the extravascular extracellular volume fraction. The solution to is
| [4] |
The signal time course from the tissue for each voxel was fit to using a nonlinear least squares approach by incorporating 1) the initial T1 values [22], 2) the fast exchange limit model with relaxivity for gadopentetate at 9.4 T = 4.8/mM/s (unpublished results) and a hematocrit value of 0.45, based on literature values [23], and 3) a population-derived vascular input function or Cp(t) derived from a cohort of 10 fox nu/nu mice found using the identical protocol described Loveless et al. [24].
Histology
On day 6 (1 day after the final imaging session), animals were dosed and killed 2 hours later using CO2. Although the histologic samples were collected 1 day after imaging, it was important to mimic the imaging timeline; this ensured that the histology and molecular markers would reflect 2 hours after dosing just as when imaging was performed. The tumor tissue was excised, placed into tissue cassettes, fixed in 10% formalin (Fisher, Pittsburgh, PA) for 24 to 48 hours, and stored in 70% ethanol. Samples were paraffin embedded and sectioned at 5-µm thickness. Immunohistochemistry was performed on the Ventana Discovery XT Autostainer. Samples were stained for phosphorylated STAT3 (pSTAT3, CST9145; Cell Signaling Technology, Danvers, MA), Ki-67 (180191Z; Invitrogen, Grand Island, NY), cleaved Parp1 (cParp, 04-576; Millipore, Billerica, MA), CD31 (sc1506; Santa Cruz Biotechnology, Santa Cruz, CA), and with hematoxylin and eosin (H&E; Ventana, Tucson, AZ).
Digital slide images were acquired at 20x magnification using the Aperio ScanScope (Vista, CA). From the CD31-stained slides, microvessel density was quantitated using the Aperio (Vista, CA) Microvessel Analysis software; briefly, the software was tuned to detect and join regions of endothelial staining. The number of vessels detected was divided by the total area of viable tumor to determine the microvessel density.
Cleaved Parp staining was measured on areas of viable tumor using the Aperio Color Deconvolution Algorithm, which quantitates pixels sufficiently stained with chromogen to be counted positive. Ki-67 expression was quantitated on areas of viable tumor using the Aperio Nuclear Algorithm, which counts nuclei based on counterstain and then calculates how many are sufficiently stained with chromogen to be counted positive.
In addition, the Aperio Color Deconvolution Algorithm was used to generate maps of intracellular and extracellular regions based on intensity thresholding on H&E-stained sections. A region was drawn to encompass the entire slice of tumor (AROI), and every pixel was analyzed to determine whether it contained enough staining to be counted. Pixels that did not reach the minimum threshold were considered nonstained (i.e., extracellular regions). Statistics summarizing the thresholding that include total region area (AROI) and total stained area (Astain) were used to determine the percentage of extracellular (EC) space as shown below:
| [5] |
Statistical Analysis
To assess group changes in Ktrans and ve, an ROI was manually drawn on the center slice for each tumor at each time point. The average signal intensity was then fit to to obtain parametric values for each animal at baseline, at day 3, and at day 5. The same ROIs were applied to the DW data and fit to as described previously. Data analysis assumed that noise contributed only random and not systematic errors. Before being placed into the appropriate treatment group for analysis, the integrity of the data was examined by using either muscle tissue (DCE-MRI) or water phantom data (DW-MRI). Briefly, if the muscle curve did not qualitatively follow characteristics of a rapid wash-in and wash-out of CA or if physiologically implausible parameters were returned, the injection was deemed unsuccessful and the DCE-MRI data were not used. Similarly, if the slice used in the DW-MRI comparison had a phantom ADC value outside the range of 0.003 mm2/s ± 15% (experimentally derived ADC of free water at approximately body temperature (37 ± 2°C) at 9.4 T) then the tumor ADC data were also not used. For each treatment group, the average and 95% confidence intervals (1.96 x SD / √n, where n is the number of samples) were computed. The percent change from baseline for each animal was also computed, and the average and 95% confidence interval were calculated. A Wilcoxon signed rank test was used to determine significant differences between each treatment group (both absolute and percent change data) [25]. Histologic data were quantified using pathologic scoring and compared with all imaging metrics.
Results
In total, n = 12/n = 10/n = 9 mice were imaged at three time points for the cediranib/AZD1480/sham groups, respectively. Animal data that exhibited a faulty injection (no contrast uptake) or an occluded catheter were not included in the DCE-MRI group analyses; similarly, animal data collected for the ADC study that were contaminated by motion (as reflected by the water phantom control measurement) were not included in further analyses. The number of mice used in analyses at each time point is indicated in Tables 1 to 3. The tumor volume changes for all animals are shown in Figure 1, where the square/solid line indicates animals treated with cediranib, the circle/dashed line indicates animals treated with AZD1480, and the triangle/dashed line represents animals in the sham group. The vertical lines are the 95% confidence interval for each respective group. There was no statistically significant difference in tumor volume among these groups at the measured time points, which is not unexpected given the relatively short, 5-day duration of drug treatment.
Table 1.
Average Absolute Ktrans Values for Each Treatment Group.
| Treatment Group | Baseline (/min) | Day 3 (/min) | Day 5 (/min) |
| AZD1480 | 0.06 ± 0.03 (n = 8) | 0.05 ± 0.05 (n = 8) | 0.04 ± 0.01 (n = 5) |
| Cediranib | 0.06 ± 0.02 (n = 7) | 0.04 ± 0.02 (n = 7) | 0.03 ± 0.02 (n = 6) |
| Sham | 0.07 ± 0.03 (n = 6) | 0.05 ± 0.02 (n = 5) | 0.04 ± 0.02 (n = 6) |
Values are presented as mean ± SD.
Table 3.
Average Absolute ve Values for Each Treatment Group.
| Treatment Group | Baseline (/min) | Day 3 (/min) | Day 5 (/min) |
| AZD1480 | 0.50 ± 0.27 (n = 8) | 0.58 ± 0.21 (n = 8) | 0.49 ± 0.29 (n = 5) |
| Cediranib | 0.41 ± 0.15 (n = 7) | 0.37 ± 0.14 (n = 7) | 0.24 ± 0.07 (n = 6) |
| Sham | 0.33 ± 0.12 (n = 6) | 0.39 ± 0.15 (n = 5) | 0.36 ± 0.10 (n = 6) |
Values are presented as mean ± SD.
Figure 1.
Volumetric data for all animals used in each treatment group. The solid square/solid line represents the cediranib group, whereas the open circle/dashed line represents the AZD1480. The sham group is represented by the triangle/dashed line with the vertical lines indicating the 95% confidence intervals for each time point. There is no significant difference between any of the groups at any time point (ncediranib = 12/nAZD1480 = 10/nsham = 9).
Group Analyses
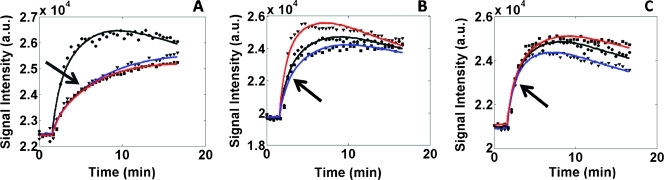
Three representative DCE-MRI curves (representing each time point) from a mouse in each treatment group are shown in Figure 2. The dynamic data and curve fit (in black) were taken from baseline, whereas the red and blue fit lines indicate days 3 and 5 data, respectively. Figure 2A shows the dramatic effect of cediranib on Ktrans (denoted by the arrow), whereas Figure 2, B and C, shows that Ktrans changes very little or without trend (as denoted by the arrows) in the AZD1480 and sham, respectively.
Figure 2.
Example DCE-MRI curves from each treatment group. (A) Subject from the cediranib treatment group, with the black line indicating the baseline, the red line indicating day 3 data, and the blue line representing day 5 data. As noted by the arrow, there is dramatic decrease in Ktrans for the cediranib group. (B) ROI curves from the AZD1480 group, with the arrow showing that no specific pattern in Ktrans can be detected. (C) Sham group showing little detectable change in Ktrans values.
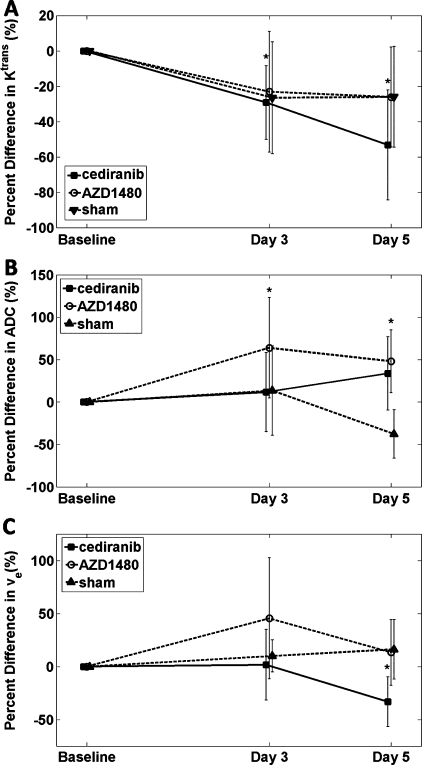
The results of the percent change in the group analyses are displayed in Figure 3. Animals treated with the VEGFR inhibitor, cediranib, showed significant decreases in Ktrans at both days 3 and 5 with an average percent difference of -29% and -53%, respectively (Figure 3A). The average percent differences in the AZD1480 treatment group were -21% and -23% for each posttreatment time points, whereas the sham group showed stable decreases of 26% at both subsequent time points. No significant differences were found in either AZD1480 or sham groups at either posttreatment time points for Ktrans. Average absolute Ktrans values for these treatment groups are listed in Table 1.
Figure 3.
(A, B, and C) Percent change from baseline for Ktrans, ADC, and ve, respectively, from a center slice ROI where cediranib (▪), AZD1480 (○), and Sham (▴), whereas the 95% confidence intervals are represented by the vertical bars. *P < .05. Statistical significance was found for Ktrans in the cediranib treatment group, whereas AZD1480 days 3 and 5 data showed significant increases in ADC. A significant decrease was found for ve in the cediranib treatment group. See text for additional details.
In Figure 3B, the percent change in ADC shows that both treatment groups demonstrate an increasing trend with a significant increase for the AZD1480 group at both days 3 (63%) and 5 (49%) after treatment. Whereas the cediranib group shows an increase on day 5 for ADC, only three animals were available at this final time point so this subgroup lacks power to achieve statistical significance. Also of note, the sham group trended negatively, suggesting that the ADC decreased as the tumor grew. Mean absolute ADC values for these treatment groups are listed in Table 2.
Table 2.
Average Absolute ADC Values for Each Treatment Group.
| Treatment Group | Baseline (x 10-4 mm2/s) | Day 3 (x 10-4 mm2/s) | Day 5 (x 10-4 mm2/s) |
| AZD1480 | 6.00 ± 1.59 (n = 9) | 7.33 ± 0.98 (n = 4) | 8.59 ± 3.02 (n = 8) |
| Cediranib | 6.51 ± 1.48 (n = 5) | 6.86 ± 2.67 (n = 5) | 8.60 ± 1.43 (n = 3) |
| Sham | 6.81 ± 1.81 (n = 7) | 6.53 ± 2.19 (n = 5) | 5.00 ± 1.60 (n = 3) |
Values are presented as mean ± SD.
In Figure 3C, the group changes for ve are given. No significant changes from baseline in ve were seen with the exception of day 5 of cediranib treatment, which reflected a significant decrease at this time point. The absolute values of ve for each treatment group are listed in Table 3.
Parametric Map Analyses
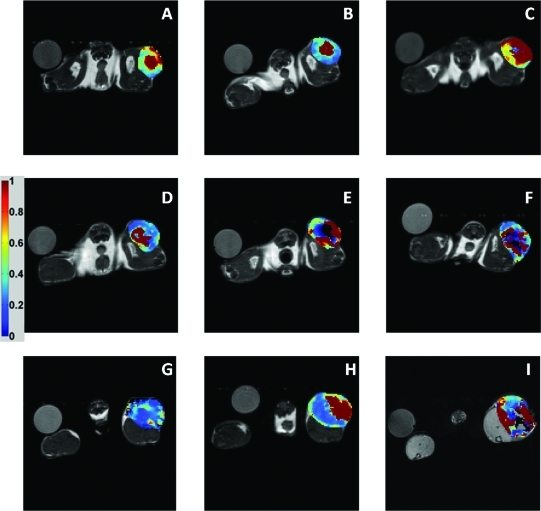
To further explore the efficacy of Ktrans, ADC, and ve to report on these treated animals, parametric maps were constructed. Figure 4 illustrates a panel of Ktrans maps for each treatment group (rows: cediranib, AZD1480, and sham) at each time point (columns: baseline, day 3, and day 5). These maps illustrate a significant decrease in Ktrans values for the cediranib treatment group, shown by the arrow (Figure 4, A, B, and C), whereas little change or trend is noted in the AZD1480 group (Figure 4, D, E, and F). Interestingly, in this sham animal, higher Ktrans values are found at the day 5 time point, as shown by the arrow (Figure 4, G, H, and I).
Figure 4.
Ktrans parametric maps for representative mice from each treatment group. The columns indicate baseline, day 3, and day 5 time points, whereas each row shows the cediranib (A–C), AZD1480 (D–F), and sham groups (G–I).
ADC maps were also constructed and displayed in Figure 5. In a similar fashion, one representative animal from each treatment group is displayed in the rows (cediranib, AZD1480, and sham groups), whereas each column represents the imaging time points (baseline, day 3, and day 5). The ADC maps do not show a dramatic difference in this particular cediranib-treated animal (Figure 5, A, B, and C), whereas there is an apparent increase in ADC values for this AZD1480-treated animal (Figure 5, D, E, and F). The sham group (Figure 5, G, H, and I) shows a decrease in ADC values that reflect the group analyses mentioned previously.
Figure 5.
ADC parametric maps for a representative mouse from each treatment group. The columns indicate baseline, day 3, and day 5 time points, whereas each row shows the cediranib (A–C), AZD1480 (D–F), and sham groups (G–I).
The final series of panels, shown in Figure 6, displays ve data from representative animals from the treatment groups in a similar fashion. No trends or correlations are evident with this parameter. Interestingly, this parameter maintains very high values at all time points for all treatments. Some voxels surpassed the physiological boundary (ve = 1), which were excluded from analyses.
Figure 6.
ve parametric maps for a representative mouse from each treatment group. The columns indicate baseline, day 3, and day 5 time points, whereas each row shows the cediranib (A–C), AZD1480 (D–F), and sham groups (G–I).
Histologic Correlation
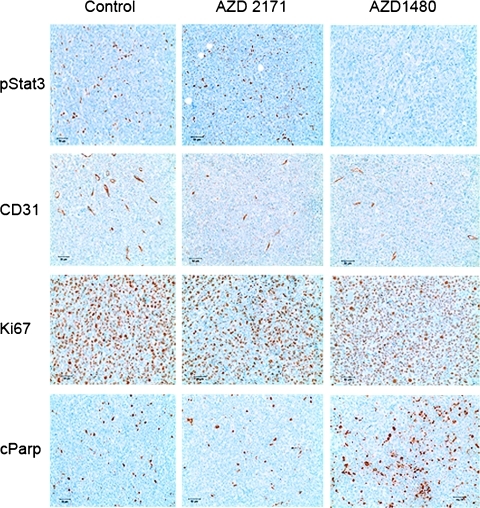
Because Jak kinase is a key regulator of STAT3 phosphorylation, tumor samples were stained for pSTAT3 2 hours after drug or sham dosing. As expected, treatment with the VEGFR inhibitor cediranib showed no effect on pSTAT3 relative to shams (staining detected in seven of eight and in six of seven tumors, respectively), whereas pSTAT3 was significantly inhibited after treatment with AZD1480 (staining in 2/11 tumors). Representative images are shown in Figure 9. To histologically assess vascularity, samples were stained for the endothelial cell marker CD31. As shown in Figure 7, no significant changes in microvessel density could be detected by CD31 staining in these samples owing to the large variation within the sham group. However, a trend toward reduction of microvessel density could be discerned in AZD1480-treated samples and more markedly in cediranib-treated samples.
Figure 9.
Sample staining for pSTAT3 (first row), CD31 (second row), Ki-67 (third row), and cParp (last row) for sham group (first column), the cediranib treatment group (second column), and the AZD1480 treatment group (last column). There is clear evidence that pSTAT3 in AZD1480 is suppressed in treated animals (no brown staining). The brown staining shown in the CD31 staining indicates a positive stain for CD31 on endothelial cells, whereas a brown staining in the Ki-67 images indicates cells that are in a proliferative state. The brown staining for the cParp images indicate cells positive for the Parp cleavage typically characterized by cells undergoing apoptosis.
Figure 7.
Mean microvessel density calculated from CD31 staining in each treatment group. Bars, SD.
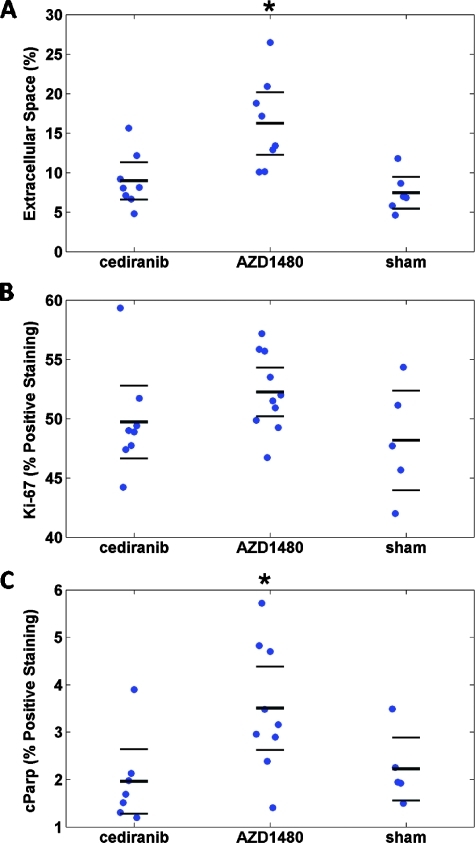
Additional immunohistochemistry analyses for cellular activity included H&E, Ki-67, and cParp staining; the results of these analyses are shown in Figure 8. Figure 8A demonstrated the extracellular space fraction (EC (%)) on H&E-stained slides, calculated as described in the Materials and Methods section, for each group. A significant (P < .05) increase in EC (%) was found between the AZD1480 treatment group and the sham group, whereas the cediranib-treated group was indistinguishable from the sham group. Ki-67 was used to identify the percentage of cells that were proliferating at the last imaging time point; the results for all three groups are shown in Figure 8B, and no statistical significance was reported between treatment and sham groups. Finally, apoptotic activity was quantified as the percentage of cells that stained positive for cParp, a marker of apoptosis. The results of this study are shown in Figure 8C with a statistically significant (P = .036) increase in cParp staining for the AZD1480 treatment group compared with the sham group. Examples of staining for CD31, Ki-67, and cParp are shown in Figure 9.
Figure 8.
(A, B, and C) Cell density, Ki-67, and cParp staining results, respectively, for each group. Each “●” represents a sample with the bold solid line being the mean and the error bars above and below indicate the 95% confidence interval. (A) Cell density results as determined by H&E staining. (B) Percentage of nuclei that stained positive for Ki-67 activity. (C) Percentage of cParp-positive pixels.
Discussion
STAT3 has been shown to promote tumor development through the activation of several oncogenic pathways, including cell proliferation, survival, and tumor angiogenesis. Jak kinase has been shown to play a central role in STAT3 activation in solid tumor cell lines [7,26–30], and inhibition of Jak1/2 with AZD1480 has been demonstrated to abrogate STAT3 phosphorylation and inhibit tumor xenograft growth [7]. One of the unique advantages of imaging tumor physiology is the ability to capture changes before any palpable/volumetric changes in tumor growth. Thus, the goal of this study was to examine the sensitivity of two imaging techniques to the antiangiogenic and antitumorigenic activity of AZD1480.
In this study, no significant change in Ktrans was noted in the AZD1480 group at any postdosing time points. However, there was a significant decrease in Ktrans for mice treated with the VEGFR inhibitor cediranib. Histologic analysis of tumor tissue stained with the endothelial cell marker CD31 did not reveal any significant changes in tumors treated with cediranib and AZD1480 relative to shams; however, significant variability in sham staining in these studies may have obscured treatment responses. Significant inhibition of vascularity has been detected in this xenograft tumor model after 6 days of treatment with AZD1480 in another study [31]. Thus, Ktrans does not seem to be sensitive enough to detect the changes in microvasculature for AZD1480-treated tumors after 5 days of treatment; alternatively, a reduced number of animals for the AZD1480 group on day 5 (n = 5) may have been too small to definitely detect a change in Ktrans from baseline measurements. ADC provided a more sensitive measure for the response to AZD1480 treatment at early time points. Significant increases in ADC were indicated in the group study and shifts were noted on a voxel-by-voxel level. In addition, although changes in tumor volume gave no significant indications of efficacy for this group, there was a significant increase in apoptosis and extracellular space (EC (%)) in this treatment group.
DW-MRI has previously been shown to correlate with cell density measures in cancer [32–35]. In this study, significant difference was found in extracellular space (indirectly identifying cell density) measures from H&E in the AZD1480 group; however, ve did not illustrate any significant trends other than demonstrating a significant decrease on day 5 for the cediranib-treated group. As cells die because of treatment, ve should, in theory, increase; however, previous studies have shown that ve has not consistently exhibited this behavior and is dependent on treatment as well as the DCE-MRI model used [17,36]. In some cases, ve reached an unphysical value (ve > 1); these voxels were excluded from further statistical analyses. The sources of error in ve could have arisen from slowly enhancing voxels in poorly perfused areas that may rely on passive diffusion for transportation of the contrast agent. In addution, the use of a population VIF can lead to parameter errors due to overestimating or underestimating the Cp in individual subjects. Recognizing this as a limitation of the study, we eliminated those voxels as including them would have skewed both ve and Ktrans measurements. With a significant increase in cleaved Parp, indicative of caspase 3-dependent apoptosis [30], and EC (%) and no difference in Ki-67 (proliferation marker), one could hypothesize that the changes in ADC are primarily induced by proapoptotic activity. As cells die, the extracellular space increases so the ADC increases.
ADC can be affected by several cellular/extracellular processes such as changes in active transport, blood flow, and tissue organization [9]. These changes can also be transient with treatment activity; for example, initial cellular swelling and reduced blood flow may lead to lower ADC values earlier in treatment [37]. As cells begin to die, edema, reduced cell density, and tissue reorganization may lead to an increase in ADC, as seen frequently during treatment of malignant tumors [9]. Thus, it is possible that the change in ADC reflects a change in tissue environment as apoptosis increases. Whereas no significant changes in ADC were noted in the cediranib group (n = 3), the ADC at day 5 demonstrated an increasing trend as did the percent extracellular space (EC (%)).
Conclusions
The goal of this study was to assess the sensitivity of DCE-MRI and DW-MRI biomarkers to the antiangiogenic and antitumorigenic activity of AZD1480. As anticipated, Ktrans, an indicator of blood flow/perfusion reported significant changes from baseline for the antiangiogenic drug, cediranib, treatment group; this finding was also supported by histology. Ktrans did not reliably report any treatment response for AZD1480, although histologic analysis showed a general decrease in microvessel density, but significant changes in ADC were measured at both days 3 and 5 after treatment, consistent with pathologic detection of significant apoptotic activity and increased extracellular fraction. Measurements of diffusion may provide a useful biomarker for early treatment response to a Jak1/2 inhibitor.
Acknowledgments
The authors thank Jeffrey Luci, Daniel Colvin, Zou Xu, Jarrod True, and the Mouse Metabolic Phenotyping Core for assistance with technical MRI and animal care.
Abbreviations
- ADC
apparent diffusion coefficient
- FOV
field of view
- Jak
Janus family of kinases
- DW-MRI
diffusion-weighted magnetic resonance imaging
- DCE-MRI
dynamic contrast-enhanced magnetic resonance imaging
- PGSE
pulsed-gradient spin-echo
- ROI
region of interest
- STAT
signal transducers and activator of transcription
- TI
inversion time
- TR
repetition time
- VEGF
vascular endothelial growth factor
Footnotes
The authors thank the National Institutes of Health through funding mechanisms NCI U24 CA126588 (Small Animal Imaging Resource Program), NCI R01 CA138599, and NCI P30 CA068485 (Cancer Center Support Grant). M.E.L. is supported on an unrestricted AstraZeneca predoctoral training grant. T.E.Y. was (partially) supported through National Institute of Biomedical Imaging and Bioengineering grant 1K25 EB005936.
References
- 1.Dy GK, Adjei AA. Systemic cancer therapy: evolution over the last 60 years. Cancer. 2008;113:1857–1887. doi: 10.1002/cncr.23651. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 3.Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, Irby R, Yeatman T, Courtneidge SA, Jove R. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci USA. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 5.Niu G, Bowman T, Huang M, Shivers S, Reintgen D, Daud A, Chang A, Kraker A, Jove R, Yu H. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene. 2002;21:7001–7010. doi: 10.1038/sj.onc.1205859. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 7.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yankeelov TE, Gore JC. Contrast enhanced magnetic resonance imaging in oncology: data acquisition, analysis, and examples. Curr Med Imaging Rev. 2007;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11:102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Einstein A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann Phys. 1905;322:549. (Ger). [Google Scholar]
- 11.Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time dependent field gradient. J Chem Phys. 1965;42:288–292. [Google Scholar]
- 12.Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology. 1988;168:497–505. doi: 10.1148/radiology.168.2.3393671. [DOI] [PubMed] [Google Scholar]
- 13.Chenevert TL, Brunberg JA, Pipe JG. Anisotropic diffusion in human white matter: demonstration with MR techniques in vivo. Radiology. 1990;177:401–405. doi: 10.1148/radiology.177.2.2217776. [DOI] [PubMed] [Google Scholar]
- 14.Anderson AW, Xie J, Pizzonia J, Bronen RA, Spencer DD, Gore JC. Effects of cell volume fraction changes on apparent diffusion in human cells. Magn Reson Imaging. 2000;18:689–695. doi: 10.1016/s0730-725x(00)00147-8. [DOI] [PubMed] [Google Scholar]
- 15.Patterson DM, Padhani AR, Collins DJ. Technology insight: water diffusion MRI—a potential new biomarker of response to cancer therapy. Nat Clin Pract Oncol. 2008;5:220–233. doi: 10.1038/ncponc1073. [DOI] [PubMed] [Google Scholar]
- 16.Thoeny HC, De Keyzer F. Extracranial applications of diffusion-weighted magnetic resonance imaging. Eur Radiol. 2007;17:1385–1393. doi: 10.1007/s00330-006-0547-0. [DOI] [PubMed] [Google Scholar]
- 17.Yankeelov TE, Lepage M, Chakravarthy A, Broome EE, Niermann KJ, Kelley MC, Meszoely I, Mayer IA, Herman CR, McManus K, et al. Integration of quantitative DCE-MRI and ADC mapping to monitor treatment response in human breast cancer: initial results. Magn Reson Imaging. 2007;25:1–13. doi: 10.1016/j.mri.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 19.Wedge SR, Kendrew J, Hennequin LF, Valentine PJ, Barry ST, Brave SR, Smith NR, James NH, Dukes M, Curwen JO, et al. AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res. 2005;65:4389–4400. doi: 10.1158/0008-5472.CAN-04-4409. [DOI] [PubMed] [Google Scholar]
- 20.Wedge SR, Ogilvie DJ, Dukes M, Kendrew J, Chester R, Jackson JA, Boffey SJ, Valentine PJ, Curwen JO, Musgrove HL, et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002;62:4645–4655. [PubMed] [Google Scholar]
- 21.Kety SS. Peripheral blood flow measurements. Pharmacol Rev. 1951;3:1–41. [PubMed] [Google Scholar]
- 22.Landis CS, Li X, Telang FW, Coderre JA, Micca PL, Rooney WD, Latour LL, Vetek G, Palyka I, Springer CS., Jr Determination of the MRI contrast agent concentration time course in vivo following bolus injection: effect of equilibrium transcytolemmal water exchange. Magn Reson Med. 2000;44:563–574. doi: 10.1002/1522-2594(200010)44:4<563::aid-mrm10>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 23.Trune DR, Kempton JB, Gross ND. Mineralocorticoid receptor mediates glucocorticoid treatment effects in the autoimmune mouse ear. Hear Res. 2006;212:22–32. doi: 10.1016/j.heares.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Loveless ME, Halliday J, Liess C, Xu L, Dortch RD, Whisenant J, Waterton JC, Gore JC, Yankeelov TE. A quantitative comparison of the influence of individual versus population-derived vascular input functions on dynamic contrast enhanced-MRI in small animals. Magn Reson Med. 2012;67(1):226–236. doi: 10.1002/mrm.22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull. 1945;1:80. [Google Scholar]
- 26.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song L, Rawal B, Nemeth JA, Haura EB. JAK1 activates STAT3 activity in non-small-cell lung cancer cells and IL-6 neutralizing antibodies can suppress JAK1-STAT3 signaling. Mol Cancer Ther. 2011;10:481–494. doi: 10.1158/1535-7163.MCT-10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreis S, Munz GA, Haan S, Heinrich PC, Behrmann I. Cell density dependent increase of constitutive signal transducers and activators of transcription 3 activity in melanoma cells is mediated by Janus kinases. Mol Cancer Res. 2007;5:1331–1341. doi: 10.1158/1541-7786.MCR-07-0317. [DOI] [PubMed] [Google Scholar]
- 30.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–2956. [PubMed] [Google Scholar]
- 31.Xin H, Herrmann A, Reckamp K, Zhang W, Pal S, Hedvat M, Zhang C, Liang W, Scuto A, Weng S, et al. Anti-angiogenic and anti-metastatic activity of JAK inhibitor AZD1480. Cancer Res. 2011;71:6601–6610. doi: 10.1158/0008-5472.CAN-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugahara T, Korogi Y, Kochi M, Ikushima I, Shigematu Y, Hirai T, Okuda T, Liang L, Ge Y, Komohara Y, et al. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging. 1999;9:53–60. doi: 10.1002/(sici)1522-2586(199901)9:1<53::aid-jmri7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Hayashida Y, Yakushiji T, Awai K, Katahira K, Nakayama Y, Shimomura O, Kitajima M, Hirai T, Yamashita Y, Mizuta H. Monitoring therapeutic responses of primary bone tumors by diffusion-weighted image: initial results. Eur Radiol. 2006;16:2637–2643. doi: 10.1007/s00330-006-0342-y. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y, Cai YQ, Cai ZL, Gao YG, An NY, Ma L, Mahankali S, Gao JH. Differentiation of clinically benign and malignant breast lesions using diffusion-weighted imaging. J Magn Reson Imaging. 2002;16:172–178. doi: 10.1002/jmri.10140. [DOI] [PubMed] [Google Scholar]
- 35.Squillaci E, Manenti G, Cova M, Di Roma M, Miano R, Palmieri G, Simonetti G. Correlation of diffusion-weighted MR imaging with cellularity of renal tumours. Anticancer Res. 2004;24:4175–4179. [PubMed] [Google Scholar]
- 36.Pellerin M, Yankeelov TE, Lepage M. Incorporating contrast agent diffusion into the analysis of DCE-MRI data. Magn Reson Med. 2007;58:1124–1134. doi: 10.1002/mrm.21400. [DOI] [PubMed] [Google Scholar]
- 37.Thoeny HC, De Keyzer F, Chen F, Vandecaveye V, Verbeken EK, Ahmed B, Sun X, Ni Y, Bosmans H, Hermans R, et al. Diffusion-weighted magnetic resonance imaging allows noninvasive in vivo monitoring of the effects of combretastatin a-4 phosphate after repeated administration. Neoplasia. 2005;7:779–787. doi: 10.1593/neo.04748. [DOI] [PMC free article] [PubMed] [Google Scholar]