Abstract
Kallikrein-related peptidase 4 (KLK4) is a glycosylated serine protease that functions in the maturation (hardening) of dental enamel. Pig and mouse KLK4 contain three potential N-glycosylation sites. We isolated KLK4 from developing pig and mouse molars and characterized their N-glycosylations. N-glycans were enzymatically released by digestion with N-glycosidase F and fluorescently labeled with 2-aminobenzoic acid. Normal-phase high-performance liquid chromatography (NP-HPLC) revealed N-glycans with no, or with one, two, or three sialic acid attachments in pig KLK4 and with no, or with one or two sialic acid attachments in mouse KLK4. The labeled N-glycans were digested with sialidase to generate the asialo N-glycan cores that were fractionated by reverse-phase HPLC, and their retention times were compared with similarly labeled glycan standards. The purified cores were characterized by mass spectrometric and monosaccharide composition analyses. We determined that pig and mouse KLK4 have NA2 and NA2F biantennary N-glycan cores. The pig triantennary core is NA3. The mouse triantennary core is NA3 with a fucose connected by an α1–6 linkage, indicating that it is attached to the first N-acetyglucosamine (NA3F). We conclude that pig KLK4 has NA2, NA2F, and NA3 N-glycan cores with no, or with one, two, or three sialic acids. Mouse KLK4 has NA2, NA2F, and NA3F N-glycan cores with no, or with one or two sialic acids.
Keywords: enamel, glycosylations, kallikrein 4, serine proteases, sialic acid
Kallikrein-related peptidase 4 (KLK4) is a glycosylated serine protease that is expressed specifically by maturation-stage ameloblasts (1, 2), with only trace or very low levels of expression in other tissues (3). Klk4 null mice have undermineralized enamel, with no apparent effects elsewhere in the body (4). A premature termination codon in both human KLK4 alleles causes a recessive hypomaturation form of amelogenesis imperfecta, with no systemic abnormalities (5). Therefore, the structural and functional features of KLK4 are presumed to be adapted to its role in the maturation of dental enamel.
When KLK4 is not expressed during tooth development, the enamel layer achieves normal thickness and mineral architecture, with decussating enamel rods (4). The enamel, however, retains enamel proteins and is progressively less mineralized with depth, so that the surface enamel is 8%, the middle enamel 15%, and the deepest enamel about 20% less mineralized than normal (6). Following eruption, the enamel rapidly fractures in the deep enamel just above the dentino–enamel junction (7). At present it is unclear how the surface enamel, without KLK4 to digest the accumulated enamel proteins, hardens to the extent observed in Klk4 null mice. It appears that superficial proteins are readily reabsorbed into ameloblasts by endocytosis, and KLK4 cleavages are required to increase the rate and ease with which protein fragments return to the enamel surface for reabsorption. The activity of membrane-bound or other secreted proteases in maturation-stage enamel has not been excluded, but should such accessory activity exist it is increasingly unable to compensate for the absence of KLK4 with distance from the ameloblast.
The activated human, pig, and mouse KLK4 enzymes all have 224 amino acids reinforced with six disulfide bridges. Pig and mouse KLK4 contain three potential N-glycan attachment sites (pig at Asn104, Asn139, and Asn184; and mouse at Asn93, Asn139, and Asn184). Human KLK4 has only one potential N-glycan attachment site (at Asn139). Recombinant human KLK4 expressed in bacteria is not glycosylated, is active, and its crystal structure has been determined (8). Human and mouse KLK4 have never been isolated from natural sources and their glycosylation status is unknown. Pig KLK4 is heavily glycosylated, and deglycosylation causes it to lose activity (9), although its three potential N-linked glycosylation sites are on the back of the enzyme, away from its active site (10).
In general, glycosylation is an important structural and functional feature that influences the physicochemical properties of proteins, such as conformational stability and solubility, protects them from proteolysis, and can affect protein–protein and protein–mineral interactions (11, 12). There are currently 16 known inherited glycosylation disorders caused by defects in the N-glycosylation pathway that cause severe protein hypoglycosylation diseases with multisystemic phenotypes and neurological impairment (13, 14). In this study we characterized the N-glycans attached to pig and mouse KLK4.
Material and methods
All experimental procedures involving the use of animals were reviewed and approved by the Institutional Animal Care and Use Program at the University of Michigan.
Sequence analysis
Human (NM_004917.3), mouse (NM_019928.1), and pig (NM_213802.1) KLK4 protein sequences were retrieved from the National Center for Biotechnology Information (NCBI) and aligned manually. Potential N-glycosylation sites were identified using NetNGlyc 1.0 linked to the Expert Protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (SIB).
Isolation of KLK4
Recombinant human KLK4 zymogen (pro-KLK4) was purchased from R&D Systems (Minneapolis, MN, USA). Porcine KLK4 was extracted and purified from the second molars of 6-month-old pigs, as described previously (15). In brief, the hard enamel was extracted with neutral buffer and subjected to a series of ammonium sulfate precipitations. The 40–65% saturation pellet was resuspended in 0.5 M acetic acid and fractionated by reverse-phase high-performance liquid chromatography (RP-HPLC). Mouse KLK4 was isolated from the first molars of 11-d-old mice. The dental pulp tissue was removed with tissue forceps, and then 50 teeth were demineralized by incubation in 3 ml of 0.17 MHCl and 0.95% formic acid containing Protease Inhibitor Cocktail Set III [1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 0.8 µM aprotinin, 50 µM bestatin, 15 µM E-64, 20 µM leupeptin, and 10 µM pepstatin; Calbiochem, San Diego, CA, USA] and 1 mM 1,10-phenanthroline for 2 h at 4°C. This solution was neutralized by dialysis against 1 l of 50 mM Tris–HCl buffer (pH 7.4) containing the same protease inhibitors for 3 h at room temperature and centrifuged for 10 min at 20,000 g at room temperature. The supernatant was directly injected onto a C18 reverse-phase column (Discovery C18, 4.6 mm × 25 cm; Supelco, Bellefonte, PA, USA). The column was eluted with a linear gradient (20–75% buffer B in 55 min) at a flow rate of 0.8 ml min−1. Buffer A was 0.05% trifluoroacetic acid (TFA); buffer B was 0.5% TFA in 80% aqueous acetonitrile. Proteins were detected by absorbance at 220 nm.
SDS-PAGE, western blots, and zymography
Mouse KLK4 anti-peptide Igs were raised against the amino-acid segments #85–99 C-HNLKGSQEPGSRMLE (Residue “C-” is added to conjugate the synthesized peptide to KLH to enhance antigenicity and is not part of the KLK4 peptide) and #187–202 YHLSMFCAGGGQDQKD and affinity purified on columns containing the immobilized peptides (Yenzyme Antibodies, San Francisco, CA, USA). Pig, mouse, and recombinant human KLK4 enzymes were separated by SDS-PAGE on Tris-glycine gels stained with Coomassie Brilliant Blue (CBB; Invitrogen, Carlsbad, CA, USA). Duplicate gels were transblotted onto Hybond-ECL membrane (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) and immunostained with polyclonal antibodies raised in rabbits against recombinant pig KLK4 (16) or mouse KLK4 anti-peptide Igs, or a commercially available N-terminal KLK4 antibody (Abcam, Cambridge, MA, USA). Zymography was performed using Novex 12% casein zymogels (Invitrogen), incubated in 50 mM Tris-HCl/10 mM EDTA containing 1 mM 1,10-phenanthrolin buffer (pH 7.4) at 37°C for 48 h.
Deglycosylation of KLK4 and labeling the N-glycans
PNGase F releases N-linked oligosaccharides from asparagine residues (17). N-glycans were enzymatically released from 2–3 µg of purified porcine or mouse KLK4 by incubation with 1 mU of N-glycosidase F (PNGase F; QA-bio, Palm Desert, CA, USA) in 50 mM sodium phosphate buffer (pH 7.5) at 37°C for 20 h. The porcine 32-kDa enamelin (90 µg) was digested as a positive control. The released N-glycans were fractionated by adding three volumes of ice-cold ethanol and the deglycosylated proteins were precipitated by centrifugation for 10 min at 14,000 g. The N-glycans in the supernatants were evaporated and labeled with 2-aminobenzoic acid (2-AA, 137.14 g mol−1) using a 2-AA labeling kit and S Cartridge (QA-bio). A labeled N-glycan standard (10 pmol; NA2F glycan; QA-bio) and the 2-AA glycans were separated by size exclusion-HPLC (SE-HPLC) using a Nanofilm-SEC 150 column (7.8 mm × 30 cm; Sepax Technologies, Newark, DE, USA) equilibrated with PBS (135 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM Na2H2PO4; pH 7.3) and eluted with the same solution at a flow rate of 1.0 ml min−1 at room temperature. The effluent was continuously monitored by a fluorescence monitor (FP-2020; JASCO, Tokyo, Japan) using an excitation wavelength of 320 nm and an emission wavelength of 420 nm. The amount of labeled N-glycans released was determined by comparing the area under its chromatographic peak with the labeled NA2F glycan standard (QA-bio).
Characterization of KLK4 N-glycans
Each of the 2-AA-labeled N-glycans (100 pmol) obtained from SE-HPLC was further separated by normal-phase (NP-HPLC) using a Supelcosil LC-NH2 column (4.6 mm × 25 cm; Supelco, Bellefonte, PA, USA). The column was eluted with a linear gradient (30–95% buffer B in 97.5 min) at a flow rate of 1.0 ml min−1. Buffer A was 2% acetic acid/1% tetrahydrofuran in 100% acetonitrile and buffer B was 5% acetic acid/1% tetrahydrofuran/3% tri-ethylamine in water. The effluent was continuously monitored by a fluorescence monitor using an excitation wavelength of 320 nm and an emission wavelength of 420 nm. The 2-AA-labeled N-glycans were collected and lyophilized. A set of 2-AA-labeled N-glycan standards having no-, mono-, di- and tri-sialic acid at the terminal position on their structures (NA2, A1, A2, and A3) (QA-bio), were also fractionated by NP-HPLC under the same conditions and their elution times were noted.
Isolation of KLK4 core-N-glycans
Core N-glycans were obtained by enzymatic removal of sialic acid. Each of the 2-AA-labeled N-glycans was incubated with 10 mU of sialidase Au (QA-bio) in 50 mM sodium phosphate buffer (pH 6.0) for 3 h at 37°C. The reaction mixture was separated by NP-HPLC, as described above. The asialo-fraction (KLK4 core N-glycans) was collected and lyophilized. This sample was fractionated by RP-HPLC using a GlycoSep R column (4.6 mm × 15 cm; ProZyme, Hayward, CA, USA), and eluted with a linear gradient (5–95% buffer B in 50 min) at a flow rate of 0.6 ml min−1. Buffer A was 50 mM ammonium acetate buffer (pH 6.0) and buffer B was 50 mM ammonium acetate in 8% acetonitrile. The effluent was continuously monitored by a fluorescence monitor using an excitation wavelength of 320 nm and an emission wavelength of 420 nm. The structures of KLK4 core N-glycans were determined by comparing their retention times with those of the 2-AA N-glycan standards (NA2, NA2F, and NA3; QA-bio). The KLK4 core N-glycans were collected, lyophilized, and used for mass spectrometric and monosaccharide composition analyses. An aliquot of the KLK4 core N-glycan sample that did not have a retention time which matched the standards (peak d) was digested with 1 mU of α-(1–6) fucosidase (QA-bio) in 50 mM sodium phosphate buffer (pH 5.0) for 20 h at 37°C. The reaction mixture was separated by RP-HPLC using a GlycoSep R column and compared with a retention time of 2-AA N-glycan standards, as described above.
Mass spectrometry
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) was performed by W.M. Keck Biotechnology Resource Laboratory at Yale University (New Haven, CT, USA) on purified KLK4 core-N-glycans.
Monosaccharide analysis of KLK4 core N-glycans
The monosaccharide compositions of each KLK4 core N-glycan were determined by digesting each N-glycan into its component monosaccharides and labeling the released monosaccharides with 2-AA, using the 2-AA Monosaccharide Release and Labeling Kit (QA-bio), and fractionating by RPHPLC. The identity of each monosaccharide was determined by comparing its retention time with 2-AA-labeled monosaccharide standards. The amount of each monosaccharide was determined by calculating the area under its chromatographic peak and normalizing against the mannose peak, as there are predictably three mannoses per N-glycan. Each purified KLK4 core N-glycan was hydrolyzed with a mixture of equal volumes of 2 M TFA and 6 M HCl at 100°C for 3 h. The solution was diluted with the water and lyophilized. The dried sample and a monosaccharide mixed standard (100 pmol) were labeled with 2-AA at 80°C for 45 min. The reaction mixture was diluted with the water and was applied onto a GlycoSep R column (4.6 mm × 15 cm; ProZyme). The column was eluted with the second step of the gradient (7–27% buffer B in 28 min and 27–100% B in 2 min) at a flow rate of 0.6 ml min−1. Buffer A was 0.2% butylamine/0.5% phosphoric acid/1% tetrahydrofuran and buffer B was 50% buffer A in 50% acetonitrile. The effluent was continuously monitored by a fluorescence monitor using an excitation wavelength of 360 nm and an emission wavelength of 425 nm.
Results
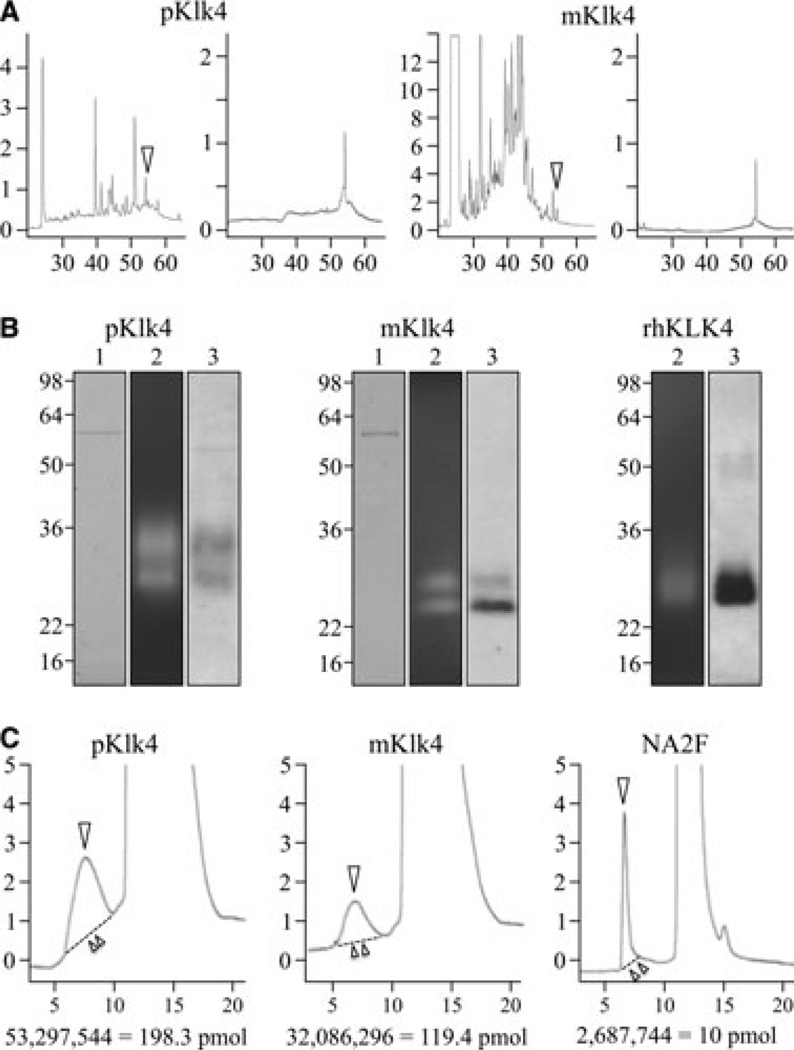
The first objective was to isolate pig, mouse, and human KLK4. Pig and mouse KLK4 fractions from developing teeth were purified by RP-HPLC, and appeared as single chromatographic peaks (Fig. 1A). The purified KLK4 proteases were characterized by SDS-PAGE, casein zymography, and western blotting (Fig. 1B). In the quantities applied KLK4 did not stain with CBB, but was readily visible on zymograms and western blots. No other protein bands besides albumin (at 57 kDa) were observed in the CBB-stained gels. Albumin is a common contaminant in KLK4 preparations, but was not a concern in this study because it is not glycosylated. These analyses suggested that KLK4 was well purified and unlikely to be contaminated with glycoproteins. Pig KLK4 was detected as a doublet at 30 and 34 kDa, mouse KLK4 at 24 and 29 kDa, and thermolysin-activated recombinant human KLK4 (which contains a C-terminal His tag) as a smear between 26 and 29 kDa. The presence of multiple KLK4 bands is probably caused by variations in glycosylation (9). Commercially available recombinant human KLK4 was our only source of the human protease, but this recombinant protein did not appear to be glycosylated (there was no change in mobility on SDS-PAGE following deglycosylation) and so no further characterization of recombinant human KLK4 was attempted.
Fig. 1.
Isolation of pig and mouse kallikrein-related peptidase 4 (KLK4) and initial characterization of pig, mouse, and recombinant human KLK4. (A) (left to right) Reverse-phase high performance liquid chromatography (RP-HPLC) chromatograms of pig neutral extract, 40–65% ammonium sulfate precipitate; pig KLK4 fraction collected at ~54 min (arrowhead); mouse acid extract; and KLK4 fraction collected at ~54 min (arrowhead). The y-axes represent 10 × the absorbance at 220 nm and the x-axes represent minutes from injection of sample. (B) (left to right) Characterization of pig, mouse, and recombinant human KLK4 samples used for N-glycan analyses. All gels: lane 1, SDS-polyacrylamide gel stained with Coomassie Brilliant Blue; lane 2, casein zymograms showing enzyme activity; lane 3, western blots probed with KLK4 antibodies. (C) Size-exclusion chromatograms of 2-aminoben-zoic acid (2-AA)-labeled N-glycans (arrowheads) being separated from excess label. The y-axes represent 100 × the fluorescence emitted at 420 nm and the x-axes represent minutes after injection of sample. The areas under the curves to the dashed lines (above double arrowhead) were used to determine the amount of labeled N-glycan based upon the value observed for a known quantity of labeled NA2F. pKLK4, pig KLK4; mKLK4, mouse KLK4; rhKLK4, recombinant human KLK4.
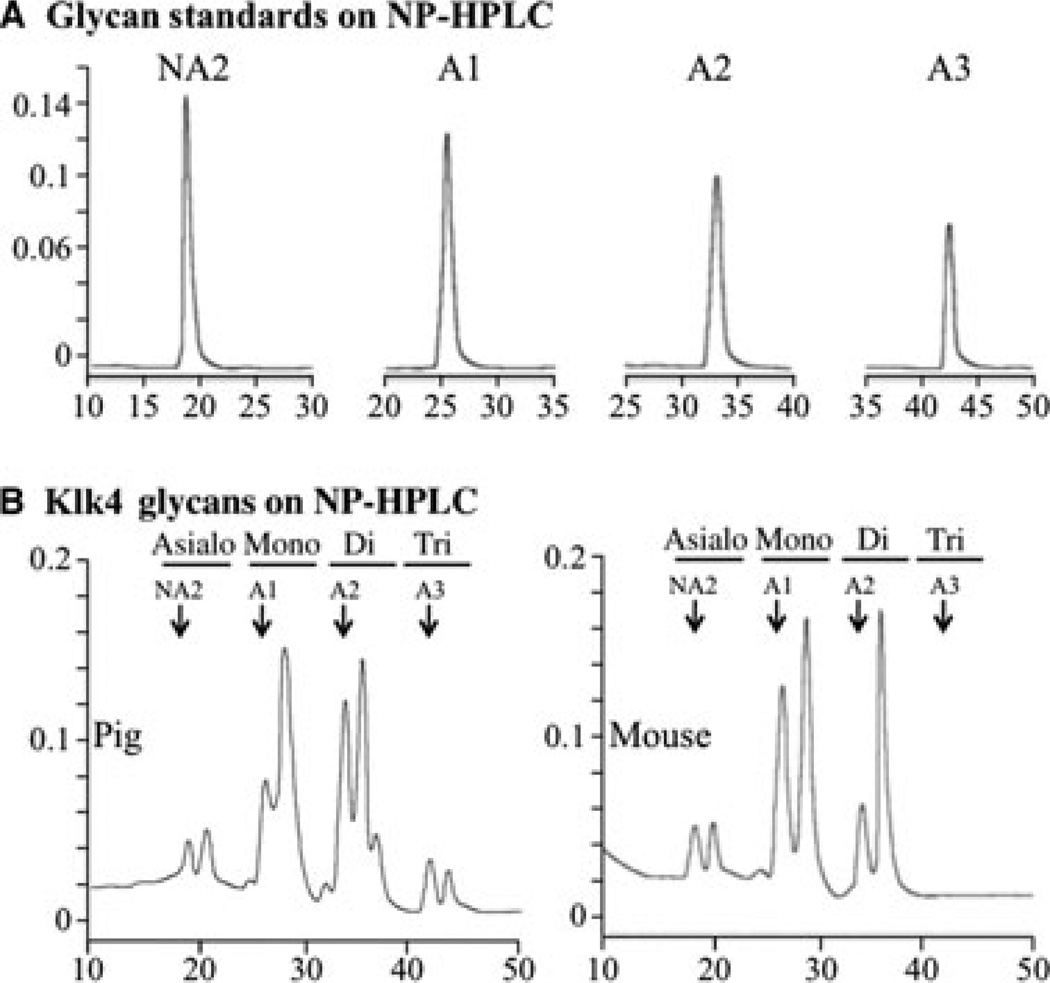
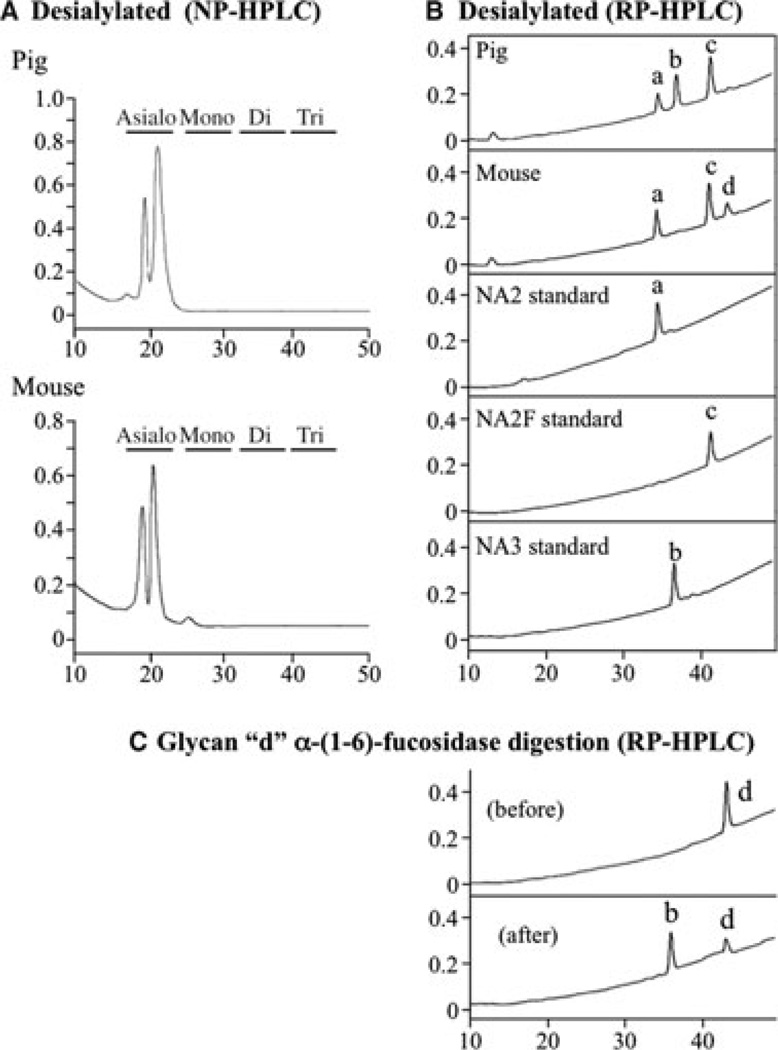
The pig and mouse KLK4 enzymes were deglycosylated, and the released N-glycans were fluorescently labeled with 2-AA (Fig. 1C) and fractionated by NP-HPLC along with similarly labeled glycan standards, which revealed significant heterogeneity in their degrees of sialylation (Fig. 2). Pig KLK4 contained N-glycans with no, or one, two or three sialic acids per N-glycan; mouse KLK4 N-glycans contained no, or one or two sialic acids per molecule (N-glycans with three sialic acids were not detected in mouse KLK4). Knowing this, the released N-glycans were reduced to their glycan ‘cores’ by enzymatic removal of all sialic acid, which was confirmed by fractionation of the sialidase digestion products by NP-HPLC (Fig. 3A). The desialylated 2-AA-labeled N-glycan cores were resolved by RP-HPLC and their retention times were compared with 2-AA-labeled N-glycan standards (Fig. 3B). Three pig N-glycan cores were observed (designated ‘a’, ‘b’, and ‘c’), which had identical retention times to the N-glycan standards NA2, NA3, and NA2F, respectively. Three mouse N-glycan cores were also observed. Two (‘a’ = NA2 and ‘c’ = NA2F) were the same as those in pig. The third (designated ‘d’) was digested by α-(1–6) fucosidase, which shifted its retention time to match N-glycan core ‘b’ (Fig. 3C), indicating that core ‘d’ was like ‘b’, but also contained fucose connected by an α-(1–6) bond, which localizes its attachment to the first N-acetylglucosamine (GlcNAc) in the N-glycan.
Fig. 2.
Sialidation of N-glycans. The y-axes represent fluorescence emitted at 420 nm and the x-axes represent minutes from injection of sample. (A) Normal-phase high-performance liquid chromatography (NP-HPLC) chromatograms of glycan standards containing no (NA2), one (A1), two (A2) and three (A3) sialic acid attachments.(B) NP-HPLC chromatograms of labeled N-glycans released from pig (left) or mouse (right) kallikrein-related peptidase 4 (KLK4). Arrows point to the retention times of the standards. Note that the N-glycans released from mouse KLK4 do not show peaks at retention times for three sialic acids.
Fig. 3.
Characterization of desialylated N-glycan cores. The y-axes represent fluorescence emitted at 420 nm and the x-axes represent minutes from injection of sample. (A) Normal-phase high-performance liquid chromatography (NP-HPLC) chromatograms of labeled N-glycans following sialidase digestion. (B) Reverse-phase high performance liquid chromatography (RP-HPLC) chromatograms of biantennary (NA2 and NA2F) and triantennary (NA3) N-glycan core standards and labeled asialo N-glycan cores from pig and mouse kallikrein-related peptidase 4 (KLK4). Note that peak ‘a’ corresponds to the NA2 standard and peak ‘c’ corresponds to the NA2F standard, and that both N-glycan cores are found in pig and mouse KLK4. Peak ‘b’ corresponds to the NA3 standard, which is found in pig KLK4 only. (C) RP-HPLC chromatograms of peak ‘d’ before and after α-(1–6)-fucosidase digestion. Note that removal of fucose converts peak ‘d’ into peak ‘b’, which corresponds to the NA3 standard.
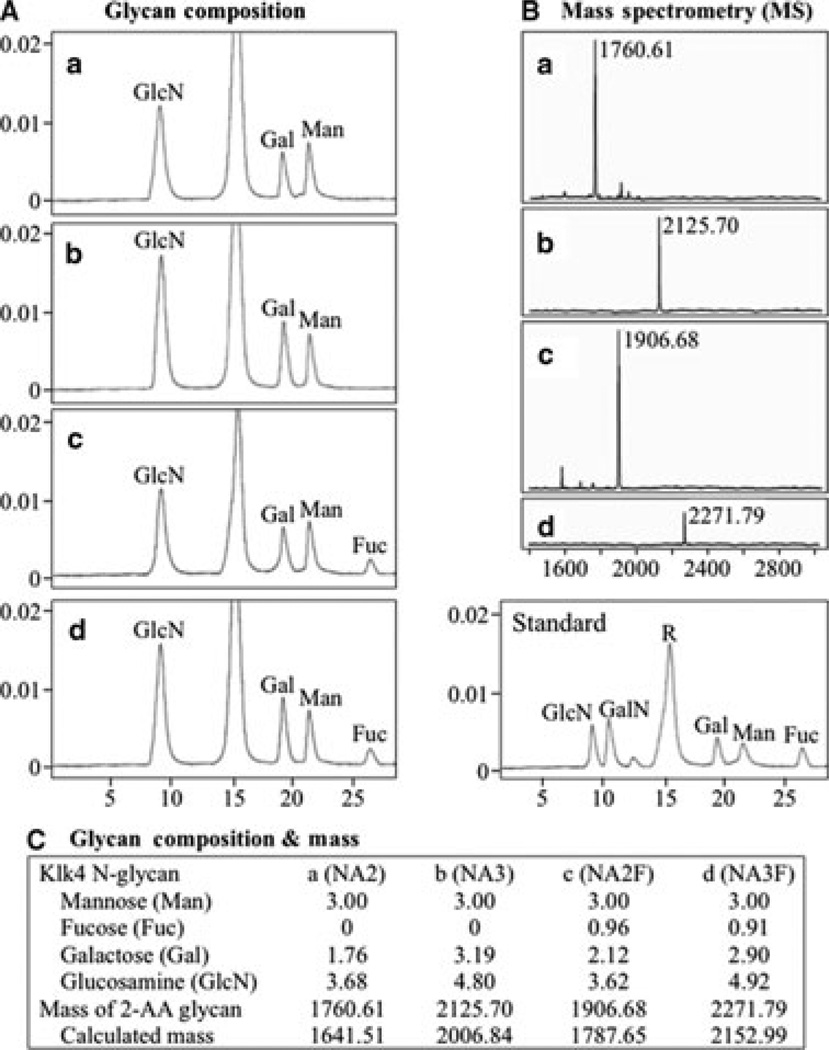
Aliquots of the four (a–d) N-glycan cores were hydrolyzed into their monosaccharide components. The labeled monosaccharides from each N-glycan core were fractionated by RP-HPLC and their retention times were compared with those of labeled monosaccharide standards, which identified the monosaccharides within each N-glycan core (Fig. 4A). By determining the areas under each chromatographic peak and normalizing to three mannoses per N-glycan core, we calculated the number of each type of monosaccharide in each N-glycan core. Depolymerizing the N-glycans into monosaccharides deacetylates GlcNAc into glucosamine (GlcN), so these monosaccharides cannot be distinguished by this analysis. For independent verification of the compositions and clarification of the GlcN/GlcNAc ambiguity, aliquots of the four 2-AA-labeled N-glycan cores were sent for mass determination (Fig. 4B). In each case the monosaccharide compositions and the N-glycan cores they suggested were strongly supported by the mass analyses, with the difference between the expected and predicted masses being smaller than half a dalton (Fig. 4C). The mass analyses showed that all of the GlcN observed in the monosaccharide composition analyses was derived from GlcNAc.
Fig. 4.
(A) Chromatograms of monosaccharide standards and labeled N-glycan cores ‘a’ to ‘d’ following their hydrolysis into monosaccharides. The areas under each chromatographic peak were calculated to determine the number of each monosaccharide per N-glycan molecule. The y-axes represent fluorescence emitted at 425 nm and the x-axes represent minutes from injection of sample. Fuc, fucose; Gal, galactose; GalN, galactosamine; GlcN, glucosamine; Man, mannose; R, reagent. (B) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) results for 2-aminobenzoic acid (2-AA)-labeled N-glycan cores ‘a’ to ‘d’. The mass numbers (x-axes) are in daltons. (C) Summary of glycan composition and mass spectrometry analyses. Mass values were calculated using GlycanMass on the ExPASy Proteomics Server (34). The calculated mass for each N-glycan core corresponds closely (within 0.32 Da) with the measured mass minus the mass of the 2-AA fluorescent attachment (119.12 Da), which is the mass of 2-AA (137.14 Da) less water (18.02 Da).
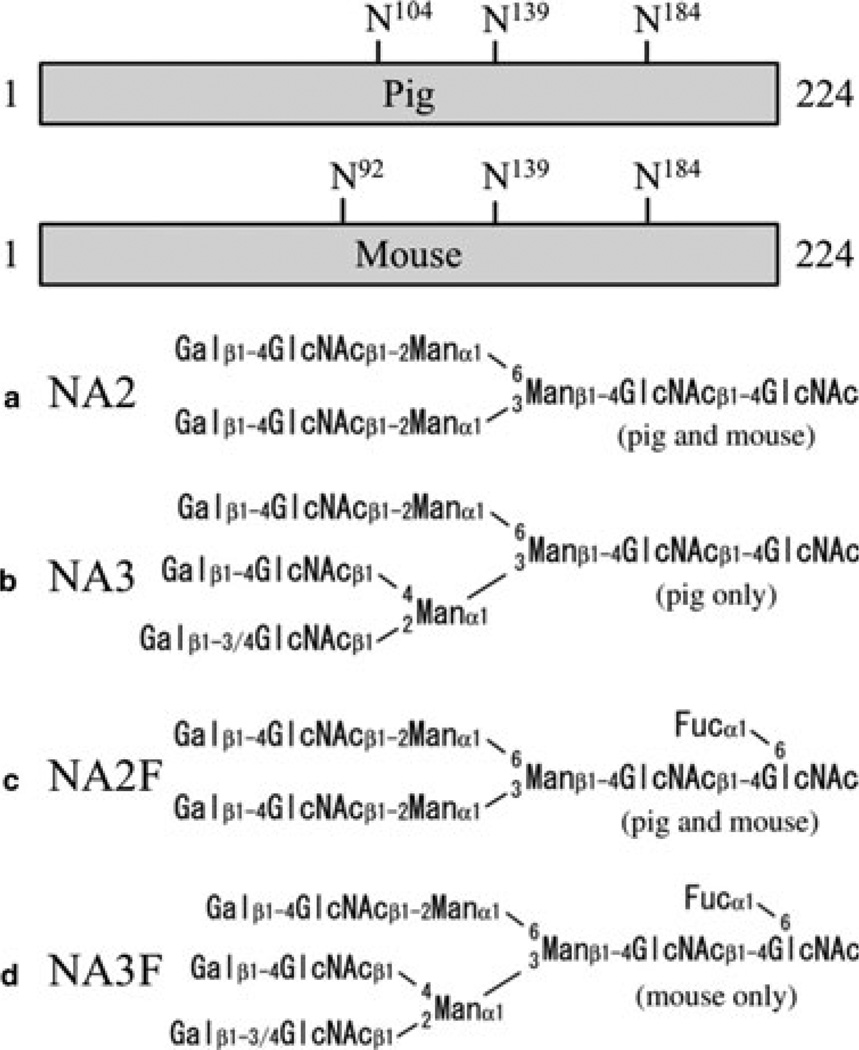
The positions of the predicted N-linked glycosylation sites on pig and mouse KLK4, along with the structures of the four N-glycan chains that modify them, are shown in Fig. 5. The differences between the N-glycan attachments for pig and mouse KLK4 are minor. Both enzymes contain three different N-glycan cores. The biantennary N-glycan cores (NA2, NA2F) are same in pig and mouse KLK4. The third core is triantennary, and shows only minor differences between pig and mouse. The mouse triantennary structure (NA3F) contains a single fucose attached to the GlcNAc at its base, while the pig structure (NA3) lacks this fucose.
Fig. 5.
Kallikrein-related peptidase 4 (KLK4) N-glycan attachment sites and core structures. Bars represent the primary sequences of the 224-amino-acid activated pig and mouse KLK4 enzymes. The positions of the three potential N-linked glycosylations are indicated. The four core glycan structures found on KLK4 are shown. The pig glycans can have 0–3 sialic acids and the mouse glycans can have 0–2 sialic acids.
Discussion
This is the first isolation of mouse KLK4 from in vivo sources and structural characterization of KLK4 glycosylations. The active pig and mouse KLK4 proteases have the same number of amino acids (n = 224) and similar calculated molecular mass values (pig = 24,117 Da; mouse = 24,179 Da) and isoelectric points (pig = 4.75; mouse = 5.54). The pig and mouse enzymes show some differences in substrate specificity: pig KLK4 is more visible on gelatin zymograms; mouse KLK4 is stronger on casein zymograms. Both enzymes are variably glycosylated with NA2 and NA2F biantennary structures, use a similar triantennary structure (pig NA3; mouse Na3F), and are heterogeneous in their sialic acid contents. Pig and mouse KLK4 both migrate as doublets on SDS-PAGE, but the apparent molecular mass of pig KLK4 is higher (Fig. 1B), suggesting that it is more glycosylated than mouse KLK4. Pig and mouse KLK4 both have three potential N-glycosylation sites, two of which are at homologous positions (Asn139 and Asn184). Our working hypothesis is that pig KLK4 can be glycosylated at all three sites, but that mouse KLK4 is glycosylated at only two, presumably at the two N-glycan attachment sites that are homologous with pig KLK4.
Human KLK4 has never been isolated from natural sources, so it is not known if it is glycosylated. Recombinant forms of human KLK4 have been characterized. Human KLK4 expressed in bacteria has a trypsin-like specificity with a strong preference for Arg at the P1 position of its substrates (18) and is inhibited by zinc (8). These findings differ from our experience with pig KLK4, which cleaves amelogenin (its natural substrate) at many sites. Among the 10 sites we characterized, only one was after arginine and one was after lysine (9). Porcine KLK4 was also active in the presence of zinc (data not shown). The commercially available recombinant human KLK4 did not appear to be glycosylated and seemed to lose activity rapidly compared with the glycosylated pig and mouse enzymes from in vivo sources. Because the pig and mouse enzymes lose activity as they are deglycosylated, it seems likely that glycosylation is important for KLK4 stability, presumably by protecting it from proteolytic degradation. The KLK4 tertiary fold is unlikely to depend upon glycosylation because it is stabilized by six disulfide bridges (10). We speculate that human KLK4 is likely to be glycosylated at Asn139, and that the glycosylated form of the enzyme will show greater stability and broader substrate specificity than the unglycosylated recombinant enzyme.
Glycosylation is an important feature of enamel proteins. Dental enamel forms in a well-defined extracellular space, the contents of which are determined by ameloblasts (19, 20). The major enamel proteins are amelogenin, ameloblastin, and enamelin (21). The enamel proteases are MMP20 and KLK4 (22, 23). Amelogenin and MMP20 are not glycosylated. Ameloblastin has O-linked glycosylations on its N-terminal (24) and C-terminal (25) domains, but has no N-linked glycosylations. Enamelin and KLK4 are both N-glycosylated. Enamelin and ameloblastin are expressed during the secretory stage and are degraded by MMP20. Their glycosylated domains preferentially accumulate in the matrix. In the pig, the 186-kDa secreted enamelin is degraded down to a 32-kDa cleavage product that is resistant to further processing by MMP20 (26), and accumulates to 1% of total enamel protein (27). Although comprised of only 104 amino acids, the 32-kDa enamelin is the most highly modified and conserved part of the enamelin (28, 29). It has three N-linked glycosylations and two phosphoserines (30, 31). The porcine 32-kDa enamelin has the same variably sialylated biantennary structure (NA2F) as KLK4. It also has a variably sialylated complex triantennary structure (G-NGA3F), which differs from the triantennary N-glycan on KLK4.
Amelogenin has a ‘tyrosyl motif’ in its primary amino acid sequence that behaves like a lectin and binds GlcNAc (32). The ability of amelogenin to bind GlcNAc has been proposed to mediate interactions between amelogenin and enamel glycoproteins. However, characterization of the N-glycosylations on enamelin and KLK4 showed that GlcNAc is never at the end of the N-glycan chains where it is accessible for amelogenin binding. Previously we demonstrated that the 32-kDa enamelin does not bind amelogenin unless it is first desialylated and then digested with β-galactosidase to remove the terminal β-galactose and expose GlcNAc at the N-glycan surface (33). There is no known glycoprotein in dental enamel with a superficial GlcNAc, so the functional significance (if any) of amelogenin binding to this monosaccharide is unknown.
Acknowledgements
We thank Mr Tom Forton, Manager of the Michigan State University Meat Laboratory, and members of the Michigan State University Department of Animal Science for their kind assistance in obtaining fresh developing molars from pigs slaughtered at their facility. We thank Kathryn Stone, Tukiet Lam, and Myron Crawford for MALDI-TOF MS analyses performed at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. This investigation was supported by USPHS Research Grants DE015846 and DE019775 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 29892.
Footnotes
All authors declare that there are no competing interests.
References
- 1.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci. 2002;110:307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 2.Simmer JP, Sun X, Yamada Y, Zhang CH, Bartlett JD, Hu JC-C. Enamelysin and kallikrein-4 expression in the mouse incisor. In: Kobayashi I, Ozawa H, editors. Biomineralization: Formation, diversity, evolution and application Proceedings of the 8th International Symposium on Biomineralization, Niigata, Jpn, Sept 25–28, 2001; Tokai University Press; Hadano, Jpn. 2004. pp. 348–352. [Google Scholar]
- 3.Simmer J, Richardson A, Smith C, Hu Y, Hu J-C. Expression of kallikrein 4 (Klk4) in dental and non-dental tissues. Eur J Oral Sci. 2011;119 Suppl. 1:226–233. doi: 10.1111/j.1600-0722.2011.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of kallikrein 4 Loss on enamel mineralization: comparison with mice lacking matrix metalloproteinase 20. J Biol Chem. 2011;286:18149–18160. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmer J, Hu Y, Richardson A, Bartlett J, Hu JC-C. Why Does Enamel in Klk4 Null Mice Break Above the Dentino-Enamel Junction? Cells Tissues Organs. 2011;194:211–215. doi: 10.1159/000324260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debela M, Magdolen V, Grimminger V, Sommerhoff C, Messerschmidt A, Huber R, Friedrich R, Bode W, Goettig P. Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site. J Mol Biol. 2006;362:1094–1107. doi: 10.1016/j.jmb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, Bartlett JD, Simmer JP. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- 10.Scully JL, Bartlett JD, Chaparian MG, Fukae M, Uchida T, Xue J, Hu CC, Simmer JP. Enamel matrix serine proteinase 1: stage-specific expression and molecular modeling. Connect Tissue Res. 1998;39:111–122. doi: 10.3109/03008209809023917. [DOI] [PubMed] [Google Scholar]
- 11.Wormald MR, Dwek RA. Glycoproteins: glycan presentation and protein-fold stability. Structure. 1999;7:R155–R160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 12.Brooks SA, Carter TM, Royle L, Harvey DJ, Fry SA, Kinch C, Dwek RA, Rudd PM. Altered glycosylation of proteins in cancer: what is the potential for new anti-tumour strategies. Anticancer Agents Med Chem. 2008;8:2–21. doi: 10.2174/187152008783330860. [DOI] [PubMed] [Google Scholar]
- 13.Jaeken J. Congenital disorders of glycosylation. Ann N Y Acad Sci. 2010;1214:190–198. doi: 10.1111/j.1749-6632.2010.05840.x. [DOI] [PubMed] [Google Scholar]
- 14.Thiel C, Korner C. Mouse models for congenital disorders of glycosylation. J Inherit Metab Dis. 2011;34:879–889. doi: 10.1007/s10545-011-9295-7. [DOI] [PubMed] [Google Scholar]
- 15.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, Arai T, Bartlett JD, Simmer JP. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu JC, Ryu OH, Chen JJ, Uchida T, Wakida K, Murakami C, Jiang H, Qian Q, Zhang C, Ottmers V, Bartlett JD, Simmer JP. Localization of EMSP1 expression during tooth formation and cloning of mouse cDNA. J Dent Res. 2000;79:70–76. doi: 10.1177/00220345000790011301. [DOI] [PubMed] [Google Scholar]
- 17.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 18.Debela M, Beaufort N, Magdolen V, Schechter NM, Craik CS, Schmitt M, Bode W, Goettig P. Structures and specificity of the human kallikrein-related peptidases KLK 4, 5, 6, and 7. Biol Chem. 2008;389:623–632. doi: 10.1515/BC.2008.075. [DOI] [PubMed] [Google Scholar]
- 19.Simmer JP, Fincham AG. Molecular mechanisms of dental enamel formation. Crit Rev Oral Biol Med. 1995;6:84–108. doi: 10.1177/10454411950060020701. [DOI] [PubMed] [Google Scholar]
- 20.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 21.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi K, Yamakoshi Y, Hu JC, Gomi K, Arai T, Fukae M, Krebsbach PH, Simmer JP. Splicing determines the glycosylation state of ameloblastin. J Dent Res. 2007;86:962–967. doi: 10.1177/154405910708601009. [DOI] [PubMed] [Google Scholar]
- 25.Yamakoshi Y, Tanabe T, Oida S, Hu CC, Simmer JP, Fukae M. Calcium binding of enamel proteins and their derivatives with emphasis on the calcium-binding domain of porcine sheathlin. Arch Oral Biol. 2001;46:1005–1014. doi: 10.1016/s0003-9969(01)00070-x. [DOI] [PubMed] [Google Scholar]
- 26.Yamakoshi Y, Hu JC-C, Fukae M, Iwata T, Simmer JP. How do MMP-20 and KLK4 process the 32 kDa enamelin? Eur J Oral Sci. 2006;114 Suppl. 1:45–51. doi: 10.1111/j.1600-0722.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 27.Tanabe T, Aoba T, Moreno EC, Fukae M, Shimizu M. Properties of phosphorylated 32 kd nonamelogenin proteins isolated from porcine secretory enamel. Calcif Tissue Int. 1990;46:205–215. doi: 10.1007/BF02555046. [DOI] [PubMed] [Google Scholar]
- 28.Al-Hashimi N, Lafont AG, Delgado S, Kawasaki K, Sire JY. The enamelin genes in lizard, crocodile, and frog and the pseudogene in the chicken provide new insights on enamelin evolution in tetrapods. Mol Biol Evol. 2010;27:2078–2094. doi: 10.1093/molbev/msq098. [DOI] [PubMed] [Google Scholar]
- 29.Al-Hashimi N, Sire JY, Delgado S. Evolutionary analysis of mammalian enamelin, the largest enamel protein, supports a crucial role for the 32-kDa peptide and reveals selective adaptation in rodents and primates. J Mol Evol. 2009;69:635–656. doi: 10.1007/s00239-009-9302-x. [DOI] [PubMed] [Google Scholar]
- 30.Yamakoshi Y. Carbohydrate moieties of porcine 32 kDa enamelin. Calcif Tissue Int. 1995;56:323–330. doi: 10.1007/BF00318054. [DOI] [PubMed] [Google Scholar]
- 31.Yamakoshi Y, Pinheiro FH, Tanabe T, Fukae M, Shimizu M. Sites of asparagine-linked oligosaccharides in porcine 32 kDa enamelin. Connect Tissue Res. 1998;39:39–46. doi: 10.3109/03008209809023910. [DOI] [PubMed] [Google Scholar]
- 32.Ravindranath R, Moradian-Oldak J, Fincham A. Tyrosyl motif in amelogenins binds N-acetyl-D-glucosamine. J Biol Chem. 1999;274:2464–2471. doi: 10.1074/jbc.274.4.2464. [DOI] [PubMed] [Google Scholar]
- 33.Yamakoshi Y, Hu J-C, Fukae M, Tanabe T, Oida S, Simmer J. Amelogenin and 32 kDa enamelin protein-protein interactions. In: Kobayashi I, Ozawa H, editors. Biomineralization (Biom2001) formation, diversity, evolution and application. Kanagawa, Jpn: Tokai University Press; 2004. pp. 338–342. [Google Scholar]
- 34.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis Tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa, NJ: Humana Press; 2005. pp. 571–608. [Google Scholar]