Abstract
Members of the golgin family of coiled-coil proteins have been implicated in the tethering of vesicles to Golgi membranes and cisternal membranes to each other. Many also bind to rab GTPases. Golgin-84 is a membrane-anchored golgin that we now show binds preferentially to the GTP form of the rab1 GTPase. It is also present throughout the Golgi stack by immuno-EM. Antibodies to golgin-84 inhibit stacking of cisternal membranes in a cell-free assay for Golgi reassembly, whereas the cytoplasmic domain of golgin-84 stimulates stacking and increases the length of re-assembled stacks. Transient expression of golgin-84 in NRK cells helps prevent the disassembly of the Golgi apparatus normally triggered by treatment with brefeldin A. Together these data suggest that golgin-84 is involved in generating and maintaining the architecture of the Golgi apparatus.
Keywords: cisterna, Golgi, golgin, GTPase, mitosis, rab
The Golgi apparatus in many eukaryotic cells has a unique and complex architecture comprising flattened cisternal membranes with dilated rims, most often, though not always, arranged in a stack. Newly synthesized proteins and lipids from the endoplasmic reticulum (ER) enter at the cis-Golgi network (CGN) and exit at the trans-Golgi network (TGN), after undergoing processing in the stack, mostly to the oligosaccharides bound to the transiting cargo molecules (1–4).
The structural basis of Golgi architecture is slowly becoming clear through analysis of membrane traffic pathways in interphase and mitotic cells. The Golgi ReAssembly Stacking Proteins (GRASPs) and golgin families of proteins are key components of Golgi structure. GRASPs were first implicated in the stacking of Golgi cisternae after mitosis (5,6). Using a cell-free system that reconstitutes many aspects of Golgi reassembly after mitosis, we were able to show that cisternal stacking was blocked in the presence of GRASP proteins rendered soluble by removal of the N-terminal signal that normally mediates addition of myristic acid, a membrane anchoring domain. Antibodies against GRASP proteins had the same effect (5,6).
Several golgins have also been implicated in the stacking of Golgi cisternae. Giantin, p115 and GM130 are thought to form a tether connecting COPl vesicles to Golgi membranes (7). Giantin is in vesicles and GM130 is on the Golgi, anchored there by a PDZ-like domain connecting it to GRASP65 (one of the GRASPs) (8). p115 links Giantin to GM130 (9). This tether has also been implicated in cisternal stacking, bringing cisternal membranes close enough together for GRASP to provide a more permanent linkage (10). Golgin-45 is bound to GRASP55, another GRASP involved in cisternal stacking (11).
Golgin-84 is a recent member of the golgin family that is structurally related to giantin and the most recent member, CASP (12). All are type II membrane proteins with their N-terminus projecting into the cytoplasm and bound to Golgi membranes by a C-terminal membrane-anchoring domain. Golgin-84 was first identified in a two-hybrid screen, as a partner for OCRL-1 (OculoCerebroRenal syndrome of Lowe), a phosphatidylinositol 4,5-bisphosphate 5-phosphatase (13).
Rab proteins are members of the ras superfamily of small GTPases that are involved in many aspects of membrane traffic processes though their precise role is still unclear. Rabs bind to a number of golgins including GM130, p115, golgin-45, golgin-245 and golgin-97 (11, 14–18).
Given these roles for golgins we decided to study golgin-84. Our data show that it binds pecifically to rab1 and contributes to Golgi structure both in vitro and in vivo.
Results and Discussion
Golgin-84 is required for the reassembly of Golgi stacks
Since several golgins have been implicated in Golgi structure, we first tested the role of golgin-84 using a cell-free assay that mimics many aspects of mitotic Golgi fragmentation and reassembly (19). In this assay, purified rat liver Golgi stacks (RLG) are disassembled, by incubation with mitotic cytosol (MC), into a collection of vesicular and tubular structures that we term mitotic Golgi fragments (MGFs). MGFs can reassemble to form stacked cisternal structures upon incubation with interphase cytosol (IC). In Figure 1, we show data from such disassembly/reassembly reactions. During the mitotic disassembly phase of the reaction, the percentage of total membrane present as cisternae was reduced by at least 2-fold (to 31 ±1% in MGFs compared to 67 ±3% in RLG). Furthermore, the percentage of membrane present in stacked cisternal structures was dramatically reduced, by more than 6-fold (to 6 ± 1 % in MGFs compared to 41 ± 2% in RLG). During the reassembly phase, incubation of MGFs with IC caused both the re-growth and restacking of cisternae. The percentage of total membrane present as cisternae in the re-assembled material was close to that in starting RLG (59 ± 1% compared to 67 ±3% in RLG). The percentage of total membrane present as stacked cisternae was also restored to a value near to that of the starting material (31 ± 2% compared to 41 ± 2% in RLG).
Figure 1. Golgin-84 is required for stacking and lateral growth of Golgi cisternae.
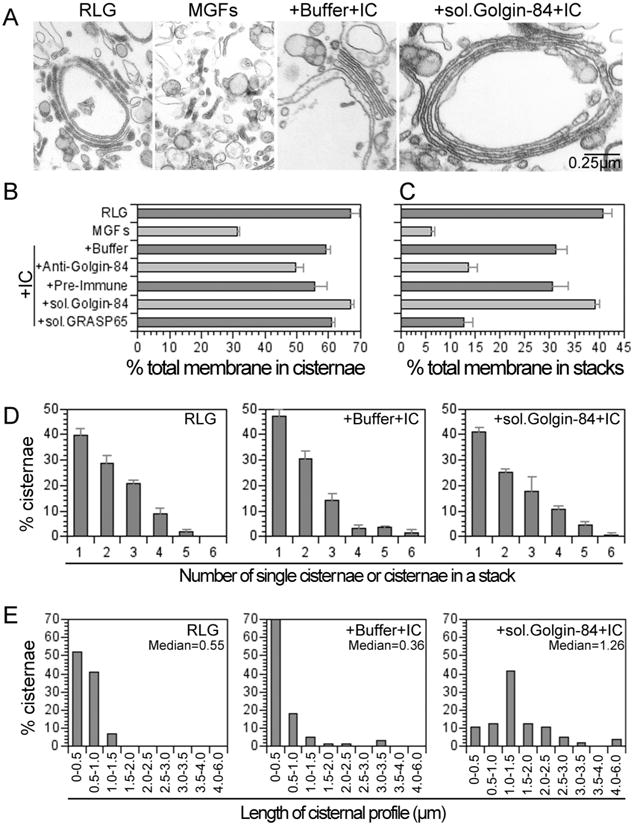
Purified rat liver Golgi (RLG) stacks were incubated with mitotic HeLa cytosol (MC) and the resulting mitotic Golgi fragments (MGFs) were isolated. After preincubation with buffer, antibodies to golgin-84 (+Anti-Golgin-84), preimmune serum (+Pre-Immune), soluble His-tagged golgin-84 (+sol.Golgin-84), or soluble His-tagged GRASP65 (+sol.GRASP65), samples were incubated with interphase cytosol (IC) for 60 min at 37 °C. Reactions were terminated by fixation, processed for EM, and quantitated. A. Images illustrating the stacked cisternal membranes found in preparations of RLG; the tubular and vesicular fragments (MGFs) after treatment with MC; the reassembly into stacked cisternal structures in the presence of IC (+Buffer+IC); and the enhancement in the length of cisternal profiles observed in the presence of recombinant soluble golgin-84 (+sol.Golgin-84 +IC). Bar = 0.25 0.25 μm. B. Percentage of total membrane in cisternae. C. Percentage of total membrane in stacks. D. Distribution of cisternae amongst single and stacked cisternae. The results in B-D are presented as the mean ±SEM (n = 4) and are representative of 3–5 independent experiments. E. The length distribution of cisternal profiles in RLG and re-assembled stacks in the absence or presence of soluble golgin-84. Median values in μm are indicated.
To test the role of golgin-84 in the reassembly of Golgi structures, we pretreated MGFs with antibodies against golgin-84 before performing the reassembly phase of the assay. After incubation with IC the percentage of membrane in stacks was much less than that obtained using preimmune serum (14 ± 2% vs. 31 ±3%) and close to that of the original MGFs (6 ± 1%) (Figure 1C). In marked contrast, these antibodies had only a modest effect on the percentage of total membrane re-assembled into cisternae (50 ± 3% compared to 56 ± 4% for the preimmune serum) (Figure 1B). Together, these data suggest that golgin-84 plays a role in cisternal stacking rather than assembling the cisternae themselves.
Soluble golgin-84 enhances the median length of re-assembled cisternae
Further experiments were carried out using a recombinant form of golgin-84, comprising the entire cytoplasmic domain. This soluble protein was preincubated with MGFs which were then incubated with IC. As a positive control we used soluble, recombinant GRASP65, which had little if any effect on cisternal re-growth (61 ± 1 % vs. 59 ± 1 % for buffer alone) (Figure 1B), but dramatically inhibited cisternal stacking (3 ± 2% vs. 31 ± 2% for buffer alone) (Figure 1C), as previously described (5,6). Soluble golgin-84 had a slight stimulatory effect on cisternal re-growth (67 ± 1% vs. 59 ± 1 % for buffer alone) (Figures 1B), but it had the opposite effect to GRASP65 in that it stimulated cisternal stacking (to 39 ± 1 % vs. 31 ± 2% for buffer alone), reaching a level close to that of the starting RLG (41 % ± 2%) (Figure 1C).
Soluble golgin-84 had little effect on the number of cisternae in the stack. The distribution of cisternae amongst single and stacked cisternae was similar for starting RLG and for stacks re-assembled with or without golgin-84 (Figure 1D). The mean value ranged from 1.9 to 2.1 cisternae/stack. Strikingly, however, soluble golgin-84 had a significant effect on the length of the re-assembled cisternae, whether they were single or in stacked structures (Figure 1A). The median length of cisternal profiles re-assembled using IC was 0.36 μm (+buffer IC) but this increased nearly 4-fold, to 1.26 μm (+sol.Golgin-84+IC), in the presence of soluble golgin-84 (Figure 1E). Furthermore, this value was more than twice that of starting RLG membranes (0.55 μm), suggesting that soluble golgin-84 might play a role in stitching stacked cisternae together into the ribbon-like structure that characterizes the Golgi apparatus in mammalian cells (20).
Golgin-84 is a novel binding partner for rab 1
The fact that soluble golgin-84 enhanced the formation of long, stacked Golgi was reminiscent of results we had obtained earlier using GTPγS (19). This nonhydrolyzable GTP analogue stimulated cisternal re-stacking when added to MGFs and IC, suggesting that a GTPase might be involved. Given that so many golgins have been found to bind to rab GTPases (11,14–18), we tested this possibility for golgin-84.
We first tested Rab1 since this Golgi-localized GTPase is known to bind to a number of golgins, such as GM130 and p115 (11,14–17). GST-rab1 was incubated with a detergent extract of RLG membranes, washed and the bound proteins fractionated and blotted for golgin-84. GST-rab1 pulled down golgin-84, which bound preferentially to the GTPγS-loaded form (Figure 2A, lanes 2 and 3). Mapping experiments were also carried out to locate the region on golgin-84 that binds Rab1, but it did not prove possible to narrow down the binding site. Almost the entire coiled-coil domain was needed for efficient binding (data not shown).
Figure 2. Golgin-84 binds directly and specifically to rab1 GTPase.
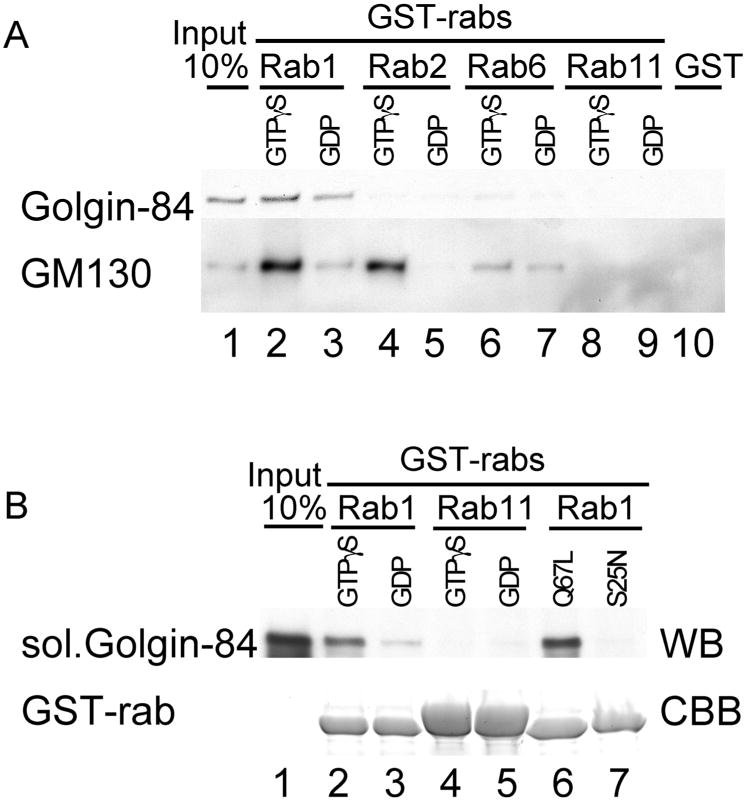
A. Detergent extracts of RLG were incubated with GST-rab1 (lanes 2 and 3), GST-rab2 (lanes 4 and 5), GST-rab6 (lanes 6 and 7), GST-rab11 (lanes 8 and 9) or GST alone (lane 10). The rabs were preloaded with either GTPγS (lanes 2, 4, 6 and 8) or GDP (lanes 3, 5, 7 and 9). The input Golgi (10%) is shown in lane 1. Bound proteins were eluted with GDP/EDTA and fractionated by SDS-PAGE followed by Western blotting for golgin-84 (upper panel) or GM130 (lower panel). B. Soluble, recombinant golgin-84 was incubated with GST-rab1 (lanes 2 and 3) or GST-rab11 (lanes 4 and 5), preloaded with either GTPγS (lanes 2 and 4) or GDP (lanes 3 and 5). Incubations were also carried out using the ‘GTP’ (rab1 Q67 L) and ‘GDP’ (rab1 S25 N) locked forms of rab1 (lanes 6 and 7). The input Golgi (10%) is shown in lane 1. Bound proteins were analyzed by SDS-PAGE followed by Western blotting for Golgin-84 (WB), or by Coomassie Brilliant Blue (CBB) staining to show the amount of rabs used.
We next tested the specificity of binding using several rab proteins known to operate in or near the Golgi apparatus. Golgin-84 bound preferentially to the GTPγS-loaded form of GST-rab1, but not to GST-rabs 2, 6 or 11, or to GST (Figure 2A), suggesting that the interaction between golgin-84 and rab 1 is specific. As a positive control, we also blotted for GM130, which is known to bind to the GTP-loaded form of rab1 and rab2 (11). GM130 bound to both these rabs as well as rab6, though to a much lesser extent (Figure 2A) and (11, 16).
To show that binding is direct, we incubated GST-rab1 with soluble golgin-84. As shown in Figure 2B, golgin-84 bound preferentially to GST-rab1 loaded with GTPγS but not to GST-rab11 loaded with either nucleotide. We also used mutated Rab1s locked in either the GTP (Q67L) or GDP (S25N) form. In this case only the former bound to golgin-84, with the GDP-locked mutant showing no detect able binding (Figure 2B). The difference between these results and those obtained using wild-type rab1 may reflect incomplete nucleotide exchange during the loading of wild-type rab1 fusion protein with GDP. Alternatively, GDP-loaded-rab1 may have a bona fide affinity for golgin-84 that is abolished by the S25N mutation.
Together, these data suggest that golgin-84 specifically binds rab1, with a clear preference for the GTP-loaded form, and that it does so through the cytoplasmic domain. This would help explain how soluble golgin-84 localizes to Golgi membranes in the absence of the membrane anchor (21). Furthermore, the stimulation of cisternal stacking by soluble golgin-84 might in part be explained by linking rab1 s in adjacent cisternae.
Golgin-84 is localized throughout the Golgi stack
Previously characterized stacking proteins were localized to the early part of the Golgi stack. GRASP65 is found in cis-cisternae and GRASP55 in cis/medial-cisternae (6). The golgin tether comprising GM130-p115-giantin is also found in cis-membranes since GM130 is bound to GRASP65. Given the role of golgin-84 in cisternal stacking suggested by the experiments detailed in Figure 1, it was of interest to determine its location at the electron microscopic (EM) level.
Ultrathin sections of NRK cells were triple-labeled with antibodies to golgin-84, rab1 and GM130 followed by Protein-A coupled to different sizes of gold particles. Cross-labeling was prevented by fixing the samples with glutaraldehyde after each labeling step. As shown in Figure 3A and quantitated in Figure 3B, GM130 was, as expected, concentrated on the cis-side of the Golgi apparatus (C and C + 1 cisternae). So too was rab1 which operates on the cis-side of the Golgi apparatus to receive incoming cargo from the ER (22).
Figure 3. Golgin-84 is localized throughout the Golgi stack using immuno-EM.
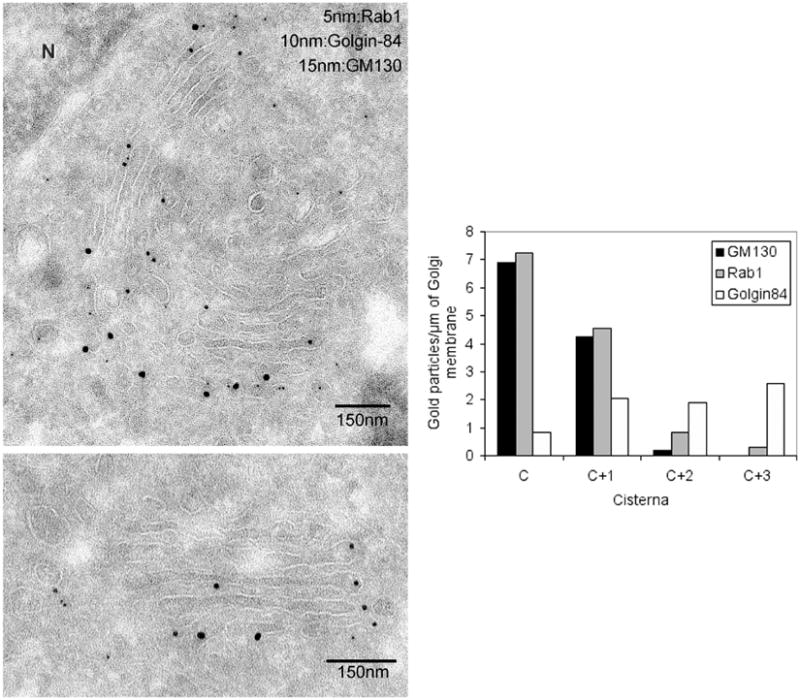
A. NRK cells were processed for cryo-EM and triple-labeled for Rab1 (5 nm gold), golgin-84 (10 nm gold) and GM130 (15 nm gold). Note the occasional concentration of golgin-84 in cisternal rims (bottom panel). N = nucleus. B. Quantitation showing that rab1 and GM130 are restricted to the cis-side of the Golgi stack (C, C + 1), whereas golgin-84 is present throughout the stack, with the lowest linear density on the cis-side (C).
Golgin-84, by way of contrast, was distributed throughout the stack and, on occasion, was found in cisternal rims (Figure 3A, bottom panel). Such a location might help explain its role in generating long cisternal structures (Figure 1E) and, by implication, the Golgi ribbon, though far more work will be needed to establish this point.
Quantitation confirmed the presence of golgin-84 in all cisternae and suggested that there might be a gradient with the least amount in cis-membranes (Figure 3B, ‘C’) and the most in trans-membranes (Figure 3B, ‘C + 3’). Such a distribution raises the interesting possibility that golgin-84 provides a means of stacking trans-cisternae and so complement the cis/medial-stacking activity of the GRASP proteins. What is still unclear is the role played by rab1 in the functioning of golgin-84. Rab1 has the opposite distribution to golgin-84, so that maximum overlap occurs in the C + 1 cisterna (Figure 3B). If golgin-84 is a rab1 effector, then this might suggest that it only functions in early Golgi cisternae. However, rab1 has other cis-binding partners, such as p115 and GM130, the latter binding more efficiently to rab1 than golgin-84 (Figure 2B). Hence the higher levels of rab1 in early Golgi cisternae might simply reflect the number of other proteins with which it can also interact. If the rab1 present in late Golgi membranes interacts only with golgin-84, then there might be sufficient for it to function there. In addition, there might be other factors that act to regulate golgin-84 function.
Expression of golgin-84 stabilizes the Golgi against brefeldin A treatment
Many members of the golgin family of coiled-coil proteins do not move back to the ER in the presence of brefeldin A (BFA) (23,24), or at least not as fast as Golgi enzymes (25). The exceptions are the membrane-anchored golgins, such as CASP, giantin and golgin-84, which behave more like Golgi enzymes and move backat similar speeds, or slightly slower (12,13,26). This contrast is illustrated in Figure 4A where treatment of NRK cells with BFA relocated golgin-84 to an ER-like distribution, whereas GM130 assumed a more dispersed, punctate appearance. Rab1 also became more dispersed, although some puncta still colocalized with GM130.
Figure 4. Transient expression of golgin-84 stabilizes the Golgi against disassembly triggered by treatment with BFA.
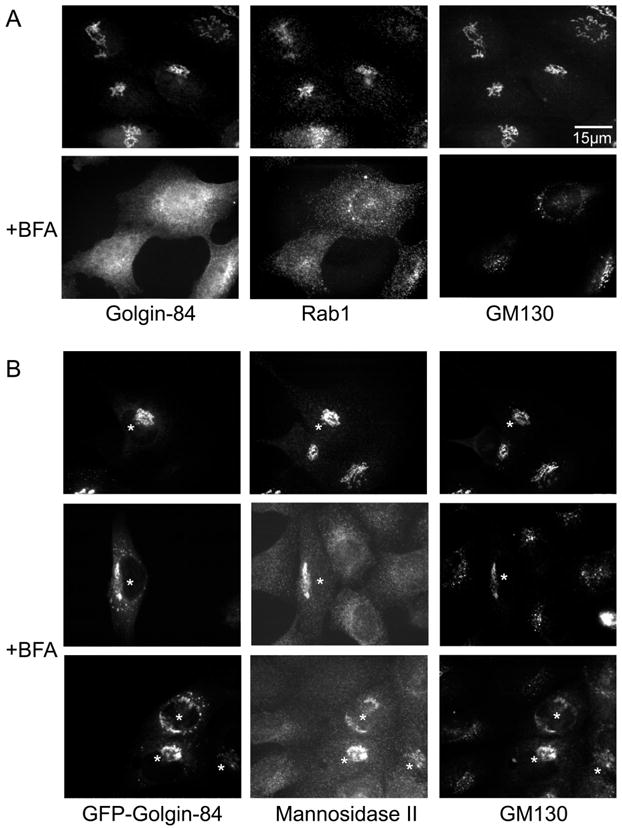
A. NRK cells were fixed before or after treatment with BFA (5 μg/ml for 30 min) then permeabilized and triple-labeled for golgin-84, rab1 and GM130. B. NRK cells expressing GFP-golgin-84 were fixed before or after treatment with BFA (5 μg/ml for 30 min), then permeabilized and labeled for mannosidase 11 and GM130. Note that cells expressing GFP-golgin-84 the Golgi were more resistant to treatment with BFA treatment than uninjected cells. Asterisks indicate micro-injected cells.
Expression of exogenous golgin-84 had an unexpected effect. The cDNA encoding golgin-84, tagged at the N-terminus with GFP, was microinjected into NRK cells and incubated for 2h. After fixation and permeabilization, a typical Golgi labeling pattern was observed which colocalized with mannosidase II and GM130. Interestingly, this pattern was largely unchanged after treatment with BFA. Some fragmentation of the Golgi was observed, but the fragments still maintained a peri-nuclear location and there was little, if any, evidence that GFP-golgin-84 was relocated to the ER. Furthermore, this same pattern was obtained after labeling for GM130 and mannosidase II (Figure 4B), suggesting that over-expression of golgin-84 somehow stabilizes the Golgi structure, making it resistant to the effects of BFA. COPl coats were still released by BFA treatment (data not shown), showing that this effect operated after disruption of COPl-mediated budding reactions.
In conclusion, our data extend the roles played by golgins in generating and maintaining Golgi architecture. Golgin-84 is involved in cisternal stacking and appears to be present throughout the stack with an increasing gradient towards the trans-side. This makes it an excellent candidate for stacking trans-cisternae, which lack the GRASP proteins previously implicated in cis/medial-stacking. Golgin-84 also enhances the length of single and stacked cisternae and stabilizes Golgi architecture in the presence of BFA. This implicates it in the process that generates and maintains the interphase Golgi ribbon. The role played by rab1 in the operation of golgin-84 is still unclear as it is in all other instances where rab proteins have been shown to bind to golgins. This will be the focus of future experiments. Nevertheless, these data add to the existing information on golgin tethers and further emphasize the importance of these proteins in Golgi structure and function (27,28).
Materials and Methods
Antibodies
Polyclonal rabbit anti-golgin-84 antibodies were raised using the purified recombinant golgin-84 as an antigen. For triple-labeling experiments, the Protein-A-purified lgG fraction of golgin-84 antibodies was conjugated to Alexa488 (Molecular Probes, Eugene, OR, USA) following the manufacturer's protocol. Polyclonal anti-GM130 (ML07) were gifts from Martin Lowe (University of Manchester, UK). Monoclonal anti-mannosidase II (53FC3) was previously described (29). Monoclonal anti-GM130 and polyclonal rabbit anti-rab1 antibodies were purchased from BD Transduction Laboratories (San Jose, CA, USA) and Santa Cruz Biotechnology (Santa Cruz, CA, USA), respectively. Goat anti-mouse lgG-Alexa594 and anti-rabbit lgG-Alexa350 were purchased from Molecular Probes; goat anti-mouse lgG-HRP and anti-rabbit lgG-HRP were purchased from Biosource (Camarillo, CA, USA).
Plasmids
GST-rab1, GST-rab2 and rab6, and GST-rab11 were kindly provided by Tommy Nilsson (EMBL, Germany), Francis A. Barr (Max-Plank-Institute, Germany) and David Sheff (Yale University, CT, USA), respectively. GST-rab1Q67L and rab1S25N were generated by site-directed mutagenesis using GST-rab1 as a template. Rat golgin-84 (accession number: AY144587) was cloned from the rat liver cDNA library (Origene, Rockville, MD, USA), and its cytoplasmic domain was subcloned into His-tag-expression vector. GFP-human golgin-84 was a gift from Robert L. Nussbaum (NIH, MD, USA). Mutants of golgin-84 were generated by site-directed mutagenesis.
Recombinant proteins
BL21 Escherichia coli was transformed with GST-rabs or His-tagged soluble golgin-84 constructs. The expression of recombinant proteins was induced by adding lPTG. The cells were sedimented and resuspended with PBS with 5 mM MgCl2, 5mM 2-mercaptoethanol and 0.2 mM GDP (for GST-rabs), or 10 mM HEPES (pH7.4) with 100 mM KCI (for His-golgin-84). The cells were lysed by sonication on ice and clarified by centrif ugation. The lysates were incubated with glutathione-Sepharose 4B (Amersham Biotech., Piscataway, NJ, USA) or Ni-NTA agarose (Qiagen, Valencia, CA, USA). His-tagged golgin-84 was eluted from Ni-NTA agarose with 500 mM imidazole, and was dialyzed against 10mM HEPES (pH7.4) with 100 mM KCI.
Golgi reassembly assay
Preparation of rat liver Golgi (RLG), mitotic HeLa cytosol (MC), interphase HeLa cytosol (IC), disassembly and reassembly reactions, were performed as previously described (19) with the following modifications. RLG stacks were incubated with MC for 20 min at 37 °C to disassemble the Golgi, and mitotic Golgi fragments (MGFs) were reisolated through a 0.4m sucrose cushion by centrifugation at 55000 r.p.m. for 30 min in a TLA55 rotor (Beckman, Fullerton, CA, USA). MGFs were incubated with IC at 37°C for 60 min. In some reactions, 2 μl of antiserum, or 2 μm recombinant His-tagged soluble GRASP65, or His-tagged soluble golgin-84 were pre-incubated with the MGFs for 30 min on ice followed by reassembly reactions in IC. Membranes were fixed and processed for EM. Cisternae were defined as membrane profiles with a length greater than four times their width, which was not greater than 80 nm. The percentage of membranes in cisternae, the number of cisternae in the stack and the median cisternal length were determined by the intersection method (19).
Rab pull-down assay
Golgi extracts were obtained by solubilization of purified RLG membranes in IP buffer (10 mM HEPES-KOH, pH 7.4, 100 mM KCI, 1 mM dithiothreitol, 5 mM MgCI2, 1% Triton X-100 and protease inhibitor cocktail from Roche, Indianapolis, IN, USA). After incubation for 30 min on ice, the extract was clarified by centrifugation at 13000 g for 20 min. The purified GST-rabs on the glutathione-beads were preloaded with GTPγS or GDP (30). The extracts (100 μg), or the recombinant, purified golgin-84 (2 μg), were incubated with the GST-rab beads (100 μg for the extracts and 20 μg for the recombinant protein) for 1 h at 4°C in the presence of 1 mg/ml of soy bean trypsin inhibitor (Sigma, St. Louis, MO, USA) and 1 mM nucleotides. Total volume of the mixtures was 250 μl for the extracts and 100 μl for the recombinant protein. After washing twice with IP buffer containing soy bean trypsin inhibitor and nucleotides, the beads were washed with IP buffer alone. The proteins bound were eluted with 10 mM EDTA and 5mM GDP in IP buffer. Proteins in the eluates or on the beads were analyzed by SDS-PAGE followed by Western blotting or Coomassie Brilliant Blue staining, respectively.
Cell culture and microinjection
NRK cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS. For microinjection and immunofluorescence, cells were cultured on 12-mm glass coverslips. Plasmid DNA encoding GFP-golgin-84 was injected into cell nuclei at a concentration of 200 μg/ml. After the microinjection, the cells were incubated for 2 h for the transient expression.
Cryoelectron microscopy and immunogold labeling
NRK cells were fixed with 8% paraformaldehyde (PFA) in 0.25m HEPES buffer, pH7.3. After washing in buffer, the cells were sedimented by centrifugation, embedded in 10% gelatin, cooled on ice, and cut into 1 mm3 blocks. The blocks were infused with 2.3 m sucrose at 4°C for at least 2 h and frozen in liquid nitrogen. ∼50 nm-thick sections were cut at −120 °C using an Ultracut T/FCS (Leica, Deerfield, IL, USA). Ultrathin sections were picked up in a mixture of 2% methyl cellulose and 2.3 m sucrose (1 : 1). For triple labeling, cryosections were collected on formvar-coated copper grids, incubated at room temperature on a drop of 100 mm NH4CI, and nonspecific binding was blocked with 2% (w/v) fish skin gelatin in PBS. Sections were incubated with the appropriate primary antibody, followed by Protein-A-gold (Department of Cell Biology, Utrecht School of Medicine, Utrecht, Netherlands). A rabbit anti-mouse Ig antibody was used as the bridging antibody when monoclonal antibodies were used. After each round of labeling, the sections were treated with 1% glutaraldehyde, followed by 100 mM NH4CI and 2% fish skin gelatin. The sections were counterstained with 2% uranyl acetate, and embedded in 1.7% methyl cellulose containing 0.75% uranyl acetate.
Immunofluorescence microscopy
NRK cells on coverslips were fixed with 3.7% PFA in PBS for 15 min, permeabilized with 0.1 % Triton X-100 in PBS for 5 min at room temperature. The cells were blocked with 3% BSA in PBS for 30 min, and then incubated for 15 min with primary antibodies diluted in 3% BSA in PBS. The cells were washed three times with PBS, and incubated for 15 min with secondary antibodies conjugated to Alexa fluorophors (Molecular Probes). After washing the cells, the coverslips were mounted on microscope slides and viewed using a Zeiss Axioplan 2 upright microscope (Carl Zeiss, Thornwood, NY, USA) equipped with an ORCA-II cooled charge-coupled device camera (1280 × 1024 pixels, Hamamatsu, Bridgewater, NJ, USA) controlled with the Openlab 3.0.8 software package (Improvision, Coventry, UK) and a Plan-Apochromat 100 × 1.4 NA objective.
Acknowledgments
We thank Ira Mellman, Derek Toomre and the entire Warren/Mellman/Toomre group for discussion and support; Drs M. Lowe, R. L. Nussbaum, T. Nilsson, W. Balch, and F. A. Barr for generous provision of reagents. This work was funded by the National Institutes of Health. A.S. was supported by the Toyobo Biotechnology Foundation.
References
- 1.Farquhar MG, Palade GE. The Golgi apparatus (complex)-l954–81-from artifact to center stage. J Cell Biol. 1981;91:77s–103s. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 3.Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 4.Mellman I, Warren G. The road taken: past and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- 5.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 6.Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sönnichsen B, Lowe M, Levine T, Jamsa E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirac-Svejstrup AB, Shorter J, Waters MG, Warren G. Phosphorylation of the vesicle-tethering protein p115 by a casein kinase Il-like enzyme is required for Golgi reassembly from isolated mitotic fragments. J Cell Biol. 2000;150:475–488. doi: 10.1083/jcb.150.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shorter J, Warren G. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Short B, Preisinger C, Körner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillingham AK, Pfeifer AC, Munro S. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol Biol Cell. 2002;13:3761–3774. doi: 10.1091/mbc.E02-06-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bascom RA, Srinivasan S, Nussbaum RL. Identification and characterization of golgin-84, a novel Golgi integral membrane protein with a cytoplasmic coiled-coil domain. J Biol Chem. 1999;274:2953–2962. doi: 10.1074/jbc.274.5.2953. [DOI] [PubMed] [Google Scholar]
- 14.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 15.Weide T, Bayer M, Köster M, Siebrasse JP, Peters R, Barnekow A. The Golgi matrix protein GM130. a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2001;2:336–341. doi: 10.1093/embo-reports/kve065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valsdottir R, Hashimoto H, Ashman K, Koda T, Storrie B, Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 2001;508:201–209. doi: 10.1016/s0014-5793(01)02993-3. [DOI] [PubMed] [Google Scholar]
- 17.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 18.Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- 19.Rabouille C, Misteli T, Watson R, Warren G. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J Cell Biol. 1995;129:605–618. doi: 10.1083/jcb.129.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambourg A, Clermont Y. Three-dimensional electron microscopy: structure of the Golgi apparatus. Eur J Cell Biol. 1990;51:189–200. [PubMed] [Google Scholar]
- 21.Misumi Y, Sohda M, Tashiro A, Sato H, Ikehara Y. An essential cytoplasmic domain for the Golgi localization of coiled-coil proteins with a COOH-terminal membrane anchor. J Biol Chem. 2001;276:6867–6873. doi: 10.1074/jbc.M010121200. [DOI] [PubMed] [Google Scholar]
- 22.Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, Der CJ, Balch WE. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seemann J, Jokitalo E, Pypaert M, Warren G. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 2000;407:1022–1026. doi: 10.1038/35039538. [DOI] [PubMed] [Google Scholar]
- 25.Miles S, McManus H, Forsten KE, Storrie B. Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J Cell Biol. 2001;155:543–555. doi: 10.1083/jcb.200103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeffer SR. Constructing a Golgi complex. J Cell Biol. 2001;155:873–875. doi: 10.1083/jcb.200109095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeffer S. Vesicle tethering factors united. Mol Cell. 2001;8:729–730. doi: 10.1016/s1097-2765(01)00371-9. [DOI] [PubMed] [Google Scholar]
- 29.Burke B, Griffiths G, Reggio H, Louvard D, Warren G. A monoclonal antibody against a 135-K Golgi membrane protein. EMBO J. 1982;1:1621–1628. doi: 10.1002/j.1460-2075.1982.tb01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christoforidis S, Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]


