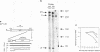Abstract
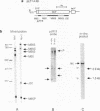
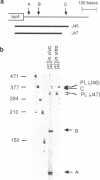
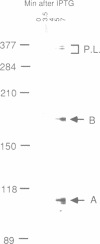
We have analyzed the processing of mRNA from the lac operon in an Escherichia coli strain carrying the lac on a multicopy plasmid. Messenger RNA was analyzed by hybridization and nuclease protection of pulse-labeled RNA and precursor-product relationships were determined by quantitating radioactivity in primary and processed transcripts at various times after induction of the lac promoter or inhibition of transcription with rifampicin. Our results support the existence of two types of processed transcripts with endpoints in the lacZ-lacY intercistronic region. One of these carries lacZ sequences and has a 3' endpoint about 30 bases downstream of this gene. The other carries lacY sequences and has a 5' end in the translation termination region of the lacZ gene. Finally, we have found evidence that transcription is continued at least 268 bases beyond the last gene (lacA) and that this 3' non-translated region is shortened by post-transcriptional processing.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumer K. J., Steege D. A. Recognition and cleavage signals for mRNA processing lie within local domains of the phage f1 RNA precursors. J Biol Chem. 1989 Dec 5;264(34):20770–20777. [PubMed] [Google Scholar]
- Blumer K. J., Steege D. A. mRNA processing in Escherichia coli: an activity encoded by the host processes bacteriophage f1 mRNAs. Nucleic Acids Res. 1984 Feb 24;12(4):1847–1861. doi: 10.1093/nar/12.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell M., Kennell D. Evidence for endonucleolytic attack in decay of lac messenger RNA in Escherichia coli. J Mol Biol. 1974 Feb 25;83(2):143–161. doi: 10.1016/0022-2836(74)90385-4. [DOI] [PubMed] [Google Scholar]
- Bremer H., Yuan D. RNA chain growth-rate in Escherichia coli. J Mol Biol. 1968 Dec 14;38(2):163–180. doi: 10.1016/0022-2836(68)90404-x. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Büchel D. E., Gronenborn B., Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980 Feb 7;283(5747):541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Evidence that the 5' end of lac mRNA starts to decay as soon as it is synthesized. J Bacteriol. 1985 Feb;161(2):820–822. doi: 10.1128/jb.161.2.820-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur J Biochem. 1989 May 1;181(2):363–370. doi: 10.1111/j.1432-1033.1989.tb14733.x. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Strominger M. B., Wice B. M., Kennell D. E. Isolating and sequencing the predominant 5'-ends of a specific mRNA in cells. I. Purification by filter hybridization. J Biochem Biophys Methods. 1985 Aug;11(2-3):153–161. doi: 10.1016/0165-022x(85)90051-x. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Mudd E. A., Krisch H. M. Transcription and messenger RNA processing upstream of bacteriophage T4 gene 32. Mol Gen Genet. 1989 Oct;219(1-2):39–48. doi: 10.1007/BF00261155. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Cashman J. S., Webster R. E., Steege D. A. Transcription of bacteriophage fl. The major in vivo RNAs. J Biol Chem. 1980 Mar 25;255(6):2554–2562. [PubMed] [Google Scholar]
- Chen C. Y., Beatty J. T., Cohen S. N., Belasco J. G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988 Feb 26;52(4):609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Enea V., Zinder N. D. Functional analysis of bacteriophage f1 intergenic region. Virology. 1981 Oct 30;114(2):463–473. doi: 10.1016/0042-6822(81)90226-9. [DOI] [PubMed] [Google Scholar]
- Fiil N. P., von Meyenburg K., Friesen J. D. Accumulation and turnover of guanosine tetraphosphate in Escherichia coli. J Mol Biol. 1972 Nov 28;71(3):769–783. doi: 10.1016/s0022-2836(72)80037-8. [DOI] [PubMed] [Google Scholar]
- Freedman L. P., Zengel J. M., Archer R. H., Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and posttranscriptional regulation. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6516–6520. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Sharp P. A. Sequences controlling in vitro transcription of SV40 promoters. EMBO J. 1983;2(12):2293–2303. doi: 10.1002/j.1460-2075.1983.tb01737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M. A., Johnson D. F., Nierlich D. P., Zabin I. DNA sequence of the lactose operon: the lacA gene and the transcriptional termination region. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6414–6418. doi: 10.1073/pnas.82.19.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen K., Shapiro J. A., Beckwith J. R. Transposition of the lac region to the gal region of the Escherichia coli chromosome: isolation of lambda-lac transducing bacteriophages. J Bacteriol. 1971 Oct;108(1):5–9. doi: 10.1128/jb.108.1.5-9.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Models for decay of Escherichia coli lac messenger RNA and evidence for inactivating cleavages between its messages. J Mol Biol. 1979 Dec 5;135(2):369–390. doi: 10.1016/0022-2836(79)90442-x. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Stabilization of the 3' one-third of Escherichia coli ribosomal protein S20 mRNA in mutants lacking polynucleotide phosphorylase. J Bacteriol. 1989 Aug;171(8):4112–4120. doi: 10.1128/jb.171.8.4112-4120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa G. J., Kwan C., Yamashita J., Nierlich D. P. Transcription and decay of the lac messenger: role of an intergenic terminator. J Bacteriol. 1991 Jan;173(1):28–36. doi: 10.1128/jb.173.1.28-36.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J., Parma D., Tuerk C., Hall D. H., Gold L. Identification of a T4 gene required for bacteriophage mRNA processing. New Biol. 1989 Oct;1(1):54–65. [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M. Cleavage by RNase III in the transcripts of the met Y-nus-A-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J Mol Biol. 1989 Nov 20;210(2):293–302. doi: 10.1016/0022-2836(89)90331-8. [DOI] [PubMed] [Google Scholar]
- Régnier P., Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986 Jan 5;187(1):23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- Schmeissner U., McKenney K., Rosenberg M., Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984 Jun 15;176(1):39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- Sekiguchi M., Iida S. Mutants of Escherichia coli permeable to actinomycin. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen V., Imamoto F., Schlessinger D. RNase III cleavage of Escherichia coli beta-galactosidase and tryptophan operon mRNA. J Bacteriol. 1982 Jun;150(3):1489–1494. doi: 10.1128/jb.150.3.1489-1494.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits M. A., Schoenmakers J. G., Konings R. N. Expression of bacteriophage M13 DNA in vivo. Isolation, identification and characterization of phage-specific mRNA species. Eur J Biochem. 1980 Nov;112(2):309–321. doi: 10.1111/j.1432-1033.1980.tb07206.x. [DOI] [PubMed] [Google Scholar]
- Steege D. A., Cone K. C., Queen C., Rosenberg M. Bacteriophage lambda N gene leader RNA. RNA processing and translational initiation signals. J Biol Chem. 1987 Dec 25;262(36):17651–17658. [PubMed] [Google Scholar]
- Subbarao M. N., Kennell D. Evidence for endonucleolytic cleavages in decay of lacZ and lacI mRNAs. J Bacteriol. 1988 Jun;170(6):2860–2865. doi: 10.1128/jb.170.6.2860-2865.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J. M., Lindahl L. Escherichia coli ribosomal protein L4 stimulates transcription termination at a specific site in the leader of the S10 operon independent of L4-mediated inhibition of translation. J Mol Biol. 1990 May 5;213(1):67–78. doi: 10.1016/S0022-2836(05)80122-6. [DOI] [PubMed] [Google Scholar]