Abstract
Matrix metalloproteinase 20 (MMP20) and kallikrein-related peptidase 4 (KLK4) are thought to be necessary to clear proteins from the enamel matrix of developing teeth. We characterized Mmp20 and Klk4 null mice to better understand their roles in matrix degradation and removal. Histological examination showed retained organic matrix in Mmp20, Klk4, and Mmp20/Klk4 double-null mouse enamel matrix, but not in the wild-type. X-gal histostaining of Mmp20 null mice heterozygous for the Klk4 knockout/lacZ knockin showed that Klk4 is expressed normally in the Mmp20 null background. This finding was corroborated by zymogram and western blotting, which discovered a 40-kDa protease induced in the maturation stage of Mmp20 null mice. Proteins were extracted from secretory-stage or maturation-stage maxillary first molars from wild-type, Mmp20 null, Klk4 null, and Mmp20/Klk4 double-null mice and were analyzed by SDS-PAGE and western blotting. Only intact amelogenins and ameloblastin were observed in secretory-stage enamel of Mmp20 null mice, whereas the secretory-stage matrix from Klk4 null mice was identical to the matrix from wild-type mice. More residual matrix was observed in the double-null mice compared with either of the single-null mice. These results support the importance of MMP20 during the secretory stage and of KLK4 during the maturation stage and show there is only limited functional redundancy for these enzymes.
Keywords: enamel, enamel maturation, kallikrein 4, proteases, proteinases, teeth
Dental enamel forms in two stages. During the secretory stage mineral ribbons lengthen along a mineralization front at the enamel surface (1, 2). By the end of the secretory stage the enamel layer has already reached its final dimensions. In the rat incisor about 9% of the volume and 36% of the weight of the enamel matrix is mineral, which equals only about 14% of the mineral present at eruption (3). During the maturation stage, crystals initially deposited during the secretory stage grow thicker and wider. By the end of the maturation stage the mineral comprises about 70% of the volume and over 90% of the weight of the enamel matrix, which is more highly mineralized than other mineralized tissues, such as bone and dentin.
Proteins are abundant in secretory-stage and early maturation-stage enamel, but are virtually absent from the late maturation-stage matrix (4). Proteins secreted during the secretory stage are degraded during both the secretory and maturation stages. Enamel protein cleavage products are reabsorbed by ameloblasts and degraded (4–7). There are two major secreted enamel proteases: matrix metalloproteinase 20 (MMP20) (8) and kallikrein-related peptidase 4 (KLK4) (9). These enzymes are necessary for enamel formation, as mutations in MMP20 (10) and KLK4 (11) cause inherited enamel malformations. A major function of enamel proteases is to facilitate the removal of enamel proteins to free up space within the enamel matrix for the enamel crystallites to grow in width and thickness (12). Several reviews on the roles of proteases in dental enamel formation are available (13–15).
Enamel protein cleavage sites have been characterized for proteins that accumulate in secretory-stage pig enamel, and MMP20 is able to catalyze the same amelogenin (16, 17) and ameloblastin (18, 19) cleavages in vitro as occur in vivo. Kallikrein-related peptidase 4 cleaves amelogenin at many sites, but the cleavage pattern is different from that produced by MMP20 (20). The amelogenin C-terminus is highly charged relative to the rest of the protein and increases the affinity of amelogenin for hydroxyapatite (21). Matrix metalloproteinase 20 removes the amelogenin C-terminus, suggesting that MMP20 may be necessary to dissociate amelogenin from the crystals. However, cleavages by KLK4 on the N-terminal side of amelogenin also decrease amelogenin binding to hydroxyapatite in vitro (22).
Mmp20 (23, 24) and Klk4 (25) null mice both have dramatic enamel phenotypes in which the hypomineralized enamel undergoes rapid attrition. The enamel in the Mmp20 null mice breaks off at the dentino–enamel junction (DEJ), while the enamel in the Klk4 null mice breaks just above the DEJ, in the deep enamel (26). Mmp20 null mice cover dentin with a rough mineral layer that is generally thin but irregular, and lacks rod and inter-rod organization (27). The enamel in Klk4 null mice has normal thickness and rod organization and is hard at the surface but is progressively less mineralized with depth (27). The enamel layers of both types of null mice retain enamel proteins, but the state of degradation of these proteins has not been characterized. In this study we analyzed the enamel proteins and proteases in wild-type, Mmp20 null, Klk4 null, and Mmp20/Klk4 double-null mouse maxillary first molars during the secretory stage, the maturation stage, and just prior to tooth eruption.
Material and methods
All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Program at the University of Michigan.
Mouse breeding
Wild-type, Klk4 null, and Mmp20 null mice were all in the C57BL/6J background. Mmp20/Klk4 (MK) double-null mice were obtained by crossing Klk4 null mice and Mmp20 null mice. The resulting Mmp20+/−/Klk4+− offspring were interbred. Then, Mmp20−/−/Klk4+/− mice were crossed to yield Mmp20−/−/Klk4−/− (MK double null) mice. Homozygous null and double-null mice were interbred to obtain litters of Klk4 null, Mmp20 null, and Mmp20/Klk4 (MK) double-null mice. All mice were fed on soft chow and no variations outside of the dentitions were noted.
Mouse genotyping
PCR genotyping was performed using genomic DNA obtained by tail biopsy. The following primer pairs were used: Klk4+ allele (5′-AACCTAAGGGACAGGGCAGT and 5′-TGAGGTGGTACACAGGGTCA; 550-bp amplicon); Klk4lacZ allele (5′-TGCCTCCAACCAGATAGGTC and 5′-GACAGTATCGGCCTCAGGAA, 595-bp amplicon) (25); Mmp20+ allele (5′-AAGTAGACTGAAGTCAGGAGAGCC and 5′-CTGTAGTGGTGACCCTAGTCATCTT, 545-bp amplicon); and Mmp20− allele (5′-CTGCGTCCCCAGACTTTTGATTT and 5′-GCTTTTCATGGCCAGAATGCTCT, ~650-bp amplicon) (23).
Histology
Wild-type, Klk4 null, and Mmp20 null mouse heads at days 5, 11, and 15 were quickly dissected of skin, cut in half, immersed in 5% paraformaldehyde + 2% sucrose fixative overnight at 4°C (pH 7.3), and then decalcified at 4°C by immersion in 1 l of 5% disodium ethylenediaminetetraacetic acid (EDTA) + 0.8% paraformaldehyde (pH 7.3) with agitation (28). Day-5 mice underwent decalcification for 3 wk, day-11 mice for 4 wk, and day-15 mice for 6 wk, with a change of fresh solution every other day. The samples were washed in PBS (135 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM Na2H2PO4; pH 7.3) + 0.8% paraformaldehyde at 4°C, four to five times (every 0.5–1 h), followed by one overnight wash, then dehydrated using a graded ethanol series followed by xylene, embedded in paraffin, sectioned at 5 µm, stained with Harris Hematoxylin and Eosin (Fisher Scientific, Waltham, MA, USA), and imaged using a Nikon Eclipse TE300 Inverted Microscope, Nikon Digital Sight DS-Ri1 camera, and NIS-Element Basic Research software (Mager Scientific, Dexter, MI, USA). Objectivity was optimized using a section from a single hemi-maxilla prepared for each figure panel.
Tissue processing for histochemistry
Mouse heads from days 5, 8, 11, 14, and 15 were quickly dissected of skin, cut in half, and immersed in 4% paraformaldehyde fixative (pH 7.3) overnight at 4°C, washed in PBS four or five times (every 0.5–1 h) at 4°C, and decalcified at 4°C by immersion in 1 l of 4.13% disodium EDTA (pH 7.3) with agitation. Day-5 mice underwent decalcification for 3 wk, days 8 and 11 mice for 4 wk, and days 14 and 15 mice for 6 wk, with a change of fresh solution every other day. The samples were washed in PBS at 4°C four or five times (every 0.5–1 h) followed by one overnight wash, then immersed in 15% sucrose (for 1–2 h) followed by 30% sucrose (for 3–4 h) at 4°C for cryoprotection, embedded in Optimal Cutting Temperature compound (OCT) and stored at −80°C. The blocks were cryosectioned at 8-µm thickness at −20 to −22°C on a Leica cryostat. The slides were stored at −80°C until staining.
X-gal staining
Slides were removed from −80°C and immediately treated with glutaraldehyde fixative [0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1.25 mM ethylene glycol tetraacetic acid (EGTA), 2 mM MgCl2, 0.5% glutaraldehyde, pH 7.3] and then washed three times, for 5 min each wash, with 0.1 M HEPES, 2 mM MgCl2 (pH 7.3). The slides were stained with X-gal solution (0.1 M HEPES, 1 mM MgCl2, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2% Triton X-100, 1 mg ml−1 X-gal substrate; pH 8.0) for 5 h at 45°C and then washed several times in PBS, counterstained with 0.1% (w/v) Nuclear Fast Red, coverslipped with Aquamount, and imaged using a Nikon Eclipse TE300 Inverted Microscope, Nikon Digital Sight DS-Ri1 camera, and NIS-Element Basic Research software.
Protein extractions
Mouse maxillary and mandibular first molars were extracted from mouse pups at days 5, 11, and 15 using a dissecting microscope. The pulp was removed from the underside of each molar, but the enamel organ epithelium (EOE) was left in place (because some enamel mineral is sometimes removed along with the EOE in the null genotypes). The mineral was rapidly dissolved by submerging the molars in 2 ml of 0.17 M HCl/0.98% formic acid for 2 h at 4°C. Undissolved material was removed by centrifugation at 3,500 g for 5 min at 4°C. The supernatant was adjusted to pH ~6 with 50–100 µl of 6 M NaOH (and checked with pH paper). The samples were then either separated by reverse-phase high-performance liquid chromatography (RP-HPLC) for zymography/western blotting or dialyzed against water overnight and then lyophilized for SDS-PAGE/western blotting.
The RP-HPLC was run on material extracted from day-11 molars in order to separate KLK4 from co-migrating amelogenins that could mask its proteolytic activity on zymograms or antibody binding on western blots. The RP-HPLC was run using a Discovery C18 column (4.6 mm · 25 cm; Supelco, Bellefonte, PA, USA) with a 20–75% gradient for 55 min at a flow rate of 0.8 ml min−1, with absorbance monitored at 230 nm. Buffer A was 0.05% trifluoroacetic acid (TFA); buffer B was 80% acetonitrile and 0.05% TFA.
SDS-PAGE, western blotting, and zymography
The amount of protein applied per lane for SDS-PAGE was normalized for each genotype on a per-tooth basis. The number of teeth from each group, their collective weights, the total milligrams of protein extracted, and the micrograms of protein extracted per tooth are provided in Table 1. Lyophilized proteins were weighed and dissolved in 1· sample buffer (32 mM Tris–HCl, pH 6.8, 13% glycerol, 1% SDS, 0.005% bromophenol blue) to achieve a final concentration of 2 µg µl−1. Then, 1 µl (2 µg) was applied in each day-5 lane. The volume applied per lane for the day-11 and day-15 samples of each genotype were adjusted to equal the same percentage of protein per tooth as applied for the day-5 sample of that genotype, and then fractionated by SDS-PAGE on 12% or 18% Tris–glycine gels and stained with Simply Blue Safe Stain (Invitrogen, Carlsbad, CA, USA). Replica gels were transblotted using a Trans-Blot SD semidry transfer cell (BioRad, Hercules, CA, USA) onto Hybond-ECL membranes (GE Healthcare Bio-Sciences, Piscataway, NJ, USA), incubated with blocking solution, immunostained using a primary antibody, then visualized using either the enhanced chemiluminescence (ECL) plus western blotting detection system (GE Healthcare) and exposure of the membrane to hyperfilm ECL (GE Healthcare) for 1 to 3 min, or using colorimetric detection with diaminobenzidine (DAB) that reacts with the anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (GE Healthcare). The three amelogenin antibodies used were: a polyclonal antibody raised against recombinant mouse amelogenin (rM179) (29), an affinity-purified custom anti-peptide antibody produced by YenZym (San Francisco, CA, USA) against the major mouse amelogenin (M180) C-terminal sequence (C-LEAWPSTDKTKREEVD) encoded by exon 6, and an affinity-purified anti-peptide antibody raised against a KLH-conjugated alternative C-terminus (C-AFSPMKWYQGMTRHP) encoded by exon 8 (30). Ameloblastin was detected using Ambn-89 (31), an affinity-purified anti-peptide antibody produced by YenZym against the KHL-conjugated peptide MRPREHETQQYEYS, which is near the ameloblastin N-terminus. Kallikrein-related peptidase 4 was detected using affinity-purified anti-peptide Igs raised by YenZym against the mouse KLK4 amino acid segment #85–99 CHNLKGSQEPGSRMLE. Matrix metalloproteinase 20 was detected using a commercial rabbit polyclonal antibody raised against a synthetic peptide corresponding to residues at the human MMP20 (Abcam, Cambridge, MA, USA) C-terminus. The incubation conditions were: rM179 (1:1,000 dilution; overnight incubation); Amel exon 6 (1:10,000 dilution; overnight incubation); Amel exon 8 (1:1,000 dilution; overnight incubation); Ambn-89 (1:1,000 dilution; overnight incubation); KLK4-85 (1:400 dilution; overnight incubation); and MMP20 (1:1,000 dilution; overnight incubation).
Table 1.
Data from protein extractions. Proteins were extracted from a minimum of 12 teeth for each genotype/age.
| Mouse age | Wild type | Mmp20 null | Klk4 null | MK Dbl Null |
|---|---|---|---|---|
| Number (mg wet weight) of teeth extracted | ||||
| Day 5 | 51 (11.59) | 46 (11.80) | 49 (9.39) | 16 (5.2) |
| Day 11 | 51 (30.67) | 52 (36.54) | 40 (34.85) | 12 (12.4) |
| Day 15 | 51 (68.29) | 53 (64.97) | 36 (45.38) | 12 (15.2) |
| Milligrams (dry weight) of protein extracted | ||||
| Day 5 | 1.90 | 1.71 | 1.55 | 0.32 |
| Day 11 | 2.69 | 2.75 | 2.86 | 0.93 |
| Day 15 | 1.10 | 1.96 | 2.32 | 0.88 |
| Micrograms (dry weight) of protein extracted per tooth | ||||
| Day 5 | 37.3 | 37.2 | 31.6 | 20 |
| Day 11 | 52.7 | 52.9 | 71.5 | 77.5 |
| Day 15 | 21.2 | 37.0 | 64.4 | 73.3 |
| Analysis | Wild type | Mmp20 null | Klk4 null | MK Dbl Null |
| Percentage of protein from a single tooth per lane | ||||
| PAGE/western | 5.3 | 5.3 | 6.3 | 10 |
| Zym/western | 4.6 teeth | 4.5 teeth | 3.4 teeth | – |
Klk4, kallikrein-related peptidase 4; MK Dbl Null, Klk4/Mmp20 (MK) double-null mice; Mmp20, matrix metalloproteinase 20; western, western blot; Zym, zymogram.
Zymograms were run on Novex 12% casein zymogels (Invitrogen) incubated at 37°C for 48 h in 50 mM Tris–HCl with either 10 mM calcium (to ensure MMP activity) or with 10 mM EDTA and 1 mM 1,10-phenanthrolin (to inhibit MMP activity).
To detect active KLK4, 1.2 mg of tooth proteins extracted from day-11 wild-type, Mmp20 null, and Klk4 null mice were separated by RP-HPLC. Three samples were collected from each run – (i) from 45–50 min, (ii) from 50 to 52.5 min; and (iii) from 52.5 to 55 min – and lyophilized. The lyophilized samples were each raised in 100 µl of 1·sample buffer and 20 µl was applied per lane.
Results
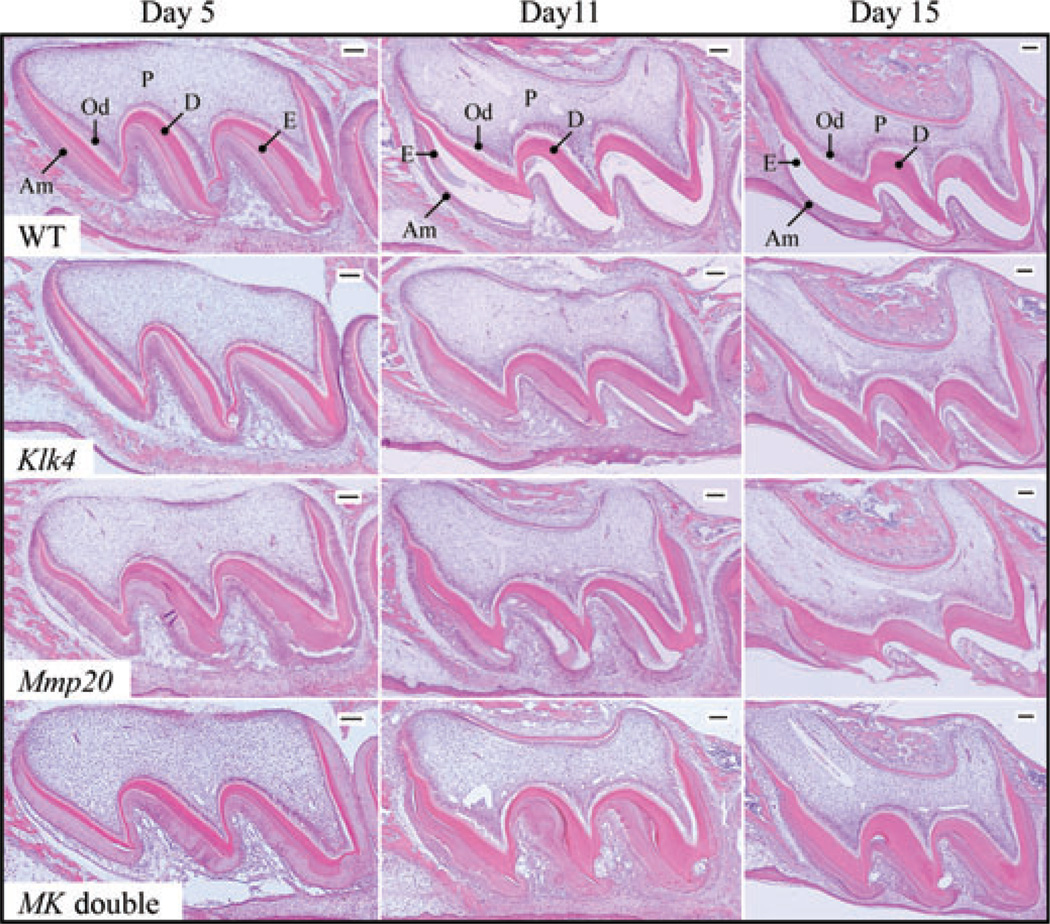
Enamel proteases are believed to be essential for the removal of proteins so that enamel can achieve its high degree of mineralization (13, 32). Our main objective in this study was to characterize and compare accumulated enamel proteins in wild-type, Mmp20 null, Klk4 null, and Mmp20/Klk4 double-null mice. First, we compared the retention of enamel proteins histologically in developing maxillary first molars at days 5, 11, and 15 (Fig. 1). At day 5, maxillary first molar ameloblasts are in the secretory stage of amelogenesis, when enamel proteins and MMP20 are secreted and accumulate in the extra-cellular matrix. Expression of KLK4 has not yet begun (26, 33, 34). At day 5, enamel proteins are observed throughout the enamel as an eosin-stained layer that is readily identified between the pink dentin and the sheet of ameloblasts. The interface between the ameloblasts and the extracellular matrix is generally linear in the wild-type and Klk4 null mice, but undulating and irregular in the Mmp20 null and Mmp20/Klk4 double-null mice. At day 11, maxillary first-molar ameloblasts are in the maturation stage (26, 33), with maturation being more advanced near the cusp tips. All or nearly all ameloblasts are positive for KLK4 expression. At day 11 in the wild-type mice, a thick layer of retained enamel proteins is observed near the cervical margin, but the layer thins and disappears near the cusp tip. The accumulated enamel proteins in the Klk4 and Mmp20 null mice do not appear to have changed from day 5 to day 11. By day 15, the maturation stage is complete and the maxillary molars are about to erupt into the oral cavity. In wild-type mice the enamel layer is clear – no protein layer is detected. In Klk4 null mice enamel proteins are retained, even at the latest stage when the tooth erupts into the oral cavity. An irregular layer of enamel proteins is observed in both the Mmp20 null and Mmp20/Klk4 double-null mice.
Fig. 1.
Histology of maxillary first molars from wild-type, kallikrein-related peptidase 4 (Klk4) null, matrix metalloproteinase 20 (Mmp20) null, and Klk4/Mmp20 (MK) double-null mice from postnatal days 5, 11, and 15. Day 5 (secretory stage): the enamel layers from all four genotypes are protein-rich and stain histologically. The Klk4 null molar is similar to that of the wild-type (WT) mice, while the Mmp20 null and MK double-null molars have irregular enamel layers that vary in thickness. Day 11 (maturation stage): a layer of retained enamel proteins is observed near the cervical margin in wild-type mice (where E is labeled), but the stained organic matrix thins and disappears near the cusp tip. The Klk4 mice molar shows a uniform thickness of enamel proteins. The Mmp20 null and MK double-null molars have a protein-rich enamel matrix that varies in thickness. Day 15 (near eruption): no protein layer is detected in the wild-type mice (the enamel space is clear, unstained). Proteins are retained throughout the enamel layer in Klk4 null and MK double-null mice. The Mmp20 null mouse molar is partially erupted, but a thick layer of protein is still visible in the enamel on the distal surface (left). Am, ameloblasts; D, dentin; E, enamel; Od, odontoblasts; P, pulp. Bars = 100 µm.
KLK4 expression in Mmp20 null mice
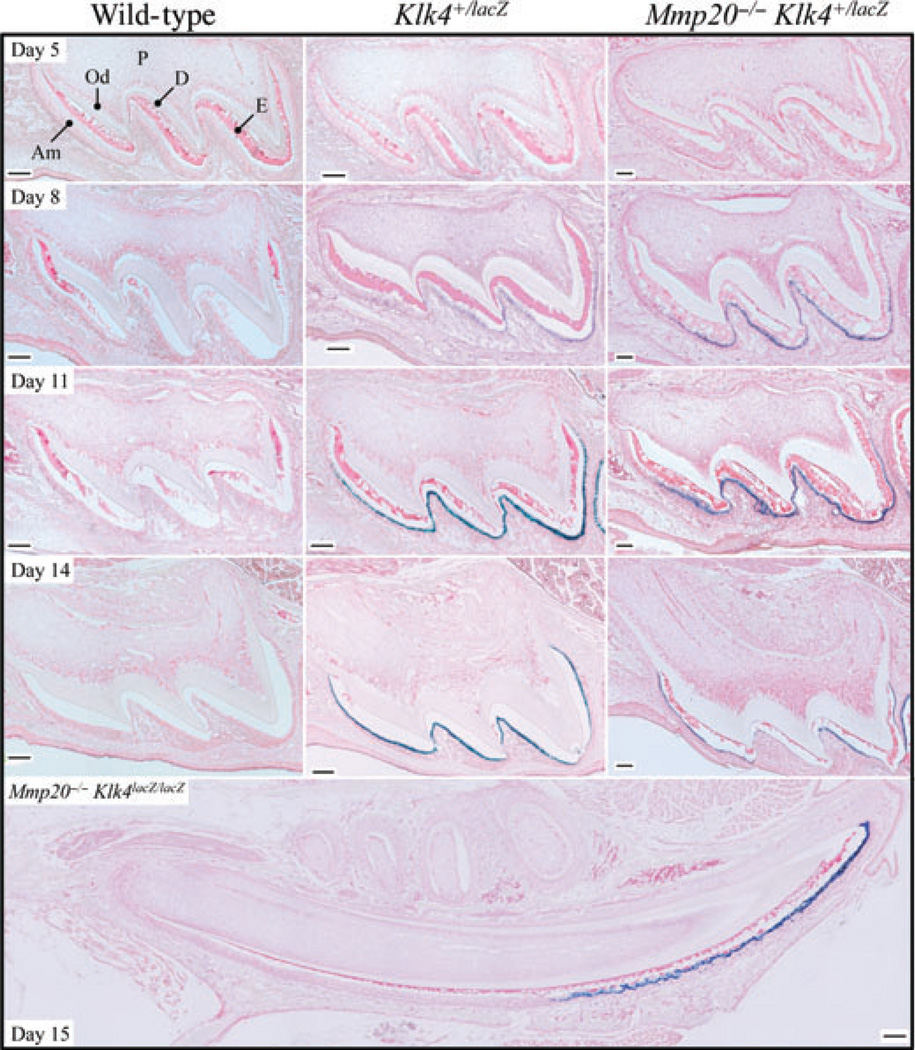
In Klk4 null mice, the bacterial lacZ gene encoding β-galactosidase fused to a mouse nuclear localization signal replaced the 5-prime KLK4 protein coding sequence. This permits visualization (by X-gal staining) of expression from the KLK4 promoter in its native context (25). We performed lacZ histochemistry on maxillary first molars from days 5, 8, 11, and 14 wild-type (negative control), Klk4 heterozygous (positive control), and Klk4 heterozygous + Mmp20 null mice (Fig. 2). No background staining was observed in wild-type molars at any day, indicating that lysosomal β-galactosidase expression was not observed under the incubation time (5 h) and conditions (pH 8) used. Klk4 promoter-driven β-galactosidase activity was observed specifically in maturation ameloblasts on days 8, 11, and 14, in Klk4 heterozygous mice in both the wild-type and Mmp20 null backgrounds. Retained enamel proteins (stained red) were not detected in the wild-type maxillary first molars at day 14, but were observed in the same molars in the Mmp20 null background, particularly near the cervical margin and near the dentin surface. LacZ histochemistry was also performed on a mandible from a day-15 Mmp20/Klk4 double-null mouse. In the longitudinally sectioned incisor, positive X-gal staining was strongly and specifically observed in maturation ameloblasts, which exactly matches previous characterizations of Klk4 expression in wild-type mice by in situ hybridization (33–35) and in Mmp20 null mice by PCR amplification (36).
Fig. 2.
Kallikrein-related peptidase 4 (KLK4) expression in a day-15 incisor and in maxillary first molars at days 5, 8, 11, and 14 in wild-type, Klk4 heterozygous (Klk4+/lacZ), and Klk4 heterozygous mice in the Mmp20 null background (Mmp20−/−Klk4+/lacZ). The enamel proteins in these sections stain pink. Blue (X-gal) stain marks cells that have expressed LacZ from the Klk4 promoter in the Klk4 knockout/lacZ knockin gene. No background X-gal staining is observed in the wild-type mice. Day 5: abundant enamel proteins are observed in all genotypes and no KLK4 expression in all genotypes. Days 8 and 11: protein levels are diminished near the cusp tips in the wild-type; and X-gal staining in the mice expressing Klk4+/lacZ is observed on the cusp tips and slopes. Day 14: X-gal staining in the mice expressing Klk4+/lacZ is observed throughout the ameloblast layer. Enamel proteins have cleared entirely from the wild-type and mostly from the Klk4+/lacZ molars in the wild-type background, but remain in the Mmp20 null condition, particularly over the dentin surface. Day 15: longitudinal section through the mandibular incisor from an Mmp20 + Klk4 double-null mouse (Mmp20−/−Klk4lacZ/lacZ). Secretory ameloblasts are negative for KLK4 expression; transition and maturation ameloblasts are positive. Enamel protein is observed throughout the enamel layer all the way to the tip of the cusp. Am, ameloblasts; D, dentin; E, enamel; Od, odontoblasts; P, pulp. Bars = 100 µm.
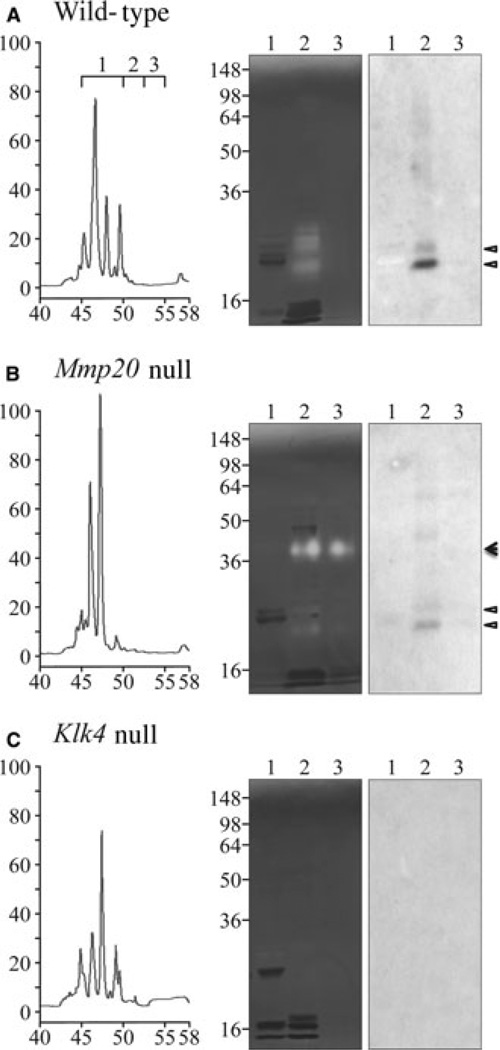
Matrix metalloproteinase 20 can activate the KLK4 zymogen in vitro, but it is not known if Mmp20 is required for KLK4 activation in vivo (20). Taking advantage of the late elution of KLK4 relative to other enamel proteins on RP-HPLC, we analyzed day-11 maxillary first molar proteins with a retention time near 50 min from wild-type (positive control), Mmp20 null, and Klk4 null mice (Fig. 3). An active KLK4 doublet was observed by casein zymography and western blotting in both the wild-type and Mmp20 null mice, but not in Klk4 null mice. The intensity of the KLK4 bands was lower in the Mmp20 null mice relative to the wild-type mice. This demonstrates that KLK4 is activated in vivo in the absence of MMP20. At present, not much significance is attached to the reduced amount of KLK4 in the Mmp20 null molars. The KLK4 samples applied to the gel from the wild-type and Mmp20 null mice were from an equivalent number of teeth (~4.5 molars), but the mineral layer covering dentin is smaller in Mmp20 null mice relative to the wild-type mice, so the apparent reduction in KLK4 activity in the Mmp20 null condition might reflect the general reduction in matrix volume per tooth. An unexpected discovery in the Mmp20 null mice was the expression of a 40-kDa protease that was not expressed in the wild-type or Klk4 null mice.
Fig. 3.
Kallikrein-related peptidase 4 (KLK4) in Mmp20 null mice. Kallikrein-related peptidase 4 elutes from a reverse-phase high-performance liquid chromatography (RP-HPLC) column between 50 and 52.5 min. Left: enamel proteins (1.2 mg) from wild-type, Mmp20 null, and Klk4 null mice were fractionated by RP-HPLC and three fractions were collected, from (i) 45–50 min, (ii) 50.0–52.5 min, and (iii) 52.5–55.0 min. Right: zymography and western blot analyses of the three fractions. Active KLK4 was detected in the Mmp20 null mice migrating as a doublet. An unknown proteolytic activity migrating at 40 kDa was observed only in the Mmp20 null mice. Triangles indicate KLK4 bands; the arrowhead marks an unidentified 40-kDa protease.
MMP20 expression in Klk4 null mice
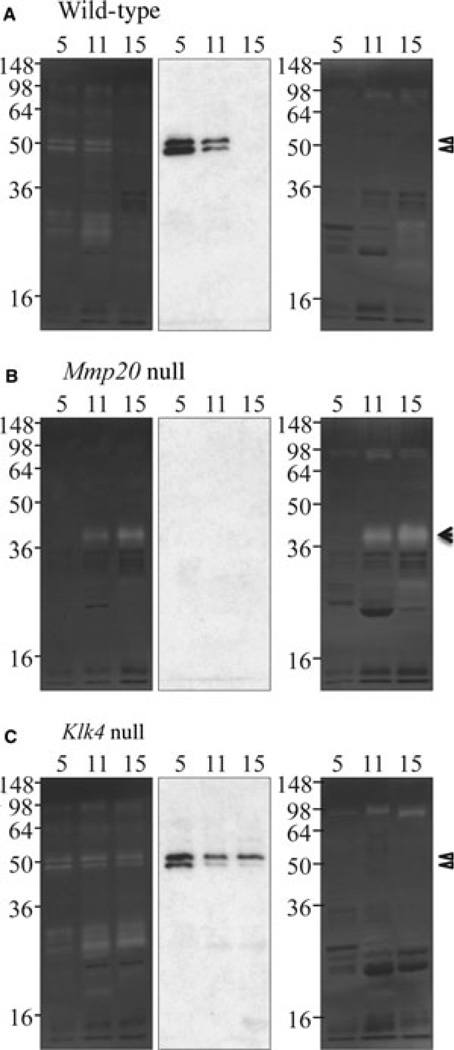
To characterize MMP20 activity in Klk4 null mice, we analyzed first molar (maxillary and mandibular) protein samples from days 5, 11, and 15 by western blotting with antibodies to MMP20 as well as by zymography with and without inhibitors of MMPs (Fig. 4). Matrix metalloproteinase 20 bands were observed as expected in the secretory and early-maturation stages in wild-type mice; however, MMP20 activity persisted in the Klk4 null mice all the way to day 15. This finding suggests that KLK4 might degrade MMP20 in vivo. The zymogram from the Mmp20 null mice showed an absence of MMP20 and demonstrated that the 40-kDa unidentified protease is up-regulated specifically in the maturation stage of Mmp20 null mice, and that this protease is not inactivated by EDTA or phenanthroline (indicating that it is not an MMP). The MMP20 knockout deleted most of intron 4 and all of exon 5, but skipping exon 5 during RNA splicing would not shift the reading frame and might generate an inactive form of MMP20 lacking the exon 5-encoded segment (23). No such protein was detected in the extracts from Mmp20 null mouse using the MMP20 C-terminal antibody, so the Mmp20 knockout appears to be a true knockout (absence of protein) and not simply a loss-of-function model.
Fig. 4.
Matrix metalloproteinase 20 (MMP20) in kallikrein-related peptidase 4 (Klk4) null mice. Matrix metalloproteinase 20 is easily detected on casein zymograms and western blots. Enamel proteins from days 5 (2 µg), 11, and 15 were analyzed on casein zymograms with calcium (left) or with EDTA and phenanthroline (MMP inhibitors) (right), and by western blotting (center) using an MMP20 C-terminal antibody. Matrix metalloproteinase 20 was not detected in the Mmp20 null mice. Day 5: MMP20 was detected in wild-type and Klk4 null mice as a doublet near 50 kDa, but not on zymograms incubated with MMP inhibitors. The MMP20 catalytic domain was responsible for a group of caseinolytic bands around 30 kDa. Day 11: MMP20 was detected in wild-type and Klk4 null mice. An unidentified 40-kDa protease was evident in the Mmp20 null mice and was active in the presence of MMP inhibitors. Day 15: MMP20 was only detected in the Klk4 null mice. The 40-kDa protease was also detected. Triangles indicate MMP20 bands and the arrowhead indicates an unidentified 40-kDa protease.
Matrix metalloproteinase 20 was readily detected in wild-type samples containing matrix from only 5% of a single day-5 or day-11 molar, whereas KLK4 was concentrated by RP-HPLC and detected in a sample derived from 4.5 teeth, suggesting that MMP20 might be 100-fold more abundant than KLK4 in developing mouse enamel. However, this difference is probably because of the greater sensitivity of casein zymograms to detect MMP20 relative to KLK4 and a more sensitive antibody for detecting MMP20 on western blots. When extracting these enzymes from pig molars in the crown-formation stage, our yields of MMP20 are only about twice those of KLK4.
Residual enamel proteins
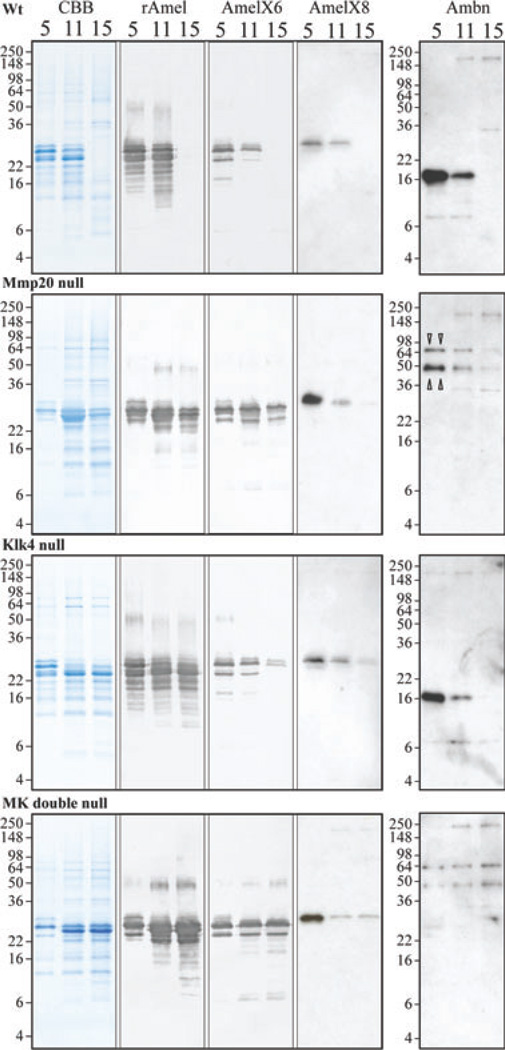
Alternative splicing generates many mouse amelogenin isoforms (30, 37–40), but only two ameloblastin isoforms (41, 42). During the secretory stage, MMP20 initially cleaves amelogenin near its carboxyl end (17, 43, 44), while MMP20 initially cleaves ameloblastin near its amino end (18, 19, 45, 46). In the absence of proteolytic processing, an amelogenin C-terminal antibody and an ameloblastin N-terminal antibody would detect the intact secreted proteins, with multiple bands being caused by alternative splicing. We analyzed accumulated enamel proteins from wild-type, Mmp20 null, Klk4 null, and Mmp20/Klk4 double-null first molars at days 5, 11, and 15 by SDS-PAGE and by western blotting using antibodies raised against full-length recombinant amelogenin (rM179), two alternative amelogenin C-terminal peptides, and an ameloblastin near the N-terminal peptide (Fig. 5).
Fig. 5.
SDS-PAGE and western blot analyses of amelogenin and ameloblastin at days 5, 11, and 15 from wild-type (Wt), matrix metalloproteinase 20 (Mmp20) null, kallikrein-related peptidase 4 (Klk4) null, and Mmp20/Klk4 (MK) double-null mice. Panels show (from left to right): Coomassie Brilliant Blue-stained SDS-polyacrylamide gels; western blots with rM179 antibody (rAmel); western blots with amelogenin C-terminal antibodies specific for the exon 6-encoded sequence (AmelX6) and for the exon 8-encoded sequence (AmelX8); and western blots with the ameloblastin (Ambn) antibody for the N-terminal domain. All day-5 lanes show 2-µg loads. Day-11 and day-15 lanes from each genotype show the same percentage of protein per tooth (see Table 1). Bands on western blots using amelogenin C-terminal antibodies are uncleaved amelogenin isoforms. Note: all amelogenin-positive bands in the day-5 MMP20 null and MK double-null mice co-migrate with intact amelogenin and the wild-type and Klk4 null samples are identical at day 5 (secretory stage). Two bands at 65 and 50 kDa in Mmp20 null and MK double-null mice are likely to be uncleaved ameloblastin (triangle pointers). All positive bands of ~200 kDa are immunoglobulins that were also detected in controls using only the secondary antibody (data not shown).
The day-5 enamel proteins from wild-type and Klk4 null mice showed virtually identical patterns of amelogenin bands using the rM179 antibody. The lower-molecular-mass amelogenins (~14–22 kDa) were not detected with either of the amelogenin C-terminal antibodies and must be cleavage products. The Mmp20 null and Mmp20/Klk4 double-null mice also produced identical amelogenin patterns using the rM179 antibody. Importantly, all of the amelogenin bands in the Mmp20 null mice co-migrate with bands also detected by the C-terminal antibodies. No amelogenin cleavage products were detected in the day-5 (secretory stage) matrix of mice lacking MMP20.
Day-5 enamel proteins from wild-type and Klk4 null first molars both contained a 17-kDa ameloblastin cleavage product that was strongly detected by the ameloblastin near N-terminal antibody. In contrast, the day-5 enamel proteins from the Mmp20 null and Mmp20/Klk4 double-null first molars both showed an ameloblastin doublet of around 65 and 50 kDa. These two bands are consistent with being the 396- and 381-amino-acid O-glycosylated ameloblastin isoforms expressed from alternatively spliced mRNA transcripts. Based upon the mouse ameloblastin cDNA sequences and characterization of the pig ameloblastin protein (47), the smaller ameloblastin has 15 fewer amino acids and lacks one potential O-linked glycosylation relative to the larger isoform. These findings show that MMP20 cleaves ameloblastin to generate a 17-kDa product that is detected by the Ambn-89 antibody. The data also support the interpretation that MMP20 further degrades this 17-kDa protein based upon the presence of smaller immunopositive bands and the diminishing of this product in the day-11 molars of the KLK4 null mice (where MMP20 is the only enamel protease). The day-5 data strongly support previous conclusions that MMP20 is the only major proteolytic activity in secretory-stage enamel.
By day 15, enamel proteins have been virtually removed from the matrix in wild-type mouse first molars. When MMP20, KLK4, or both enzymes are missing from the matrix, high-molecular-weight uncleaved enamel proteins are retained in the enamel layer. Kallikrein-related peptidase 4 is not able to compensate for the absence of MMP20 in Mmp20 null mice, and high-molecular-weight amelogenins are retained in the matrix. Kallikrein-related peptidase 4 degrades some of the intact enamel proteins that accumulate in the absence of MMP20, as the retention of enamel proteins is clearly increased in the Mmp20/Klk4 double-null mice when compared with Mmp20 null mice. Similarly, continued expression of MMP20 during the maturation stage cannot compensate for the absence of KLK4. The effects of MMP20 activity during the maturation stage are evident in the Klk4 null mice by the reduced amounts of intact amelogenins recognized by the C-terminal antibodies and by the loss of the 17-kDa ameloblastin from the matrix as maturation progresses to day 15. Although the full-length amelogenins diminish during the maturation stage in the Klk4 null mice, the cleaved products appear to remain as the pattern of accumulated amelogenins in enamel matrix recognized by the rM179 antibody does not change appreciably after the secretory stage.
Discussion
Substantial evidence supports the conclusion that MMP20 is the predominant, if not the exclusive, extracellular protease in secretory-stage enamel. Matrix metalloproteinase 20 has been shown to be expressed by secretory-stage ameloblasts by in situ hybridization (33, 35, 48) and immunohistochemistry (49, 50). Matrix metalloproteinase 20 is readily isolated from developing pig enamel scrapings as a doublet migrating at 41 and 46 kDa on SDS-PAGE and casein zymograms (49, 51). Enamel protein cleavage products have been isolated from developing pig enamel, and the cleavage sites that generated them from parent proteins have been characterized. Matrix metalloproteinase 20 cleaves amelogenin and ameloblastin in vitro at exactly the same sites as in vivo (16–19). Furthermore, MMP20 has low activity against the major cleavage products that accumulate to abundance in secretory-stage enamel, such as the tyrosine- and leucine-rich amelogenin peptides (TRAP and LRAP, respectively) (17, 52) and the 32-kDa enamelin (53). Here we demonstrate that the accumulated enamel matrix of secretory-stage mouse molars in Mmp20 null and Mmp20/Klk4 double-null mice comprises higher-molecular-weight intact amelogenin isoforms that retain their original C-terminal peptides. Absent from the matrix are lower-molecular-weight amelogenin cleavage products that are observed in the wild-type and Klk4 null mice. We also observed protein bands that probably correspond to the two ameloblastin-secreted isoforms in the Mmp20 null and Mmp20/Klk4 double-null mice. These ameloblastins were absent from both the wild-type and Klk4 null mice that express MMP20 and in their place was a 17-kDa ameloblastin cleavage product that is similar in size to a porcine ameloblastin cleavage product from the same part of the protein (46). These findings strongly support the conclusion that MMP20 is the predominant protease in secretory-stage enamel and that no other matrix protease contributes significantly to the hydrolysis of secreted enamel proteins during the secretory stage.
In addition to the substantial evidence demonstrating that MMP20 is the predominant secretory-stage enamel protease, there is additional strong evidence that KLK4 is not expressed or secreted into secretory-stage enamel, but is solely expressed and secreted into the enamel matrix by transition and maturation-stage ameloblasts. Active KLK4 enzyme is readily isolated from developing pig teeth, and its ability to cleave enamel proteins in vitro has been determined experimentally. Kallikrein-related peptidase 4 cleaves amelogenin at many sites, and the positions of the cleavage sites are complementary to those of MMP20 (20). Kallikrein-related peptidase 4 cleaves at many sites on the N-terminal side of amelogenin. If present during the secretory stage, KLK4 would hydrolyze the relatively stable amelogenin-cleavage products that normally accumulate in enamel and lead to an altered pattern of residual amelogenins, which is not observed. Klk4 mRNA has been shown by in situ hybridization to initially appear in transition-stage ameloblasts (33–35). This finding has been supported by X-gal histochemistry in Klk4 knockout/lacZ knockin mice (26, 54) and by immunohistochemistry (55). Kallikrein-related peptidase 4 in situ hybridization sometimes showed a weak signal in odontoblasts, which supports the hypothesis that KLK4 might be secreted through odontoblastic processes into the deep enamel at the DEJ (56). However, X-gal histochemistry results clearly show only trace, sporadic expression of KLK4 by odontoblasts that could not be sufficient to influence protein accumulation in enamel. The enamel in Klk4 null mice, although hypomineralized, was as thick as enamel in wild-type mice and had normal rod architecture, indicating proper secretory-stage development (25). In this study we present SDS-PAGE and western blot analyses which show that the accumulated amelogenin and ameloblastin proteins in day-5 (secretory stage) molars are identical in wild-type and Klk4 null mice. In the Klk4 null mice the accumulated enamel proteins in late maturation (day 15) look the same as those in the secretory stage (day 5). In contrast, there are no residual amelogenin or ameloblastin proteins in the day-15 wild-type mice. These results strongly support the conclusion that KLK4 is the predominant matrix protease in the maturation stage and that no other maturation-stage enzyme is able to digest enamel proteins in a way that facilitates their efficient removal from the matrix.
Analysis of the proteins and proteases in MMP20- and/or KLK4-deficient mice yielded several surprises. The first was finding a 40-kDa protease in maturation-stage tooth extracts specifically when MMP20 is lacking. The identity of this protease is currently under investigation. Its presence may be part of a response by ameloblasts to the faulty array of inputs they receive from the defective extracellular matrix. Perhaps understanding its induction could provide information about normal feedback mechanisms that allow ameloblasts to adjust to the dynamic requirements of enamel deposition.
We have shown that KLK4 is active in the absence of MMP20 and conversely that MMP20 is active in the absence of KLK4. Matrix metalloproteinase 20 may persist for longer in the matrix in the absence of KLK4. The activity of MMP20 during the maturation stage may help to explain why the superficial enamel hardens to near-normal levels in Klk4 null mice. If so, this would represent the full extent of the functional overlap of the two enzymes. There may also be some functional synergy. By day 15 in wild-type mice, KLK4 was able to clear a large amount of enamel proteins that had accumulated by day 11. In contrast, KLK4 was not able to clear the uncleaved enamel proteins that had accumulated in the Mmp20 null mice. Perhaps KLK4 is less efficient at cleaving intact enamel proteins in vivo (although it readily cleaves full-length amelogenin or ameloblastin in vitro) or has trouble accessing enamel proteins in the disorganized matrix that forms in the absence of MMP20. However, KLK4 certainly cleaves some enamel proteins in the absence of MMP20. When both enzymes are absent, the level of accumulated enamel proteins is higher than when either of the enzymes alone is missing. In the double-null mouse at the time of eruption (day 15) there appears to be even more organic matrix than earlier in the maturation stage (day 11).
Acknowledgements
This investigation was supported by USPHS Research Grants DE015846, DE019775, and DE016276 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 29892, USA.
Footnotes
Conflicts of interest – All authors declare that there are no competing interests.
References
- 1.Ronnholm E. The amelogenesis of human teeth as revealed by electron mircoscopy I. The fine structure of the ameloblasts. J Ultrastruct Res. 1962;6:229–248. doi: 10.1016/s0022-5320(62)90055-2. [DOI] [PubMed] [Google Scholar]
- 2.Boyde A. The development of enamel structure. Proc R Soc Med. 1967;60:923–928. doi: 10.1177/003591576706000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 4.Smith CE, Pompura JR, Borenstein S, Fazel A, Nanci A. Degradation and loss of matrix proteins from developing enamel. Anat Rec. 1989;224:292–316. doi: 10.1002/ar.1092240219. [DOI] [PubMed] [Google Scholar]
- 5.Nanci A, Slavkin HC, Smith CE. Application of high-resolution immunocytochemistry to the study of the secretory, resorptive, and degradative functions of ameloblasts. Adv Dent Res. 1987;1:148–161. doi: 10.1177/08959374870010020301. [DOI] [PubMed] [Google Scholar]
- 6.Nanci A, Slavkin HC, Smith CE. Immunocytochemical and radioautographic evidence for secretion and intracellular degradation of enamel proteins by ameloblasts during the maturation stage of amelogenesis in rat incisors. Anat Rec. 1987;217:107–123. doi: 10.1002/ar.1092170202. [DOI] [PubMed] [Google Scholar]
- 7.Smith CE. Ameloblasts: secretory and resorptive functions. J Dent Res. 1979;58:695–707. doi: 10.1177/002203457905800221011. [DOI] [PubMed] [Google Scholar]
- 8.Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- 9.Simmer JP, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, Margolis HC, Shimizu M, DeHart BC, Hu CC, Bartlett JD. Purification, characterization, and cloning of enamel matrix serine proteinase 1. J Dent Res. 1998;77:377–386. doi: 10.1177/00220345980770020601. [DOI] [PubMed] [Google Scholar]
- 10.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukae M, Yamamoto R, Karakida T, Shimoda S, Tanabe T. Micelle structure of amelogenin in porcine secretory enamel. J Dent Res. 2007;86:758–763. doi: 10.1177/154405910708600814. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 14.Simmer JP, Hu JC. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, Simmer JP. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 17.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, Arai T, Bartlett JD, Simmer JP. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, Simmer JP. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86:153–157. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- 19.Chun YH, Yamakoshi Y, Yamakoshi F, Fukae M, Hu JC, Bartlett JD, Simmer JP. Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J Dent Res. 2010;89:785–790. doi: 10.1177/0022034510366903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, Bartlett JD, Simmer JP. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- 21.Ryu OH, Hu CC, Simmer JP. Biochemical characterization of recombinant mouse amelogenins: protein quantitation, proton absorption, and relative affinity for enamel crystals. Connect Tissue Res. 1998;38:207–214. doi: 10.3109/03008209809017038. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z, Fan D, Fan Y, Du C, Moradian-Oldak J. Enamel proteases reduce amelogenin-apatite binding. J Dent Res. 2008;87:1133–1137. doi: 10.1177/154405910808701212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (matrix metalloproteinase-20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 24.Bartlett JD, Beniash E, Lee DH, Smith CE. Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage. J Dent Res. 2004;83:909–913. doi: 10.1177/154405910408301204. [DOI] [PubMed] [Google Scholar]
- 25.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmer J, Hu Y, Richardson A, Bartlett J, Hu JC-C. Why does enamel in Klk4 null mice break above the dentino–enamel junction? Cells Tissues Organs. 2011;194:211–215. doi: 10.1159/000324260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of kallikrein 4 loss on enamel mineralization: comparison with mice lacking matrix metalloproteinase 20. J Biol Chem. 2011;286:18149–18160. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bronckers AL, Lyaruu DM, Jansen ID, Medina JF, Kellokumpu S, Hoeben KA, Gawenis LR, Oude-Elferink RP, Everts V. Localization and function of the anion exchanger Ae2 in developing teeth and orofacial bone in rodents. J Exp Zool B Mol Dev Evol. 2009;10:10. doi: 10.1002/jez.b.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmer JP, Lau EC, Hu CC, Aoba T, Lacey M, Nelson D, Zeichner-David M, Snead ML, Slavkin HC, Fincham AG. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- 30.Bartlett JD, Ball RL, Kawai T, Tye CE, Tsuchiya M, Simmer JP. Origin, splicing, and expression of rodent amelogenin exon 8. J Dent Res. 2006;85:894–899. doi: 10.1177/154405910608501004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun YH, Lu Y, Hu Y, Krebsbach PH, Yamada Y, Hu JC, Simmer JP. Transgenic rescue of enamel phenotype in Ambn null mice. J Dent Res. 2010;89:1414–1420. doi: 10.1177/0022034510379223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suga S. Histochemical observation of proteolytic enzyme activity in the developing dental hard tissues of the rat. Arch Oral Biol. 1970;15:555–558. doi: 10.1016/0003-9969(70)90109-3. [DOI] [PubMed] [Google Scholar]
- 33.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci. 2002;110:307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 34.Hu JC, Zhang C, Sun X, Yang Y, Cao X, Ryu O, Simmer JP. Characterization of the mouse and human PRSS17 genes, their relationship to other serine proteases, and the expression of PRSS17 in developing mouse incisors. Gene. 2000;251:1–8. doi: 10.1016/s0378-1119(00)00203-1. [DOI] [PubMed] [Google Scholar]
- 35.Simmer JP, Sun X, Yamada Y, Zhang CH, Bartlett JD, Hu JC-C. Enamelysin and kallikrein-4 expression in the mouse incisor. In: Kobayashi I, Ozawa H, editors. Biomineralization: formation, diversity, evolution and application Proceedings of the 8th International Symposium on Biomineralization; Sept 25–28, 2001; Niigata, Japan. Hadano, Japan: Tokai University Press; 2001. pp. 348–352. [Google Scholar]
- 36.Tye CE, Sharma R, Smith CE, Bartlett JD. Altered ion-responsive gene expression in Mmp20 null mice. J Dent Res. 2009;89:1421–1426. doi: 10.1177/0022034510384625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau EC, Simmer JP, Bringas P, Hsu D, Hu CC, Zeichner-David M, Thiemann F, Snead ML, Slavkin HC, Fincham AG. Alternative splicing of the mouse amelogenin primary RNA transcript contributes to amelogenin heterogeneity. Biochem Biophys Res Commun. 1992;188:1253–1260. doi: 10.1016/0006-291x(92)91366-x. [DOI] [PubMed] [Google Scholar]
- 38.Simmer JP. Alternative splicing of amelogenins. Connect Tissue Res. 1995;32:131–136. doi: 10.3109/03008209509013715. [DOI] [PubMed] [Google Scholar]
- 39.Hu CC, Ryu OH, Qian Q, Zhang CH, Simmer JP. Cloning, characterization, and heterologous expression of exon-4-containing amelogenin mRNAs. J Dent Res. 1997;76:641–647. doi: 10.1177/00220345970760020401. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Mathews C, Gao C, DenBesten PK. Identification of two additional exons at the 3𠄲 end of the amelogenin gene. Arch Oral Biol. 1998;43:497–504. doi: 10.1016/s0003-9969(98)00013-2. [DOI] [PubMed] [Google Scholar]
- 41.Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada K, Yamada Y. Full-length sequence, localization, and chromosomal mapping of ameloblastin: a novel tooth-specific gene. J Biol Chem. 1996;271:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 42.Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Sheathlin: cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. J Dent Res. 1997;76:648–657. doi: 10.1177/00220345970760020501. [DOI] [PubMed] [Google Scholar]
- 43.Fincham AG, Moradian-Oldak J. Amelogenin post-translational modifications: carboxy-terminal processing and the phosphorylation of bovine and porcine “TRAP” and “LRAP” amelogenins. Biochem Biophys Res Commun. 1993;197:248–255. doi: 10.1006/bbrc.1993.2468. [DOI] [PubMed] [Google Scholar]
- 44.Fincham AG, Moradian-Oldak J. Comparative mass spectrometric analyses of enamel matrix proteins from five species suggest a common pathway of post-secretory proteolytic processing. Connect Tissue Res. 1996;35:151–156. doi: 10.3109/03008209609029186. [DOI] [PubMed] [Google Scholar]
- 45.Uchida T, Murakami C, Wakida K, Dohi N, Iwai Y, Simmer JP, Fukae M, Satoda T, Takahashi O. Sheath Proteins: synthesis, secretion, degradation and fate in forming enamel. Eur J Oral Sci. 1998;106:308–314. doi: 10.1111/j.1600-0722.1998.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 46.Fukae M, Kanazashi M, Nagano T, Tanabe T, Oida S, Gomi K. Porcine sheath proteins show periodontal ligament regeneration activity. Eur J Oral Sci. 2006;114 Suppl 1:212–218. doi: 10.1111/j.1600-0722.2006.00309.x. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi K, Yamakoshi Y, Hu JC, Gomi K, Arai T, Fukae M, Krebsbach PH, Simmer JP. Splicing determines the glycosylation state of ameloblastin. J Dent Res. 2007;86:962–967. doi: 10.1177/154405910708601009. [DOI] [PubMed] [Google Scholar]
- 48.Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 49.Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, Wakida K, Simmer JP, Yamada Y, Bartlett JD. Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. J Dent Res. 1998;77:1580–1588. doi: 10.1177/00220345980770080501. [DOI] [PubMed] [Google Scholar]
- 50.Bourd-Boittin K, Septier D, Hall R, Goldberg M, Menashi S. Immunolocalization of enamelysin (matrix metalloproteinase-20) in the forming rat incisor. J Histochem Cytochem. 2004;52:437–445. doi: 10.1177/002215540405200402. [DOI] [PubMed] [Google Scholar]
- 51.Yamada Y, Yamakoshi Y, Gerlach R, Hu C, Matsumoto K, Fukae M, Oida S, Bartlett J, Simmer J. Purification and characterization of enamelysin from secretory stage pig enamel. Arch Comp Biol Tooth Enam. 2003;8:21–25. [Google Scholar]
- 52.Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Dental enamel matrix: sequences of two amelogenin polypeptides. Biosci Rep. 1981;1:771–778. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- 53.Yamakoshi Y, Hu JC-C, Fukae M, Iwata T, Simmer JP. How do MMP-20 and KLK4 process the 32 kDa enamelin? Eur J Oral Sci. 2006;114 Suppl 1:45–51. doi: 10.1111/j.1600-0722.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 54.Simmer J, Richardson A, Smith C, Hu Y, Hu J-C. Expression of kallikrein 4 in dental and non-dental tissues. Eur J Oral Sci. 2011;119 Suppl. 1:226–233. doi: 10.1111/j.1600-0722.2011.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu JC, Ryu OH, Chen JJ, Uchida T, Wakida K, Murakami C, Jiang H, Qian Q, Zhang C, Ottmers V, Bartlett JD, Simmer JP. Localization of EMSP1 expression during tooth formation and cloning of mouse cDNA. J Dent Res. 2000;79:70–76. doi: 10.1177/00220345000790011301. [DOI] [PubMed] [Google Scholar]
- 56.Fukae M, Tanabe T, Nagano T, Ando H, Yamakoshi Y, Yamada M, Simmer JP, Oida S. Odontoblasts enhance the maturation of enamel crystals by secreting EMSP1 at the enamel–dentin junction. J Dent Res. 2002;81:668–672. doi: 10.1177/154405910208101003. [DOI] [PubMed] [Google Scholar]