SUMMARY
The altered metabolism of tumors has been considered a target for anti-cancer therapy. However, the relationship between distinct tumor-initiating lesions and anomalies of tumor metabolism in vivo has not been addressed. We report that MYC-induced mouse liver tumors significantly increase both glucose and glutamine catabolism, whereas MET-induced liver tumors use glucose to produce glutamine. Increased glutamine catabolism in MYC-induced liver tumors is associated with decreased levels of glutamine synthetase (Glul) and the switch from Gls2 to Gls1 glutaminase. In contrast to liver tumors, MYC-induced lung tumors display increased expression of both Glul and Gls1 and accumulate glutamine. We also show that inhibition of Gls1 kills cells that over-express MYC and catabolize glutamine. Our results suggest that the metabolic profiles of tumors are likely to depend on both the genotype and tissue of origin and have implications regarding the design of therapies targeting tumor metabolism.
INTRODUCTION
Increased consumption and altered metabolism of two major nutrients, glucose and glutamine, are often observed in tumors and cancer cell lines (reviewed in (Baggetto, 1992; Gatenby and Gillies, 2004; Medina, 2001). Pathways of glucose and glutamine metabolism fuel processes vital for cellular proliferation and survival and can be controlled by several oncogenes involved in human cancers. For example, the MYC proto-oncogene regulates the expression of several enzymes of glycolysis, the Krebs cycle, mitochondrial respiration and nucleotide synthesis (http://www.myccancergene.org), as well as glutamine transporters and glutaminase (Gao et al., 2009; Wise et al., 2008), the first enzyme of glutamine catabolism; hepatocyte growth factor (HGF)/scatter factor and its tyrosine kinase receptor MET regulate carbohydrate metabolism (Kaplan et al., 2000; Perdomo et al., 2008); PI3K/AKT and RAS/MAPK signaling pathways, which are initiated by HGF binding to MET (Vogelstein and Kinzler, 2004), are well-known regulators of cellular metabolism (reviewed in (Yuneva, 2008); β-catenin, a central player in the Wnt pathway (Klaus and Birchmeier, 2008), regulates genes responsible for glutamine synthesis (Cadoret et al., 2002); and β-catenin activation in mouse liver is associated with altered expression of proteins regulating glycolysis and mitochondrial activity (Chafey et al., 2009). These reports provide important clues to understanding how expression of oncogenes promote metabolic changes and indicate that such changes might vary among tumors carrying distinct oncogenic lesions. However, most of these studies were either performed in cell lines or were limited mainly to evaluating protein expression profiles that do not necessarily reflect the activity of metabolic pathways.
In this study, we used stable isotope-based metabolic analysis to evaluate how glucose and glutamine metabolism is altered during tumorigenesis initiated by individual oncogenes in vivo. We also assessed the expression and activity of the main enzymes regulating the metabolism of these two nutrients. In order to compare the effect of two distinct oncogenes, we used well-characterized mouse models of liver cancer induced by tissue-specific over-expression of either human MYC (Shachaf et al., 2004) or MET (Wang et al., 2001). Mouse liver tumors induced by MYC resemble immature hepatoblastomas with high mitotic activity (Shachaf et al., 2004; Tward et al., 2005), whereas tumors induced by MET resemble differentiated human hepatocellular carcinomas (HCC) (Tward et al., 2005; Wang et al., 2001). The tumors induced by MET carry spontaneous activating mutations of β-catenin (Tward et al., 2007). Abnormal expression and activity of MYC (Kaposi-Novak et al., 2009; Thorgeirsson and Grisham, 2002), activating mutations and increased expression of MET (Kaposi-Novak et al., 2006; Ueki et al., 1997) and activating mutations of β-catenin (de La Coste et al., 1998; Tward et al., 2007) occur frequently in human liver cancers.
To evaluate whether the effect of oncogenic transformation on metabolism depends on the tissue context, we used a model of lung cancer induced by over-expression of MYC (Allen et al., 2011). Increased expression of MYC family members is observed in some human lung cancers (Gazzeri et al., 1994; Yokota et al., 1988), and MYC over-expression in the lungs of mice causes adenocarcinomas (Allen et al., 2011; Tran et al., 2008).
We found that glucose and glutamine metabolism in tumors varies with both the nature of the initiating lesion (MYC or MET) and the tissue of origin. Moreover, inhibition of Gls1, an isoform of glutaminase over-expressed in MYC-induced tumors, causes apoptosis in cells that have high levels of MYC and use glutamine to fuel the Krebs cycle. We conclude that metabolic profiling will be essential to determine the nature of metabolic alterations in tumors and to choose proper therapeutics aimed at these alterations.
RESULTS
MYC and MET Have Different Effects on Glucose Metabolism in the Liver
One of the hallmarks of cancer is aerobic glycolysis (the Warburg effect) associated with increased consumption of glucose and increased levels of lactate, a product of glucose catabolism (Warburg, 1956). Therefore, we first examined whether glucose metabolism was changed in mouse tumors induced by either MYC or MET transgenes expressed specifically in the liver of FVBN mice under the control of a Tet-repressible promoter (Shachaf et al., 2004; Wang et al., 2001). Expression of the transgenes and tumor formation were induced by the removal of doxycycline from a diet. Transgenic animals kept on a doxycycline diet and wild type FVBN mice were used as controls.
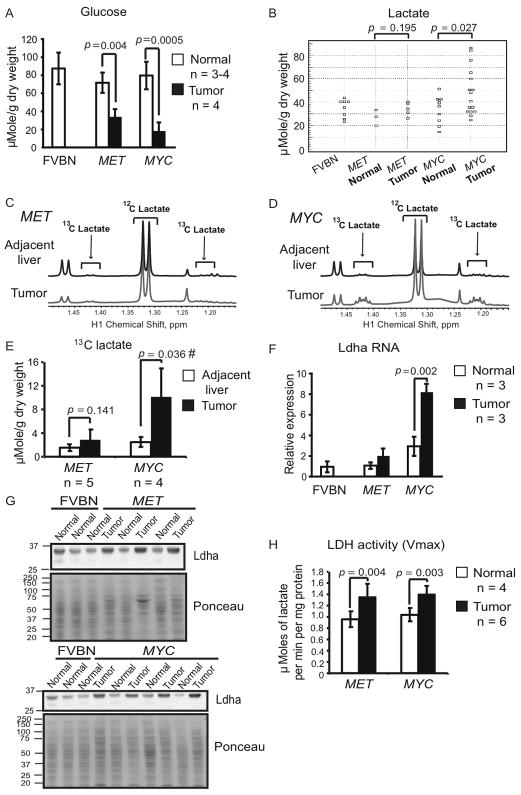
We found that the levels of glucose were below normal in tumors induced by either MYC or MET (Figure 1A). Consistent with this, the expression of glucose-6-phosphatase, one of the key enzymes of glucose biosynthesis, was decreased in both types of tumors (Figures S1A and S1B), suggesting that glucose synthesis might be deficient in both types of tumors. In MYC-induced tumors decreased levels of glucose were frequently associated with increased levels of lactate in comparison with normal liver (Figure 1B). Moreover, lactate levels in MYC-induced tumors were significantly higher than in the ostensibly normal liver tissue adjacent to tumors (Figures S1C and S1D). Although in three out of seven MET-induced tumors we observed levels of lactate higher than in the adjacent tissue (data not shown), overall, MET-induced tumors did not demonstrate a statistically significant increase of lactate levels in comparison with either normal liver (Figure 1B) or adjacent liver tissue (Figures S1C and S1D).
Figure 1. Lactate Levels are Significantly Increased in MYC-induced but not in MET-induced Murine Liver Tumors.
(A and B) Glucose and lactate levels in tumor and normal liver tissue extracts measured by enzyme-based assays. Normal tissue was from wild type FVBN mice and transgenic mice kept on doxycycline. (C and D) Representative profiles of 1H NMR performed on extracts from tumors and adjacent liver tissues from animals injected with [U-13C]-glucose (number of animals analyzed is indicated in 1E). Two satellite peaks at 1.21 and 1.42 ppm represent fully-labeled 13C-lactate. The profiles were normalized to the initial dry weight of tissue. (E) Quantification of 13C-lactate concentration in the samples from 1C and 1D. #, p = 0.050 for MET-induced tumors vs. MYC-induced tumors. (F) Ldha RNA levels estimated by real-time PCR in the samples outlined in A and B. Mouse β-actin served as an internal control. One of the FVBN normal livers was used as a reference. (G) Ldha expression estimated by immunoblotting of whole tissue lysates of normal livers and tumors. Ponceau staining of membranes was used to confirm the protein loading. (H) LDH activity measured in the cytosolic fractions of tumors and normal livers from transgenic mice kept on doxycycline. All values are given as the mean ± s.d. See also Figure S1.
In order to test if glucose catabolism contributes to increased lactate levels in MYC-induced tumors we administered uniformly labeled 13C-glucose, [U-13C]-glucose, to tumor-bearing animals. Administration of [U-13C]-glucose resulted in increase of 13C-lactate levels in liver tumors induced by MYC (Figures 1D, 1E and 4C). In contrast, MET-induced tumors displayed only slight or no increase in 13C-lactate levels, which overall was not statistically significant (Figures 1C, 1E and 4C). These results indicate that increased lactate levels in MYC-induced tumors may be, at least in part, the result of increased lactate production from glucose.
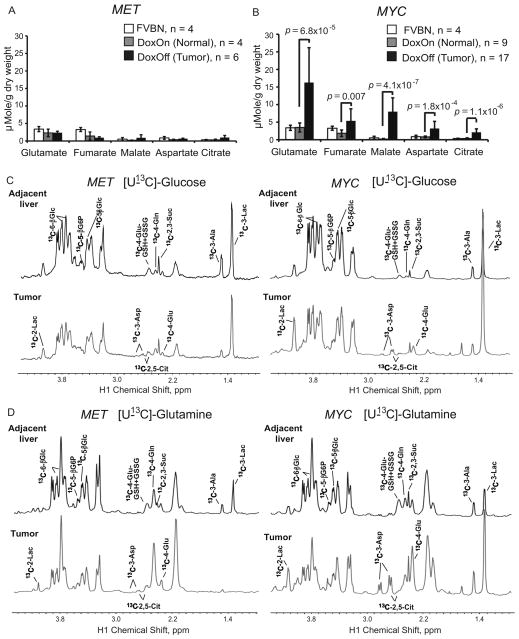
Figure 4. Catabolism of Glucose and Glutamine Contributes to Increased Synthesis of Krebs Cycle Intermediates in MYC-induced Liver Tumors.
(A and B) The levels of Krebs cycle intermediates measured in tissue extracts by GC-MS. Values are given as the mean ± s.d. (C and D) The representative spectra of 1H-13C HSQC NMR performed on extracts of tumor-adjacent and tumor tissues from animals injected with either [U-13C]-glucose (C) or [U-13C]-glutamine (D) (at least 3 animals per group). The profiles were normalized to the initial dry weight. Glc, glucose; G6P, glucose-6-phosphate; Glu, glutamate; GSH, reduced glutathione; GSSG, oxidized glutathione; Gln, glutamine; Asp, aspartate; Suc, succinate; Ala, alanine; Lac, lactate. See also Figures S4.
Lactate is produced from pyruvate, the end product of glucose catabolism through glycolysis, by lactate dehydrogenase (LDH). Increased activity of LDH and increased expression of its A subunit (Ldha), which favors rapid conversion of pyruvate into lactate, are observed in human cancers (Goldman et al., 1964). Unexpectedly, although the levels of Ldha RNA were increased only in MYC-induced tumors (Figure 1F), the levels of Ldha protein (Figure 1G) and total LDH activity (Figure 1H) were increased to the same extent in both types of tumors. This demonstrates that increased LDH activity is not the only factor responsible for the difference in lactate levels between tumors induced by the two oncogenes.
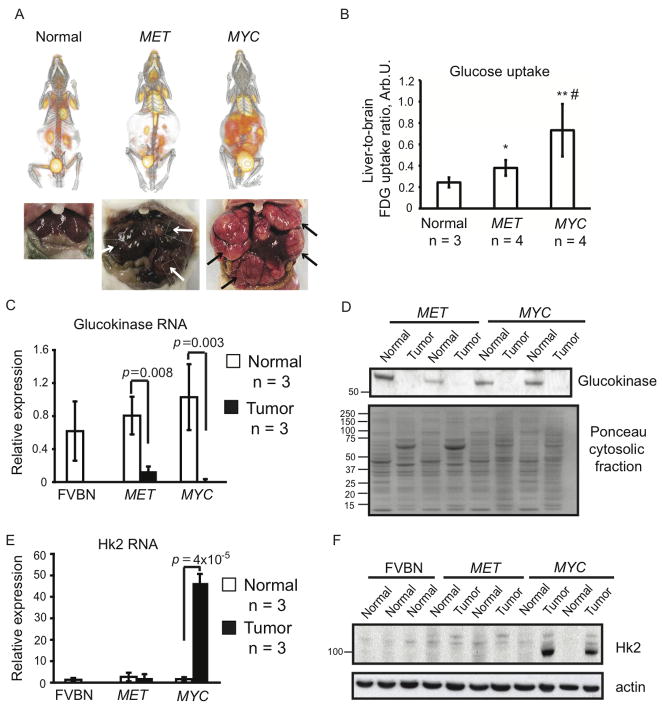
To understand why increased LDH activity was accompanied by higher levels of glucose-derived lactate in MYC-induced tumors than in MET-induced tumors, we compared glucose transport and catabolism in both types of tumors. To analyze glucose transport we used 18F-fluorodeoxyglucose (FDG) and positron emission tomography/computed tomography (PET/CT). Although glucose transport was above normal in both types of tumors, the intensity of FDG uptake was significantly higher in MYC-induced tumors than in MET-induced tumors (Figures 2A and 2B). The significantly higher glucose transport and comparably lower glucose levels in MYC-induced tumors suggest that these tumors catabolize glucose faster than MET-induced tumors.
Figure 2. Glucose Levels are Controlled Differently in Tumors Induced by either MYC or MET.
(A) PET/CT imaging of 18F-FDG uptake performed on normal and tumor-bearing animals. After imaging tumor presence was confirmed as demonstrated by photographs under each scan image. Tumors are indicated by arrows. Images of representative animals are shown. (B) 18F-FDG uptake quantified as described in Experimental Procedures. The normal group contained one FVBN mouse, and one each of mice carrying either MYC or MET transgenes and kept on doxycycline. *, p = 0.038 for MET vs. normal, **, p = 0.023 for MYC vs. normal and #, p = 0.036 for MET vs. MYC. (C and E) RNA levels for glucokinase (C) and Hk2 (E) measured in normal and tumor tissues using mouse β-actin as an internal control. Results are normalized as in Fig. 1F. Values are given as the mean ± s.d. (D) Glucokinase expression estimated in the cytosolic fractions of normal and tumor tissues by immunoblotting. Ponceau staining was used to confirm the protein loading. (F) Hk2 protein levels measured in whole tissue lysates from normal and tumor samples by immunoblotting using β-actin as a loading control. See also Figure S2 and Table S1.
We next compared the regulation of the first step of glucose catabolism, glucose phosphorylation, in the two types of tumors. In normal hepatocytes glucose phosphorylation is catalyzed by glucokinase, which has a high Km (low affinity) for glucose and, in its active form, is localized in the cytosol (Bustamante et al., 2005). We found that glucokinase expression was decreased in MYC-induced tumors (Figures 2C and 2D). Consistent with this, we detected glucose-6-phosphorylating activity with a high Km (17 mM) in the cytosolic fraction of normal liver but not in the cytosolic fraction of MYC-induced tumors (Table S1). In contrast to MYC-induced tumors, decreased expression of glucokinase in MET-induced tumors (Figures 2C and 2D) was associated with the emergence of glucose-6-phosphorylating activity in the cytosolic fraction of these tumors that had both decreased Km and Vmax compared to normal liver (Table S1). These results suggest that, although MET-induced tumors may have less glucokinase in comparison with normal liver, it may be a more efficient form of glucokinase.
Hexokinase II (Hk2) is a hexokinase isoform that has a low Km (high affinity) for glucose and can be over-expressed in human tumors and cancer cell lines (Bustamante et al., 1981). Hk2 localizes both in the cytosol and at the mitochondrial membrane. Mitochondrial localization of Hk2 may increase its efficiency (Bustamante et al., 1981). We found that Hk2 expression was significantly increased in tumors induced by MYC but not in those induced by MET (Figures 2E and 2F). Consistent with this, glucose-6-phosphorylating activity with a low Km (0.01–0.08 mM) appeared in the mitochondrial fraction and increased 10-fold in the cytosolic fraction of MYC-induced tumors, while staying unchanged in MET-induced tumors (Table S1).
In addition to Hk2, tumors induced by MYC, but not MET, had increased expression of liver- and platelet-specific subunits of phosphofructokinase (Figure S2), one of the key glycolytic enzymes. Higher abundance of these subunits in the holo-enzyme contributes to acceleration of glucose catabolism (Vora et al., 1985).
Altogether our results suggest that glucose transport and catabolism may be increased in both types of tumors in comparison with normal liver. However, MYC-induced liver tumors have significantly higher glucose uptake and catabolism than MET-induced liver tumors, which may explain why MYC-induced tumors have significantly increased levels of lactate whereas MET-induced tumors do not.
MYC and MET Differently Affect Glutamine Metabolism in the Liver
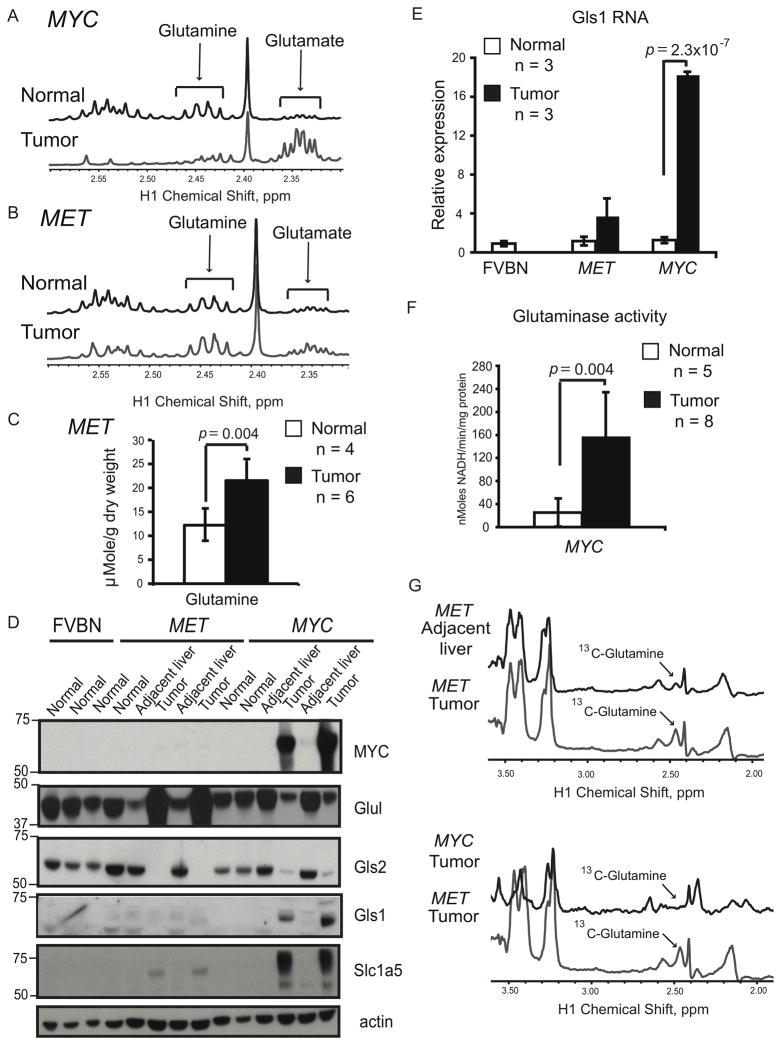
Glutamine is another major nutrient whose metabolism can be altered during tumorigenesis (Marquez et al., 1989). Nuclear magnetic resonance (NMR) analysis revealed that glutamine levels were undetectable in MYC-induced tumors (Figure 3A), whereas glutamine levels increased almost 2-fold in MET-induced tumors compared to normal liver (Figures 3B and 3C).
Figure 3. Glutamine Metabolism is Different in Liver Tumors Induced by either MYC or MET.
(A and B) Glutamine levels determined by NMR in tumors and normal livers from transgenic mice kept on doxycycline. Representative 1H NMR profiles of normal livers and tumors are shown (at least four animals per group). Glutamine peaks are in 2.42–2.47 ppm region and glutamate peaks are in 2.32–2.37 ppm region. The profiles were normalized to the initial dry weight. (C) Glutamine concentration quantified in normal livers and MET-induced tumors based on 1H-NMR data in B. (D) Expression of indicated proteins evaluated by immunoblotting of whole tissue lysates using β-actin as a loading control. (E) Gls1 RNA levels measured in normal and tumor tissues using mouse β-actin as an internal control. (F) Phosphate-dependent glutaminase activity measured in the mitochondrial fraction of MYC-induced tumors and normal livers from transgenic mice kept on doxycycline. All values are given as the mean ± s.d. (G) Detection of 13C-labeled glutamine by 1H-13C heteronuclear single quantum coherence spectroscopy (HSQC) NMR performed on extracts of tumor-adjacent and tumor tissues from animals injected with [U-13C]-glucose (5 animals per group). The representative HSQC spectra are shown. 13C-4-glutamine peak is at 2.45 ppm. Consistent with glutamine depletion a corresponding 13C-4-glutamine peak was not observed in MYC-induced tumors. The profiles were normalized to the initial dry weight of tissue. See also Figure S3.
To reconcile the differences in the levels of glutamine between tumors induced by the two oncogenes, we evaluated the expression of two enzymes: glutamine synthetase (Glul), which catalyzes the ligation of ammonia and glutamate to produce glutamine; and phosphate-dependent glutaminase, which is responsible for glutamine catabolism into glutamate. Liver-type glutaminase (Gls2) is expressed in adult periportal hepatocytes, brain and pancreas, whereas kidney-type glutaminase (Gls1) is expressed in embryonic hepatocytes, in most adult tissues with the exception of the postnatal liver, and in some cancer cell lines (Curthoys and Watford, 1995; Perez-Gomez et al., 2005). We found that in MYC-induced tumors the expression of Glul was suppressed in comparison with normal liver (Figures 3D and S3A) and the expression of the Gls2 isoform of glutaminase was replaced by the Gls1 isoform (Figures 3D, 3E and S3B). Importantly, the shift in glutaminase isoforms was associated with an increase in glutaminase activity (Figure 3F). Finally, MYC-induced tumors exhibited increased expression of the high-affinity glutamine transporter Slc1a5 (Figures 3D and S3C), which is thought to supply cancer cell lines with amounts of glutamine sufficient for its increased catabolism (Bode, 2001). These results indicate that glutamine synthesis is decreased in MYC-induced tumors, whereas its transport and catabolism are increased.
In contrast to MYC-induced tumors, MET-induced tumors had increased expression of Glul (Figures 3D and S3A), which was consistent with the presence of activating mutations in β-catenin (Tward et al., 2007), a transcriptional activator for Glul (Cadoret et al., 2002). Moreover, in MET-induced tumors Gls2 expression was decreased (Figures 3D and S3B), Gls1 was not expressed (Figures 3D and 3E) and the expression of Slc1a5 was only slightly higher than normal (Figures 3D and S3C). Consistent with the expression pattern of glutamine-metabolizing enzymes, administration of uniformly labeled 13C-glutamine, [U-13C]-glutamine, to tumor-bearing mice resulted in significant build-up of 13C-glutamine in MET-induced tumors (Figure 4D, right panel), indicating that glutamine was transported into tumors, but its catabolism was significantly decreased in comparison with normal liver.
One of the main substrates for glutamine biosynthesis is glucose. Administration of [U-13C]-glucose resulted in increased levels of 13C-glutamine in MET-induced tumors (Figure 3G and Figure 4C, left panel). Together these results indicate that in MET-induced tumors, increased consumption of glucose contributes to increased glutamine biosynthesis, and the elevated levels of glutamine could result from both increased synthesis and repressed catabolism. We conclude that glutamine catabolism is increased in MYC-induced tumors, whereas glutamine production is increased in MET-induced tumors.
MYC-induced Tumors Have Increased Synthesis of Krebs Cycle Intermediates
Since glucose and glutamine are the main fuels of the Krebs cycle, we compared total levels of the Krebs cycle intermediates (fumarate, malate and citrate), and related metabolites (glutamate and aspartate) in the two types of tumors and normal liver. The levels of all these compounds were increased in MYC-induced but not in MET-induced tumors (Figures 3A, 3B, 4A and 4B). Moreover, administration of either [U-13C]-glucose or [U-13C]-glutamine resulted in increased levels of 13C-citrate, 13C-glutamate and 13C-aspartate in MYC-induced tumors (Figures 4C, 4D, S4A and S4B). This contrasted with MET-induced tumors, which displayed either insignificant or no changes (Figures 4C, 4D, S4A and S4B). These results suggest that increased transport and catabolism of both glucose and glutamine in MYC-induced tumors contribute to the synthesis of Krebs cycle intermediates, which is significantly enhanced in comparison with normal liver and MET-induced tumors.
Injection of [U-13C]-glutamine also resulted in increased incorporation of 13C into lactate in MYC-induced tumors (Figure 4D, right panel), indicating that increased catabolism of glutamine may contribute to the increased lactate levels observed in these tumors. However, currently we cannot discern whether labeled lactate was produced from glutamine through glutaminolysis (DeBerardinis et al., 2008) or was the result of increased catabolism of glucose that was derived from [U-13C]-glutamine through gluconeogenesis.
Tissue Context Affects the Outcome of Metabolic Reprogramming by MYC
To evaluate if metabolic changes induced by oncogenic transformation are tissue-specific, we analyzed the main pathways of glucose and glutamine metabolism in lung adenocarcinomas induced by MYC (Allen et al., 2011). Expression of MYC was driven specifically in pneumocytes of FVBN mice by a Tet-activated promoter by keeping mice on a doxycycline diet (Allen et al., 2011; Tran et al., 2008). Normal lungs from wild type mice and mice fed a diet without doxycycline were used as controls.
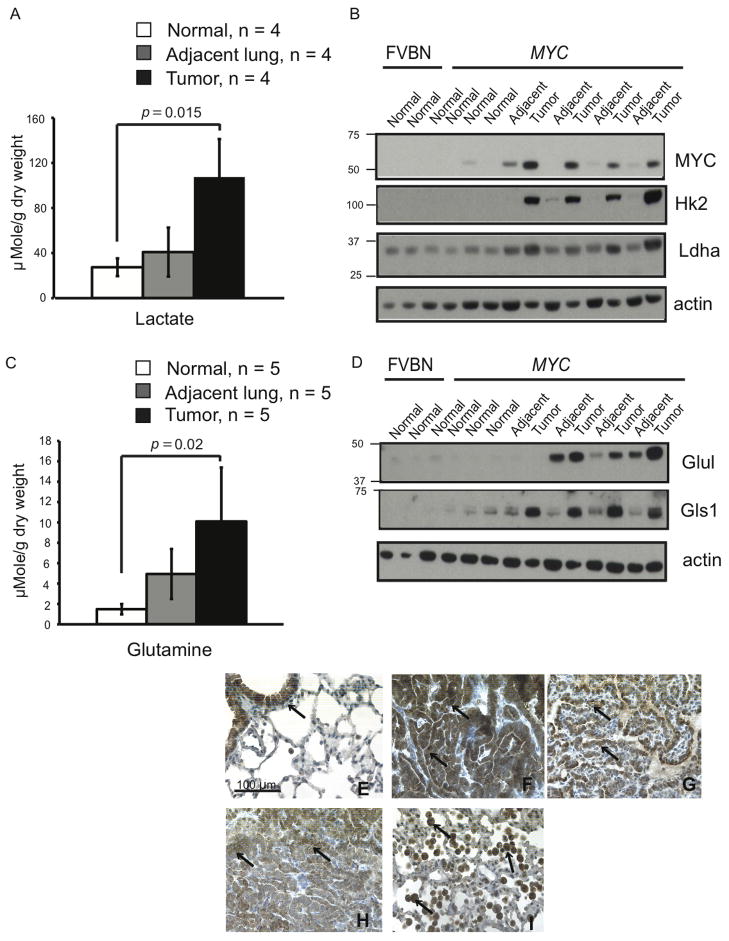
We found that the levels of lactate (Figure 5A) as well as expression of both Hk2 and Ldha (Figure 5B) were increased in lung tumors induced by MYC. This suggested that, similar to liver tumors, MYC-induced transformation was associated with increased glucose catabolism into lactate in lung tumors. In contrast to liver tumors, however, lung tumors induced by MYC had elevated levels of glutamine (Figure 5C).
Figure 5. Metabolism of MYC-induced Lung Tumors Differs from Metabolism of MYC-induced Liver Tumors.
(A) Lactate levels measured by 1H NMR in lung tumors induced by MYC (Tumor) and either adjacent lung lobes from the same animals (Adjacent lung) or normal lungs from mice with MYC transgene kept without doxycycline (Normal). Values are given as the mean ± s.d. (B and D) The levels of indicated proteins estimated by immunoblotting in whole tissue lysates using β-actin as a loading control. (C) Glutamine levels in tissue extracts determined by GC-MS. Values are given as the mean ± s.d. (E–I) Expression of Glul examined by immunochemistry in normal lung (E), MYC-induced tumors (F–H) and adjacent lung (I). Expression of Glul is apparent in bronchial epithelium of normal lung, in tumor cells and in infiltrating immune cells of tumor-adjacent lungs (indicated by arrows).
Although expression of Gls1 was increased in MYC-induced lung tumors, expression of Glul also appeared to be above normal in three out of four lung tumors (Figure 5D). Immunohistochemical analysis revealed that in normal lungs Glul expression was restricted to bronchial epithelium (Figure 5E), whereas in lung tumors Glul was highly expressed in tumor cells (Figures 5F–H). The expression of Gls2 in normal lungs was lower than in the liver and did not change in lung tumors (Figure 6A and data not shown). By analogy with previous experiments conducted on human lung cancer patients infused with [U-13C]-glucose (Fan et al., 2009), increased glutamine production in MYC-induced mouse lung tumors might be the result of increased glucose conversion into glutamine. Although glutamine catabolism may also be increased in lung tumors due to the increased Gls1 expression, increased Glul expression could override increased Gls1 expression and account for increased net glutamine levels.
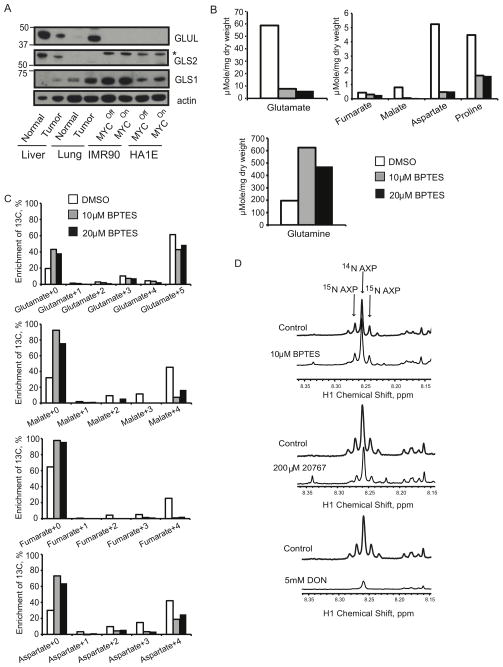
Figure 6. GLS1 Inhibitors Affect Glutamine Catabolism through the Krebs Cycle.
(A) IMR90-ERMYC and HA1E-MYCER cells were incubated for 24 hours with either OHT (MYCOn) or 0.1% ethanol (MYCOff). Expression of GLUL, GLS2 and GLS1 was detected in whole lysates of these cells, normal liver and lung as well as of MYC-induced liver and lung tumors by immunoblotting using β-actin as a loading control. *, indicates cross-reactive band recognized by anti-Gls2 antibody in human cell lines. (B and C) IMR90-ERMYC cells with induced MYC activity were incubated with 2 mM [U-13C]-glutamine and indicated concentrations of BPTES for 12 hours. 0.1% DMSO was used as a vehicle control. Absolute concentrations (B) and 13C isotopologue enrichment (C) of indicated compounds were measured by GC-MS. In C metabolate+0 represents monoisotopic or unlabeled isotopologue while metabolite+1 to 5 are isotopologues with 1 to 5 13C atoms. The results were referenced to the initial dry weight of cell pellet. Average of two replicate samples is presented. The experiment was repeated twice. (D) IMR90-ERMYC cells with induced MYC were incubated with 2 mM 15N2-glutamine and indicated concentrations of BPTES, 20767 or DON. Incorporation of 15N into adenine nucleotides (AXP) was evaluated by 1H NMR. The profiles are referenced to the dry weight of cell pellet and the internal standard concentration. The representative profiles from two independent experiments are presented. See also Figure S5 and S6.
Increased levels of Glul protein and glutamine were also observed in ostensibly normal lung tissue adjacent to mouse tumors (Figures 5C and 5D). Immunohistochemical analysis showed that Glul was expressed in immune cells infiltrating the normal tissue (Figure 5I), which could account at least in part for the increased levels of Glul and glutamine.
Together these data demonstrate that, although some mechanisms that regulate metabolic pathways in MYC-induced liver tumors are similarly altered in MYC-induced lung tumors, tissue-specific differences in the outcome do exist.
MYC Sensitizes Cells to Inhibition of Glutaminase
We have previously demonstrated that glutamine metabolism through the Krebs cycle is required for the survival of cells that over-express MYC (Yuneva et al., 2007). Since the shift from Gls2 to Gls1 accompanies the increase in glutamine catabolism through the Krebs cycle in MYC-induced mouse liver tumors (Figures 3D and 4D), we reasoned that Gls1 could be a plausible target for eliminating cells that catabolize glutamine and over-express MYC. To test this possibility, we used normal human lung fibroblasts (IMR90) and immortalized human kidney epithelial cells (HA1E) expressing conditional versions of MYC (IMR90-ERMYC and HA1E-MYCER) that can be activated by 4-hydroxytamoxifen (OHT) (Yuneva et al., 2007), as well as HA1E cells that constitutively over-express wild-type MYC (HA1E-MYC). We found that all of these lines had metabolic features resembling those of MYC-induced liver tumors, irrespective of the presence or absence of active ectopic MYC, namely: (i) levels of GLS1 comparable to those in the tumors (Figures 6A and 7D); (ii) prevalence of GLS1 over GLS2 expression (Figure 6A); (iii) low levels of GLS2 and GLUL (Figure 6A). Moreover, all of the cell lines had glucose as the primary source of lactate (Figures S5A, S5C, and data not shown) and glutamine as the principal fuel for the Krebs cycle (Figures S5B, S5D and data not shown). We attribute this phenotype to the relative abundance of endogenous MYC in the cell lines (Figure 7D and data not shown), which could represent adaptation to sustained propagation in vitro. It remains the case, however, that a further increase in MYC activity increases the sensitivity of HA1E and IMR90 cells to glutamine deprivation (Yuneva et al., 2007). These results indicate that different levels of MYC might be required to change a metabolic phenotype of the cells and to make them dependent on glutamine for survival. We proceeded to test the response of IMR90 and HA1E cells to inhibition of GLS1 in the absence and presence of augmented MYC activity.
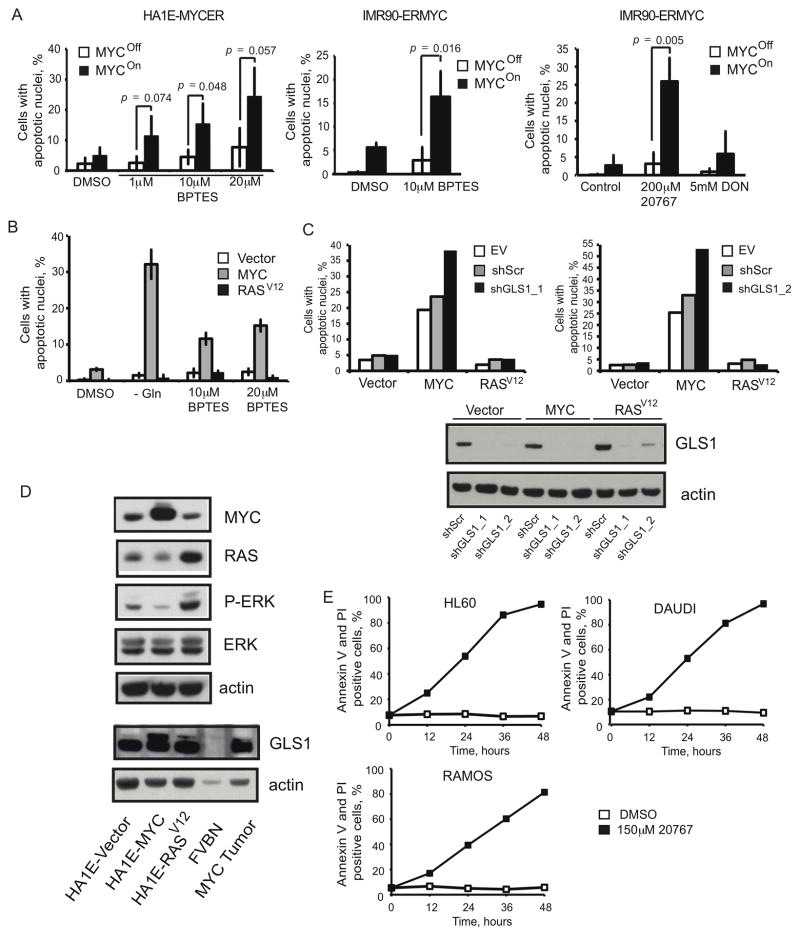
Figure 7. GLS1 Inhibitors Induce MYC-dependent Apoptosis.
(A) MYC activity was induced in HA1E-MYCER and IMR90-ERMYC cells as described in Fig. 6A. Cells then were treated with indicated concentrations of BPTES or 20767 or with 5 mM DON. 0.1% DMSO was used as a vehicle control for the experiments with BPTES. HA1E-MYCER cells were treated for 36 hours and IMR90-ERMYC cells were treated for 48 hours and percentage of apoptotic nuclei was scored. The average of three independent experiments is presented ± s.d. (B) HA1E cells transduced with either empty vector or vectors carrying either MYC or H-RASV12 were either incubated in glutamine-free media or treated with indicated concentrations of BPTES for 24 hours. Apoptosis was scored in three independent replicate samples. The values are given as the mean ± s.d. A representative of two independent experiments is shown. (C) HA1E-derived cell lines were infected with lentiviruses expressing either a scrambled shRNA (shScr) or two independent shRNAs targeting GLS1 (shGLS1_1 and shGLS1_2). Cells were collected 36 or 48 hours after the infection, GLS1 expression was evaluated by immunoblotting, and apoptosis was scored by counting apoptotic nuclei. For each shRNA the experiment was repeated at least twice. Transduction with empty pLKO.1 vector (EV) was used to account for the exceptional level of cell death induced by lentiviral infection observed specifically in HA1E cells with ectopic expression of MYC. (D) The ectopic expression of MYC and H-RASV12, as well as expression of ERK, P-ERK and GLS1 was estimated by western blotting in whole cell lysates. Increased expression of PERK indicates increased activity of RAS. (E) Indicated Burkitt’s lymphoma cell lines were cultured in the presence of 150 μM 20767. Cell death was analyzed by Annexin V and PI staining. A representative of two independent experiments is shown.
We used an inhibitor of GLS1, bis-2-(5-phenylacetoamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) (Robinson et al., 2007), and its water-soluble analog, 20767 (Erdmann et al., 2007). Together with increasing the levels of glutamine, both BPTES and 20767 decreased the levels of glutamate and other Krebs cycle intermediates and their products (Figures 6B, S6A and data not shown), as well as the incorporation of glutamine-derived 13C into these compounds (Figures 6C, S6B and data not shown) in IMR90-ERMYC cells incubated either with or without OHT. Importantly, unlike 6-diazo-5-oxo-L-norleucine (DON), a glutamine analog and inhibitor of both glutaminase and amidotransferases, neither BPTES nor 20767 inhibited the incorporation of glutamine-derived 15N into adenine nucleotide species (Figure 6D). These results indicate that both BPTES and 20767 inhibit glutaminase activity but spare amidotransferase activities involved in nucleotide biosynthesis.
Both BPTES and 20767 induced apoptosis in IMR90-ERMYC and HA1E-MYCER cells in a MYC-dependent manner (Figure 7A). Moreover, HA1E-MYC cells were more sensitive to BPTES treatment than HA1E transduced with a vector control (HA1E-Vector) (Figure 7B). Silencing GLS1 expression by two independent shRNAs in HA1E-MYC and HA1E-Vector cells confirmed that the BPTES effect on the viability of cells with ectopically-expressed MYC was due to inhibition of GLS1 activity (Figure 7C). Treatment with 20767 also killed human Burkitt’s lymphoma cell lines that over-express MYC as a consequence of well-characterized translocations of MYC (Figure 7E). In contrast to MYC, ectopic expression of H-RASV12 did not sensitize HA1E cells to either glutamine deprivation or inhibition of GLS1 activity (Figures 7B and 7C).
We conclude that MYC but not H-RASV12 exhibits a synthetic lethal interaction with the inhibition of GLS1 activity and that over-expression of MYC may be a valuable biomarker for sensitivity to such inhibition. Moreover, specific inhibition of glutaminase might be less toxic than a therapy employing glutamine analogs that may interfere with nucleotide biosynthesis and thus induce DNA damage (Hastak et al., 2008).
DISCUSSION
Determinants of Tumor Metabolism
Here we demonstrate that liver tumors induced by either MYC or MET have distinct metabolic phenotypes. Moreover, the metabolism of MYC-induced liver tumors differs from the metabolism of MYC-induced lung tumors. Some of the metabolic changes that are observed during MYC-induced tumorigenesis in both liver and lung, including increased expression of Hk2, Ldha and Gls1, may be the result of MYC over-expression itself (Gao et al., 2009; Osthus et al., 2000; Wise et al., 2008). On the other hand, there are differences between liver and lung tumors induced by MYC that may be due to other tumorigenic events or determined by tissue-specific functions. For example, MYC-induced lung tumors are distinguished from those in the liver by increased expression of Glul. Whereas, to the best of our knowledge, regulation of Glul expression by MYC has never been reported, Glul is a well-known transcriptional target of β-catenin (Cadoret et al., 2002), which is activated in a subset of human lung cancers (Sunaga et al., 2001). However, our immunohistochemical analysis revealed that in MYC-induced lung tumors, β-catenin is located at the cytoplasmic membrane rather than in the nucleus (data not shown), indicating that the protein had not been activated by mutation and is not likely the cause of Glul over-expression. During inflammation, expression of Glul can be stimulated by glucocorticoids in lung epithelial cells (Abcouwer et al., 1996). This suggests that increased Glul expression in MYC-induced lung tumor cells may be induced by tumor-associated inflammation.
Although in liver tumors the expression of both Gls1 and Glul is regulated at the RNA level (Figures 3 and S3A), the expression of these proteins in lung tumors is regulated at the protein level (data not shown). This observation is consistent with the ability of MYC to regulate protein expression post-transcriptionally (Chang et al., 2008) and with variations in the regulation of Gls1 expression by MYC among cell lines of different origin (Gao et al., 2009; Wise et al., 2008). It also suggests that the mechanism of regulation of metabolic enzyme expression either by MYC or other factors involved in tumorigenesis is context-specific.
Increased Lactate Levels and LDH Activity do not Always Correlate in Tumors
Increase in glucose consumption and lactate production even in the presence of normal oxygenation is a widely recognized characteristic of tumor metabolism. Our results show that, although the activity of LDH, the enzyme responsible for pyruvate conversion into lactate, was increased to the same extent in liver tumors induced by either MYC or MET (Figure 1H), a significant increase in lactate levels was only observed in MYC-induced tumors (Figures 1B and S1C). This demonstrates that increased LDH activity does not necessarily correlate with increased lactate levels in tumors.
In both MYC and MET-induced liver tumors, the activities of glucose transport and glucose phosphorylation were higher than in normal liver (Figure 2 and Table S1), indicating that glycolytic flux and steady-state levels of glucose-derived pyruvate may be increased in both types of tumors (Marin-Hernandez et al., 2006). In contrast to MYC-induced liver tumors, however, the increase of glucose transport and catabolism in most of MET-induced tumors may not be sufficient to produce amounts of pyruvate and NADH that would saturate the enzymes that compete for these substrates with LDH (Spriet et al., 2000). Moreover, increased glutamine catabolism observed in MYC-induced tumors may also contribute to creating a surplus of pyruvate available to LDH (DeBerardinis et al., 2008); and MYC and MET may have different effects on the activity of pyruvate-utilizing enzymes that compete with LDH, as well as the redox state of the cells. Nevertheless, increased expression of LDH in MET-induced tumors may serve to insure that once any surplus of pyruvate becomes available, it is efficiently converted into lactate regenerating NAD+ required for the turnover of glycolysis. The increased expression of LDH observed in either MYC or MET-induced tumors may also play a role independent of its metabolic function (Ronai, 1993).
The Role of Metabolic Changes in MYC and MET-induced Tumorigenesis
The combination of glucose catabolism into lactate and glutamine catabolism through the Krebs cycle was suggested to support rapid proliferation of tumor cells by simultaneously supplying them with ATP and biosynthetic precursors (DeBerardinis et al., 2008; McKeehan, 1982). Indeed, MYC-induced liver tumors, which have increased lactate production from glucose and increased catabolism of both glucose and glutamine through the Krebs cycle, demonstrate higher proliferation than MET-induced liver tumors (Figure S7A), which do not significantly increase glucose catabolism into lactate and produce rather than catabolize glutamine.
Elevated intracellular levels of glutamine were proposed to prime cells for activation of the mammalian target of rapamycin complex 1 (mTORC1), one of the major regulators of cell growth and proliferation (Nicklin et al., 2009). Thus, increased GLUL expression and the resulting elevated intracellular glutamine levels in MET-induced liver tumors and MYC-induced lung tumors might serve to ensure mTORC1 activity in the absence of exogenous glutamine and give tumor cells an advantage in nutrient-restricted conditions. Consistent with this hypothesis, glutamine deprivation did not affect phosphorylation of S6 (P-S6), a marker of mTORC1 activity, in Huh7 or HEPG2 cells, which have high levels of GLUL (Figure S7B). In contrast, glutamine deprivation caused a reduction of P-S6 in SK-HEP-1 cells, which do not have detectable GLUL expression (Figure S7B).
Synthetic Lethal Interaction between MYC and Inhibition of GLS1
Our present work demonstrates that inhibitors of Gls1 might eliminate tumor cells that overexpress MYC and catabolize glutamine to feed the Krebs cycle. In most of the MYC-induced lung tumors we examined, however, increased expression of Gls1 was accompanied by increased expression of Glul (Figure 5), which indicates that these tumors may both catabolize and produce glutamine. Increased expression of Glul can be sufficient to protect cells from glutamine deprivation (Kung et al., 2011). However, whether it can render cells that over-express MYC resistant to GLS1 inhibition, remains to be investigated. The contribution of various anaplerotic pathways that can contribute to the Krebs cycle (Cheng et al., 2011) should also be taken into account when identifying malignancies that might benefit from inhibiting glutaminase activity.
Since the GLS2 isoform of glutaminase is responsible for glutamine catabolism in normal liver, selective inhibition of GLS1 offers an opportunity to target glutamine catabolism of liver tumors that over-express MYC without affecting the metabolism of normal liver. On the other hand, GLS1 inhibition may be toxic to other normal tissues that express GLS1 such as kidney, intestine, lung and brain. However, our results demonstrate that expression of GLS1 in IMR90 and HA1E cells does not expose the cells to the lethal effect of GLS1 inhibition unless high levels of MYC are present, indicating that normal tissues with normal MYC levels may not be affected by GLS1 inhibitors. One of the well-known consequences of MYC overexpression is the induction of apoptosis by shifting the balance between pro-survival and pro-death signals (Meyer and Penn, 2008). The reduction in the Krebs cycle activity in response to GLS1 inhibition (Figures 6 and S6) may represent a stress signal that can unleash the pro-apoptotic activity of MYC when the gene is expressed above a certain threshold (Murphy et al., 2008).
Glutamine is a substrate for multiple metabolic reactions and glutaminase catalyzes only one of them. Indeed, complete deprivation of glutamine is more efficient in killing cells with high levels of MYC than inhibition of glutaminase (Figure 7B). This suggests that other pathways of glutamine metabolism may provide the opportunity for additional synthetic lethal interactions with the overexpression of MYC (DeBerardinis and Cheng, 2010; Mates et al., 2006). Identification of such interactions might provide additional therapeutic opportunities.
EXPERIMENTAL PROCEDURES
Animals
Oncogene-driven liver (Shachaf et al., 2004; Wang et al., 2001) and lung (Allen et al., 2011; Tran et al., 2008) tumors were generated as described previously. See the Supplemental Experimental Procedures for more details.
For isotopomer analysis of either glucose or glutamine metabolism tumor-bearing mice were injected with either 20 mg of [U-13C]-glucose (Cambridge Isotope Laboratories) (Fan et al., 2011) or with three consecutive injections (with a 15-minute interval) of [U-13C]-glutamine, 7mg each (Cambridge Isotope Laboratories) i.v. Mice were sacrificed 15 minutes after the last injection. The percent of 13C enrichment in either glucose or glutamine was comparable between tumor-free liver tissue and tumors from animals with either MYC or MET-induced liver tumors. The percent of 13C enrichment in glucose was 6 ± 2 %. The percent of 13C enrichment in glutamine varied between 13 and 42 % depending on an individual animal. Samples of tumor and adjacent liver tissue were snap-frozen in liquid nitrogen. All experiments were performed according to protocols approved by the Committee on Animal Research at the University of California, San Francisco.
Cell Culture
Primary IMR90 human lung fibroblasts and immortalized HA1E human kidney epithelial cells expressing ER-MYC or MYC-ER respectively, were described previously (Yuneva et al., 2007). HA1E cells with ectopic expression of MYC or H-RASV12 were generated by transduction with the retroviral vector pMaRX-puro (Hannon et al., 1999) carrying either MYC or H-RASV12 as described previously (Faleiro and Lazebnik, 2000). See the Supplemental Experimental Procedures for the detailed description of culture, treatment conditions and sequence of shRNAs.
Metabolite Profiling
All frozen tissue samples for metabolite analysis were pulverized and lyophilized. Frozen cell pellets were lyophilized. Metabolites were extracted and analyzed by NMR and gas chromatography-mass spectrometry (GC-MS) as described previously (Fan, 2010; Fan et al., 2009). See the Supplemental Experimental Procedures for more details.
18F-FDG PET/CT Imaging
The animal handling and preparation protocols followed established procedures to obtain consistent PET results (Fueger et al., 2006) (See the Supplemental Experimental Procedures for more details). For quantitative assessment, volumes of interest (VOIs) were placed in liver (cylinder having 5 mm diameter with 5 mm height) involving tumors when present, and in brain (cylinder having 3 mm diameter with 3 mm height) mostly involving cerebellum. The uptake in the liver VOI was normalized to the uptake in the brain VOI.
Enzyme Assays
A mitochondrial fraction was isolated from freshly-excised normal livers or tumors as described previously (Frezza et al., 2007). Cytosol was further separated from microsomes by centrifugation at 103,000 g for 60 minutes. LDH activity was measured in the cytosolic fraction, glucose-6-phosphorylating activities were measured in cytosolic and mitochondrial fractions, phosphate-dependent glutaminase activity was measured in mitochondrial fractions. See the Supplemental Experimental Procedures for more details.
Quantitative PCR Analysis and Western Blotting
Protocols, TaqMan® Gene Expression Assays and antibodies are described in Supplemental Experimental Procedures.
Statistical Analysis
Statistical analysis was performed applying a two-tailed Student’s t-test. For the experiments in which concentration of total and 13C-labeled lactate and glutamate were compared in tumors and adjacent liver tissue (Figure 1E, S1C and S4) paired Student’s t-test was used. For all other analysis un-paired Student’s t-test was used.
Supplementary Material
Highlights.
MYC and MET affect glucose and glutamine metabolism differently in the liver.
Tissue context affects the outcome of metabolic reprogramming by MYC.
MYC over-expression sensitizes cells to inhibition of Gls1 glutaminase.
Acknowledgments
We thank Dr. A. Lane, Dr. S. Arumugam, J. Tan and R. Burra (University of Louisville, Louisville, KY) for help with metabolic profiling analysis and Dr. A. Lane for valuable comments on the manuscript; Drs. A. Goga and A. Balakrishnan (University of California San Francisco, CA) for useful discussion of preliminary results; L. Urisman (University of California San Francisco, CA) for assistance with animal husbandry; and members of the Bishop Laboratory for valuable discussions and comments.
NMR analyses were supported in part by NIH Grant P20RR018733 from the National Center for Research Resources and NSF EPSCoR grant EPS-0447479. Metabolite profiling analysis was supported by NSF EPSCoR grant EPS-0447479, NCI grants 1R01CA118434-01A2 and 3R01 CA118434-02S1; University of Louisville CTSPGP award (T.W.M.F.). The work was also supported by UCSF/G.W. Hooper Research Foundation Endowment Funds and by NCI Grant R35 CA44338 (J.M.B.); UCSF Hellen Diller Family Comprehensive Cancer Center Pilot Grant and the UCSF Department of Radiology and Biomedical Imaging Seed Grant 10-09 (X.C. and Y.S.); the Johns Hopkins Brain Science Institute through the NeuroTranslational program (T.T. and D.V.F.); by NIH Grant R21NS074151 (to T.T.); Ministerio de Ciencia y Tecnología SAF2010-17573 and Junta de Andalucía, Proyectos de Investigación de Excelencia, Convocatoria 2010, CVI-6656, Spain (J.M.M. and F.J.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abcouwer SF, Lukaszewicz GC, Souba WW. Glucocorticoids regulate glutamine synthetase expression in lung epithelial cells. Am J Physiol. 1996;270:L141–151. doi: 10.1152/ajplung.1996.270.1.L141. [DOI] [PubMed] [Google Scholar]
- Allen TD, Zhu CQ, Jones KD, Yanagawa N, Tsao MS, Bishop JM. Interaction between MYC and MCL1 in the genesis and outcome of non-small-cell lung cancer. Cancer Res. 2011;71:2212–2221. doi: 10.1158/0008-5472.CAN-10-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggetto LG. Deviant energetic metabolism of glycolytic cancer cells. Biochimie. 1992;74:959–974. doi: 10.1016/0300-9084(92)90016-8. [DOI] [PubMed] [Google Scholar]
- Bode BP. Recent molecular advances in mammalian glutamine transport. J Nutr. 2001;131:2475S–2485S. doi: 10.1093/jn/131.9.2475S. discussion 2486S–2477S. [DOI] [PubMed] [Google Scholar]
- Bustamante E, Morris HP, Pedersen PL. Energy metabolism of tumor cells. Requirement for a form of hexokinase with a propensity for mitochondrial binding. J Biol Chem. 1981;256:8699–8704. [PubMed] [Google Scholar]
- Bustamante E, Pediaditakis P, He L, Lemasters JJ. Isolated mouse liver mitochondria are devoid of glucokinase. Biochem Biophys Res Commun. 2005;334:907–910. doi: 10.1016/j.bbrc.2005.06.174. [DOI] [PubMed] [Google Scholar]
- Cadoret A, Ovejero C, Terris B, Souil E, Levy L, Lamers WH, Kitajewski J, Kahn A, Perret C. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- Chafey P, Finzi L, Boisgard R, Cauzac M, Clary G, Broussard C, Pegorier JP, Guillonneau F, Mayeux P, Camoin L, et al. Proteomic analysis of beta-catenin activation in mouse liver by DIGE analysis identifies glucose metabolism as a new target of the Wnt pathway. Proteomics. 2009;9:3889–3900. doi: 10.1002/pmic.200800609. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, Deberardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Erdmann N, Zhao J, Lopez AL, Herek S, Curthoys N, Hexum TD, Tsukamoto T, Ferraris D, Zheng J. Glutamate production by HIV-1 infected human macrophage is blocked by the inhibition of glutaminase. J Neurochem. 2007;102:539–549. doi: 10.1111/j.1471-4159.2007.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faleiro L, Lazebnik Y. Caspases disrupt the nuclear-cytoplasmic barrier. J Cell Biol. 2000;151:951–959. doi: 10.1083/jcb.151.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW. Metabolomics-Edited Transcriptomics Analysis (Meta) In: McQueen CA, editor. Comprehensive Toxicology. Oxford: Academic Press; 2010. pp. 685–706. [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Yan J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics. 2011;7:257–269. doi: 10.1007/s11306-010-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- Fueger BJ, Czernin J, Hildebrandt I, Tran C, Halpern BS, Stout D, Phelps ME, Weber WA. Impact of animal handling on the results of 18F-FDG PET studies in mice. J Nucl Med. 2006;47:999–1006. [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gazzeri S, Brambilla E, Caron de Fromentel C, Gouyer V, Moro D, Perron P, Berger F, Brambilla C. p53 genetic abnormalities and myc activation in human lung carcinoma. Int J Cancer. 1994;58:24–32. doi: 10.1002/ijc.2910580106. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Kaplan NO, Hall TC. Lactic Dehydrogenase in Human Neoplastic Tissues. Cancer Res. 1964;24:389–399. [PubMed] [Google Scholar]
- Hannon GJ, Sun P, Carnero A, Xie LY, Maestro R, Conklin DS, Beach D. MaRX: an approach to genetics in mammalian cells. Science. 1999;283:1129–1130. doi: 10.1126/science.283.5405.1129. [DOI] [PubMed] [Google Scholar]
- Hastak K, Paul RK, Agarwal MK, Thakur VS, Amin AR, Agrawal S, Sramkoski RM, Jacobberger JW, Jackson MW, Stark GR, Agarwal ML. DNA synthesis from unbalanced nucleotide pools causes limited DNA damage that triggers ATR-CHK1-dependent p53 activation. Proc Natl Acad Sci U S A. 2008;105:6314–6319. doi: 10.1073/pnas.0802080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan O, Firon M, Vivi A, Navon G, Tsarfaty I. HGF/SF activates glycolysis and oxidative phosphorylation in DA3 murine mammary cancer cells. Neoplasia. 2000;2:365–377. doi: 10.1038/sj.neo.7900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Coulouarn C, Conner EA, Factor VM, Roskams T, Thorgeirsson SS. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–2782. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Hernandez A, Rodriguez-Enriquez S, Vital-Gonzalez PA, Flores-Rodriguez FL, Macias-Silva M, Sosa-Garrocho M, Moreno-Sanchez R. Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J. 2006;273:1975–1988. doi: 10.1111/j.1742-4658.2006.05214.x. [DOI] [PubMed] [Google Scholar]
- Marquez J, Sanchez-Jimenez F, Medina MA, Quesada AR, Nunez de Castro I. Nitrogen metabolism in tumor bearing mice. Arch Biochem Biophys. 1989;268:667–675. doi: 10.1016/0003-9861(89)90335-4. [DOI] [PubMed] [Google Scholar]
- Mates JM, Segura JA, Alonso FJ, Marquez J. Pathways from glutamine to apoptosis. Front Biosci. 2006;11:3164–3180. doi: 10.2741/2040. [DOI] [PubMed] [Google Scholar]
- McKeehan WL. Glycolysis, glutaminolysis and cell proliferation. Cell Biol Int Rep. 1982;6:635–650. doi: 10.1016/0309-1651(82)90125-4. [DOI] [PubMed] [Google Scholar]
- Medina MA. Glutamine and cancer. J Nutr. 2001;131:2539S–2542S. doi: 10.1093/jn/131.9.2539S. discussion 2550S–2531S. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Perdomo G, Martinez-Brocca MA, Bhatt BA, Brown NF, O’Doherty RM, Garcia-Ocana A. Hepatocyte growth factor is a novel stimulator of glucose uptake and metabolism in skeletal muscle cells. J Biol Chem. 2008;283:13700–13706. doi: 10.1074/jbc.M707551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gomez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sanchez P, Gonzalez ME, Marquez J, Mates JM. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–542. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, Hansen JC, Curthoys NP. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronai Z. Glycolytic enzymes as DNA binding proteins. Int J Biochem. 1993;25:1073–1076. doi: 10.1016/0020-711x(93)90123-v. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Spriet LL, Howlett RA, Heigenhauser GJ. An enzymatic approach to lactate production in human skeletal muscle during exercise. Med Sci Sports Exerc. 2000;32:756–763. doi: 10.1097/00005768-200004000-00007. [DOI] [PubMed] [Google Scholar]
- Sunaga N, Kohno T, Kolligs FT, Fearon ER, Saito R, Yokota J. Constitutive activation of the Wnt signaling pathway by CTNNB1 (beta-catenin) mutations in a subset of human lung adenocarcinoma. Genes Chromosomes Cancer. 2001;30:316–321. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1097>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Tran PT, Fan AC, Bendapudi PK, Koh S, Komatsubara K, Chen J, Horng G, Bellovin DI, Giuriato S, Wang CS, et al. Combined Inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS One. 2008;3:e2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tward AD, Jones KD, Yant S, Cheung ST, Fan ST, Chen X, Kay MA, Wang R, Bishop JM. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc Natl Acad Sci U S A. 2007;104:14771–14776. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tward AD, Jones KD, Yant S, Kay MA, Wang R, Bishop JM. Genomic progression in mouse models for liver tumors. Cold Spring Harb Symp Quant Biol. 2005;70:217–224. doi: 10.1101/sqb.2005.70.058. [DOI] [PubMed] [Google Scholar]
- Ueki T, Fujimoto J, Suzuki T, Yamamoto H, Okamoto E. Expression of hepatocyte growth factor and its receptor c-met proto-oncogene in hepatocellular carcinoma. Hepatology. 1997;25:862–866. doi: 10.1002/hep.510250413. [DOI] [PubMed] [Google Scholar]
- Vora S, Halper JP, Knowles DM. Alterations in the activity and isozymic profile of human phosphofructokinase during malignant transformation in vivo and in vitro: transformation- and progression-linked discriminants of malignancy. Cancer Res. 1985;45:2993–3001. [PubMed] [Google Scholar]
- Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153:1023–1034. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota J, Wada M, Yoshida T, Noguchi M, Terasaki T, Shimosato Y, Sugimura T, Terada M. Heterogeneity of lung cancer cells with respect to the amplification and rearrangement of myc family oncogenes. Oncogene. 1988;2:607–611. [PubMed] [Google Scholar]
- Yuneva M. Finding an “Achilles’ heel” of cancer: the role of glucose and glutamine metabolism in the survival of transformed cells. Cell Cycle. 2008;7:2083–2089. doi: 10.4161/cc.7.14.6256. [DOI] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.