Abstract
The proteasome recognizes its substrates via a diverse set of ubiquitin receptors, including subunits Rpn10/S5a and Rpn13. In addition, shuttling factors, such as Rad23, recruit substrates to the proteasome by delivering ubiquitinated proteins. Despite the increasing understanding of the factors involved in this process, the regulation of substrate delivery remains largely unexplored. Here we report that Rpn10 is monoubiquitinated in vivo and that this modification has profound effects on proteasome function. Monoubiquitination regulates the capacity of Rpn10 to interact with substrates by inhibiting Rpn10’s ubiquitin interacting motif (UIM). We show that Rsp5, a member of NEDD4 ubiquitin-protein ligase family, and Ubp2, a deubiquitinating enzyme, control the levels of Rpn10 monoubiquitination in vivo. Notably, monoubiquitination of Rpn10 is decreased under stress conditions, suggesting a mechanism of control of receptor availability mediated by the Rsp5-Ubp2 system. Our results reveal an unanticipated link between monoubiquitination signal and regulation of proteasome function.
Keywords: proteasome, ubiquitin, monoubiquitination, Rpn10 (S5a), Rsp5 (Nedd4), Ubp2
Introduction
A crucial aspect of the ubiquitin-proteasome pathway is the regulation of productive interaction between polyubiquitinated substrates and the proteasome (Finley, 2009). A failure of this regulation may lead to proteasome dysfunction and to protein accumulation, events observed in multiple pathologies (Ciechanover and Brundin, 2003). The proteasome is composed of a core particle (CP, or 20S particle) and a regulatory particle (RP, 19S, or PA700; Glickman et al., 1998). The CP is a barrel-shaped complex, which contains multiple proteolytic active sites facing its interior. The RP recognizes, unfolds and translocates targeted substrates into the CP. There are several factors involved in the recruitment of targeted proteins to the RP of the proteasome. Among them, Rpn10/S5a has been shown to play a role in binding ubiquitin conjugates by means of a ubiquitin interacting motif (UIM; Deveraux et al., 1994; Van Nocker et al., 1996; Fu et al., 1998). For example, Rpn10 mediates the targeting to the proteasome of cyclin B1, Sic1, Gic2 and Gcn4 (Verma et al., 2004; Hanna et al., 2006; Seong et al., 2007). Mutation of RPN10 UIM produces a decrease in the proteolytic capacity of the proteasome (Elsasser et al., 2004; Verma et al., 2004) and is lethal in mouse (Hamazaki et al., 2007). Recently, roles of extraproteasomal Rpn10 in controlling ubiquitin chain synthesis and regulating Dsk2, another ubiquitin receptor, have been proposed (Matiuhin et al., 2008; Kim et al., 2009; Zhang et al., 2009). Rpn13, another subunit of the base of the RP which contains a PRU domain, efficiently binds ubiquitin and promotes the recruitment of ubiquitin conjugates (Husnjak et al., 2008).
Ubiquitin chain recognition by the proteasome also involves UBL-UBA domain proteins such as Rad23, Ddi1, and Dsk2, which shuttle ubiquitinated substrates to the proteasome. Proteasomes deficient in Rad23 show a decrease in association of ubiquitin conjugates (Elsasser et al., 2004) and in protein degradation (Verma et al., 2004). Rad23 functions exhibit partial redundancy with respect to Rpn10. For instance, RPN10 and RAD23 mutations have additive effects in proteolytic stress phenotypes (Saeki et al., 2002; Elsasser et al., 2004).
Monoubiquitination is a molecular event different from polyubiquitination that drives the recruitment of proteins containing ubiquitin binding domains (UBDs). Monoubiquitination thus provides a signalling mechanism that regulates important cellular pathways such as DNA repair, histone function and endocytosis (Kirkin and Dikic, 2007). Although multiple roles of monoubiquitin and polyubiquitin signals in intracellular proteolysis in eukaryotes have been established, there is no evidence of involvement of monoubiquitin signals in the regulation of protein degradation by the proteasome.
Here we show that Rpn10 is regulated by monoubiquitination. Monoubiquitination strongly inhibits the capacity of Rpn10 to interact with substrates, thus decreasing proteasome activity. We show that Rsp5 and Ubp2 control the levels of Rpn10 monoubiquitination, at K71, K84 and K99, located within the VWA domain, and in K268, located at the C-terminus of the protein. We provide genetic evidence that link monoubiquitination of Rpn10 with proteasome function. In addition, cold shock, heat shock and cadmium reduce Rpn10 monoubiquitination. We propose that Rpn10 monoubiquitination acts as a stress sensitive mechanism that controls the recruitment of substrates to the proteasome.
Results
Rpn10 is Monoubiquitinated In vivo
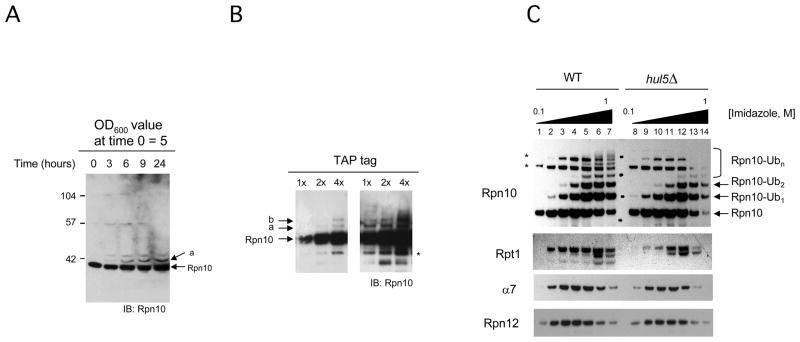
In a recent study, it was shown that Rpn10 is degraded by the proteasome and that the ubiquitin ligase Hul5 is involved in this process (Crosas et al., 2006). To further examine the physiological significance of ubiquitination of Rpn10 we analyzed the status of Rpn10 protein in exponentially growing cultures. In direct analysis of cell extracts, an additional Rpn10 inmunoreactive band with slower mobility was observed, suggestive of postranslational modification by ubiquitin (Figure 1A, b and a). The band was not observed in cultures from cells carrying a deletion of the RPN10 gene (Figure S1A). We then purified Rpn10 expressed from an inducible vector (Figure S1B) and from its own chromosomal locus by means of an integrated C-terminal Tandem Affinity Purification (TAP) tag (Figure 1B). By western blot analysis of purified samples it was observed that Rpn10 showed band ‘a’ again and an additional form (band ‘b’) that suggested modification by two ubiquitin groups (Figure 1B). Band ‘a’ was analyzed by MS and two unique abundant proteins, Rpn10 and ubiquitin, were identified (Figure S1C). These approaches were not successful in detecting polyubiquitinated Rpn10, probably reflecting a relative low abundance of longer ubiquitin intermediates of Rpn10. Using a strain that expresses ubiquitin with a 6xHIS N-terminal tag, pulldown assays of total cell ubiquitin conjugates were performed. With this approach we observed, in addition to the most abundant form corresponding to monoubiquitinated Rpn10 (mUb-Rpn10; Figure 1C, lanes 2 to 7), diubiquitinated Rpn10 (lanes 3 to 7) and polyubiquitinated Rpn10 (lanes 6 and 7). Synthesis of polyubiquitined forms of Rpn10 is catalyzed by the chain elongating factor Hul5, which associates with the proteasome (Crosas et al., 2006). We performed the same ubiquitin conjugate purification procedure using a strain that carries a deletion of HUL5 gene. We observed that, in the absence of Hul5, polyubiquitinated Rpn10 virtually disappeared (Figure 1C, lanes 13 and 14; see also Crosas et al., 2006), but levels of mono and diubiquitinated Rpn10 were not affected (lanes 9 to 14), suggesting that a Hul5-independent ubiquitin ligating activity is responsible for Rpn10 mono- and diubiquitination.
Figure 1. Rpn10 is monoubiquitinated in vivo.
(A) Logarithmically growing wild-type yeast cells (see strain list in Table S1) analyzed by western blotting using Rpn10 antibody. Time points were taken as shown.
(B) Purified TAP-tagged Rpn10, analyzed by western blotting. Increasing amounts of fractions were visualized by short (left) and long film exposures. Bands ‘a’ and ‘b’ represent putative mono and diubiquitinated Rpn10. Asterisk, Rpn10 breakdown product.
(C) Native 6xHis-Ubiquitin conjugates eluted at different imidazole concentrations (lanes 1 to 7), analyzed by western blotting with Rpn10 antibody. The same purification and analysis procedure was performed using a hul5Δ strain (lanes 8 to 14). Immunoblots with antibodies against proteasome subunits Rpt1, α7 and Rpn12 are included. Asterisks, unspecific bands detected with Rpn10 antibody.
Monoubiquitinated Rpn10 is Found in Both Proteasomal And Non-Proteasomal Contexts
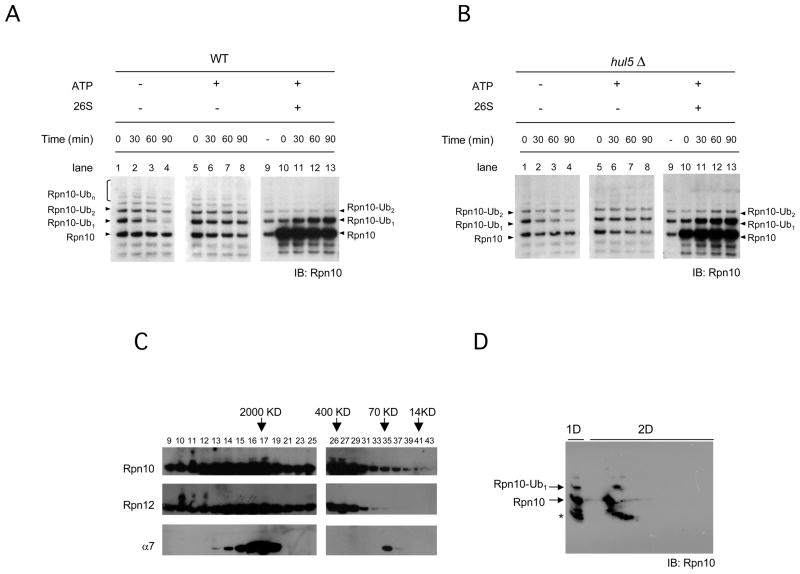
Since a significant fraction of cellular Rpn10 is not bound to the proteasome (Fu et al., 1998; Hiyama et al., 1999; Matiuhin et al., 2008; Kim et al,. 2009; this work) we sought to determine the cellular context of Rpn10 monoubiquitination. We isolated protein fractions rich in enzymatic factors of the ubiquitin-proteasome system, in ubiquitin-protein conjugates, and in Rpn10 (fractions UR8 and UR10; see Figure S2A and supplemental data). We used fraction UR8 as a catalytically active extract. Incubations of UR8 fraction with MG132 in the absence of ATP showed that mUb-Rpn10 decreased rapidly whereas, strikingly, the rest of ubiquitin modified forms of Rpn10 remained constant (Figure 2A, lanes 1 to 4). These results suggested the involvement of a deubiquitinating enzyme highly specific for mUb-Rpn10. When the fraction was incubated in the presence of ATP, mUb-Rpn10 did not decline; instead it remained stable (Figure 2A, lanes 5 to 8), suggesting that an ATP-dependent activity was counteracting Rpn10 deubiquitination by catalyzing Rpn10 monoubiquitination or by inhibiting deubiquitination. To address this question we added to the reaction conventionally purified proteasomes, which contain unmodified Rpn10, and we observed that added proteasomal Rpn10 was robustly monoubiquitinated (Figure 2A, lanes 10 to 13). We performed the same assays shown in Figure 2A but using protein fractions and proteasomes purified from strains carrying a deletion of the HUL5 gene. Using hul5Δ samples we observed the same behaviour as with wild-type (Figure 2B). Proteasomes used in these assays were unable to catalyze Rpn10 monoubiquitination by themselves, but simply adding fraction UR8, ten fold diluted, reconstituted the reaction efficiently, indicating a high turnover rate of the Rpn10-ubiquitin ligating activity contained in this extract (Figure S2B). These findings suggest that levels of mUb-Rpn10 are controlled in vivo by dynamically opposed ubiquitin ligase and deubiquinating activities, and that substoichiometric amounts of the ubiquitin ligase bound to the proteasome are sufficient to promote Rpn10 monoubiquitination.
Figure 2. Monoubiquitination of proteasomal Rpn10 is dynamically regulated.
(A–B) Fraction UR8 (see supplemental data and Figure S2A) was incubated without ATP (lanes 1 to 4), with 5mM ATP (lanes 5 to 8) or with 5 mM ATP and purified proteasomes (lanes 10 to 13), at 30 °C, and time points were taken. In B, assays were performed using UR8 fraction and proteasomes from hul5Δ strains.
(C) Total cell lysates from a wild-type strain were applied to a Superose 6 column. Fractions obtained were analyzed by western blotting of Rpn10, Rpn12 or α7 proteasome components.
(D) Proteasomes purified in the presence of 5 mM ATP and 5 μM of MG132 were analyzed by two dimensioned electrophoresis (isoelectrofocusing and 4–12% gradient SDS-PAGE) followed by Rpn10 immunodetection. Asterisk, Rpn10 breakdown product.
To corroborate the observation of physiological Rpn10 monoubiquitination we performed a fractionation of a whole cell extract by Superose 6 chromatography. In this analysis, mUb-Rpn10 was present both in fractions corresponding to the proteasome elution peak, analyzed by immunodetection of Rpn12, a component of the lid of the proteasome, and α7, a core particle subunit (Figure 2C, lanes 16 and 17), and in the non-proteasomal peak of Rpn10 (Figure 2C, lanes 26 and 27). Moreover, proteasomes isolated using Rpn11-Protein A tag (Leggett et al., 2002), in the presence of ATP and MG132, contained mUb-Rpn10 (Figure 2D). These results provide evidence of in vivo Rpn10 monoubiquitination.
Rsp5 Ubiquitin Ligase and Ubp2 Deubiquitinating Enzyme Control Rpn10 Monoubiquitination
We searched for enzymatic factors involved in controlling the levels of mUb-Rpn10. Among likely candidates were ubiquitin ligases that interact with the proteasome, such as Ufd4 and Ubr1 (Xie and Varshavsky, 2000). We purified proteasomes from ufd4Δ and ubr1Δ strains and found that the levels of mUb-Rpn10 were identical to those observed in proteasomes purified from a wild-type strain (Figure S3A), suggesting that neither Ufd4 nor Ubr1 were ubiquitin ligases for Rpn10.
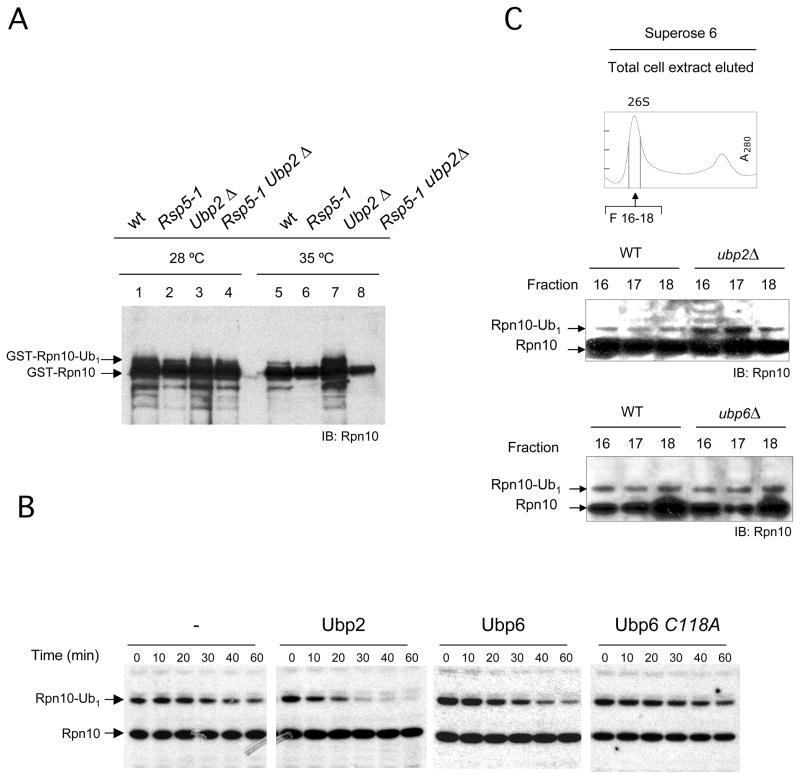
In addition to a functional interaction with the proteasome, a signature feature of the E3 enzyme that we were searching for might be efficient catalysis of monoubiquitination. An E3 that fits with this second requirement is Rsp5, orthologue of NEDD4.2 mammalian enzyme (Dupré et al., 2004), which is involved in many cellular processes in Saccharomyces cerevisiae and part of a large family of proteins that control analogous processes in mammalian cells (Hicke, 2001). Rsp5 is founding a complex with the deubiquitinating enzyme Ubp2, which exhibits antagonistic activity (Kee et al., 2005). Recently, Rpn10 was a positive hit in a proteomic screen for Rsp5 substrates (Lu et al., 2008). Therefore, we analyzed the putative involvement of Rsp5 and Ubp2 in Rpn10 monoubiquitination. Wild-type, the rsp5–1 thermosensitive mutant, ubp2Δ, and double mutant rsp5–1 ubp2Δ strains were used, all of them carrying a GST-Rpn10 expressing plasmid. Pulled down fractions from cultures grown under galactose induction and restrictive temperature (see supplemental data and Figure S3B) were analyzed by anti-Rpn10 western blotting (Figure 3A). Strikingly, the presence of mUb-Rpn10 was completely dependent on Rsp5 activity (Figure 3A, lanes 6 and 8), suggesting that Rsp5 is the major E3 for Rpn10 in vivo. In addition, levels of mUb-Rpn10 were strongly increased in the absence of Ubp2 (Figure 3A, lane 7). To corroborate the involvement of Rsp5 in Rpn10 monoubiquitination in vivo, we used a rsp5Δ strain and analyzed the status of endogenous Rpn10. Cultures of rsp5Δ strains, grown in the presence of 1M sorbitol to rescue growth (Figure S3C; Kee et al., 2005), showed a strong decrease of endogenous mUb-Rpn10 levels as compared to a wild-type strain (Figure S3D).
Figure 3. Levels of monoubiquitinated Rpn10 are controlled by Rsp5-Ubp2 enzymatic system in vivo.
(A) Wild-type, rsp5-1, ubp2Δ and double rsp5-1 ubp2Δ strains, carrying GST-Rpn10 galactose inducible plasmid (plasmids are listed in Table S2), were expressed and shifted to restrictive temperature. GST-Rpn10 was pulled down and analyzed by Rpn10 western blotting. Lanes 1 to 4, cultures grown at 28 °C. Lanes 5 to 8, cultures grown at 35 °C. Induction levels are shown in Figure S3B.
(B) Monoubiquitinated Rpn10 from a ubp6Δ strain (see Figure S3E and supplemental data) was incubated with equimolar amounts of Ubp2, Ubp6 and Ubp6C118A. Reactions were analyzed by Rpn10 western blotting.
(C) Total cell lysates from wild-type, ubp2Δ and ubp6Δ strains were applied to a Superose 6 chromatography. Fractions that contain eluted proteasome were analyzed by Rpn10 western blotting.
To characterize deubiquitination of proteasomal mUb-Rpn10, we considered, in addition to Ubp2, the putative activity of Ubp6, which is involved in substrate deubiquitination in the proteasome and is related to Hul5 (Leggett et al., 2002; Hannah et al., 2006; Crosas et al., 2006). We performed assays using recombinant Ubp2, Ubp6, the inactive mutant Ubp6C118A, and proteasome fractions containing mUb-Rpn10, from ubp6Δ strains (Figure S3E). mUb-Rpn10 was processed by both Ubp2 and Ubp6, but Ubp2 was more efficient in the reaction (Figure 3B). To observe the effect of Ubp2 and Ubp6 on the levels of proteasomal mUb-Rpn10 in vivo, cellular extracts from ubp2Δ and ubp6Δ strains were fractionated by Superose 6 chromatography and the proteasomal peak was analyzed by western blotting. It was observed that mUb-Rpn10 was significantly increased in proteasomes from ubp2Δ cells with respect to wild-type proteasomes (Figure 3C). However, fractions from ubp6Δ cellular extracts showed levels of proteasomal mUb-Rpn10 identical to the ones of extracts from wild-type cells (Figure 3C). These results suggest a physiological role of Ubp2, but not of Ubp6, in Rpn10 deubiquitination. Therefore, our data suggest that the catalytic cycle that controls homeostasis of mUb-Rpn10 is largely independent of ubiquitin chain processing by Hul5 and Ubp6 in the proteasome.
Monoubiquitination of Rpn10 In vitro
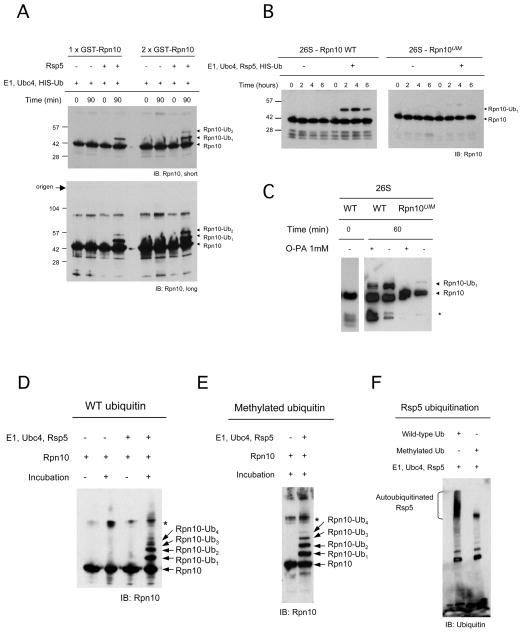
To establish the direct involvement of Rsp5 in the catalysis of Rpn10 monoubiquitination we attempted to reconstitute the enzymatic reaction in vitro. Incubations of free or proteasomal Rpn10 with recombinant E1, Ubc4, Rsp5 and 6xHIS-ubiquitin showed that catalysis was promoted only in the presence of Rsp5, excluding an E3-independent monoubiquitination (Hoeller et al., 2007), and that monoubiquitination was efficiently promoted for both free and proteasomal Rpn10 (Figure 4A and 4B, left panel). A remarkable feature of these reactions is that they are highly specific in producing mUb-Rpn10, and only synthesis of Rpn10-Ub2 was observed secondarily (Figure 4A, long exposure). These results recapitulate the observations of Rpn10 monoubiquitination with endogenous extracts (Figure 2A and 2B) and in hul5Δ samples in vivo (Figure 1C), suggesting that the reaction in vitro largely reproduces the physiological conditions.
Figure 4. Reconstitution of the reaction of Rpn10 monoubiquitination in vitro.
(A) Recombinant Rsp5, Rpn10, E1, Ubc4 and ubiquitin were incubated as indicated. Right lanes show a reaction in which Rpn10 input was doubled. Lower panel, longer exposures which show diubiquitinated Rpn10 (Rpn10-Ub2) more clearly.
(B) Proteasomes (wild-type and Rpn10UIM) were incubated with Rsp5, E1, Ubc4 and ubiquitin.
(C) Proteasomes (wild-type and Rpn10UIM) were incubated with a cell lysate fraction. An inhibitory effect of o-phenanthroline was observed when 1 mM final concentration of this compound was added to the reaction. Asterisk, Rpn10 breakdown product.
(D–E) Scaled-up reactions of Rpn10 monoubiquitination in vitro, using wild-type (D) or methylated ubiquitin (E). Asterisk, unspecific band also observed in 4A.
(F) Autoubiquitinated Rsp5 using wild-type or methylated ubiquitin, from reactions similar to (D) and (E), respectively.
Rpn10 UIM is Required for Rpn10 Monoubiquitination
The NEDD4 ubiquitin-protein ligase family detects substrates and protein adaptors that contain PPY or UIM motifs (Polo et al., 2002; Hicke and Dunn, 2003; Dupré et al., 2004; Hoeller et al., 2006). The latter type of protein-protein interaction usually requires a functional UIM and promotes the so-called ‘coupled’ monoubiquitination (Hoeller et al., 2006). Since Rpn10 is a UIM protein, we assessed whether this motif is required for monoubiquitination. We purified proteasomes from a strain that carries the Rpn10UIM mutant (Elsasser et al., 2004) and performed Rpn10 monoubiquitination reactions. We observed a dramatic inhibition of the synthesis of mUb-Rpn10 in Rpn10UIM proteasomes, with respect to wild-type proteasomes (Figure 4B). In addition, when the activity of cell fraction UR8 towards wild-type and Rpn10UIM proteasomes was challenged, it was observed again that monoubiquitination was not promoted in the Rpn10UIM mutant (Figure 4C). These results show that the UIM is necessary for Rpn10 monoubiquitination.
Monoubiquitination Takes Place in Distinct Lysines of Rpn10 and is Required to Rescue Proteasome Function
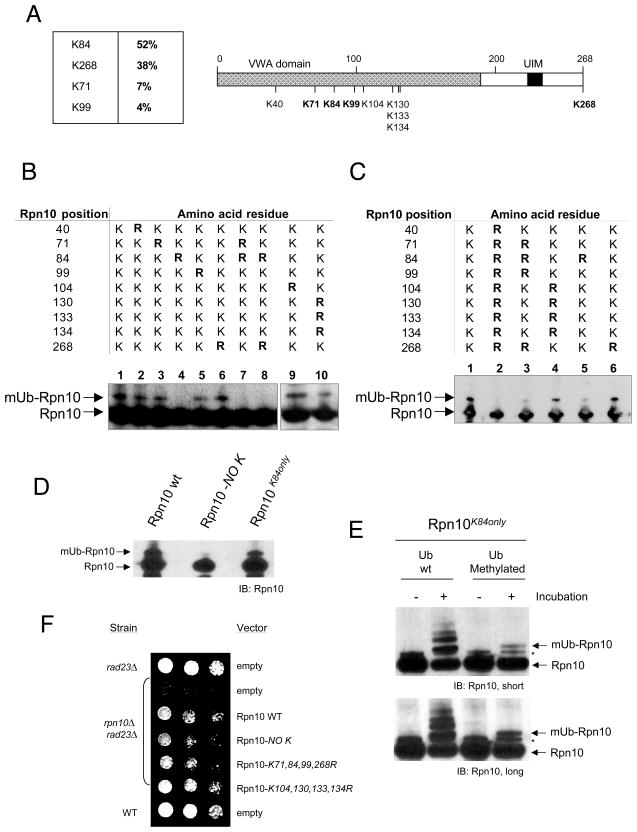
We scaled up the in vitro reaction of Rpn10 monoubiquitination in order to produce higher amounts of mUb-Rpn10 for MS analysis. In optimized reactions, large amounts of mono and diubiquitinated forms of Rpn10 were produced, including tri- and tetraubiquitinated Rpn10 at lower levels, but no polyubiquitinated Rpn10 (Figure 4D). However, with this assay we could not distinguish the topology of the ubiquitin linkage formed because multi-monoubiquitination and short ubiquitin chains produce a similar electrophoretic mobility shift. To address this important issue we performed the reaction using methylated ubiquitin, which cannot form Ub chains (Hershko and Heller, 1985). Strikingly, this reaction (Figure 4E) produced the same pattern of ubiquitination observed using wild-type ubiquitin (Figure 4D), including four bands of Rpn10 modification, which is similar to the pattern observed in purified endogenous ubiquitinated Rpn10 (Figure 1C, lane 14; se also Crosas et al., 2006). In addition, a notable observation is that Rsp5, although inactive in the synthesis of polyubiquitinated forms of Rpn10, undergoes auto-polyubiquitination (Figure 4F), likely generating K63-linked ubiquitin chains (Saeki et al., 2009). These results suggest that Rpn10 is multi-monoubiquitinated at four distinct lysine residues. To detect the residues modified by ubiquitin in this reaction, the two major produced bands corresponding to Rpn10-Ub1 and Rpn10-Ub2 (Figure S4A) were excised from the gel and analyzed by LC-MS/MS. The analysis revealed that monoubiquitination could engage K71, K84, K99 and K268 (Figure 5A and Figure S4B). K71, K84, and K99 are located within the VWA domain, whereas K268 is situated at the very C-terminus of the protein. In Rpn10-Ub2 band only modifications at K84 and K268 were identified and, in the whole analysis, K84 was the most abundantly modified, representing the 52% of the GlyGly-containing peptides (Figure 5A).
Figure 5. Analysis of Rpn10 lysines modified by ubiquitin.
(A) Summary of mass spectrometry analysis of Rpn10. Left, relative abundance of the Gly-Gly peptides found. Right, scheme of Rpn10 protein including all lysines contained in the sequence. Modified lysines are shown in bold.
(B–D) GST-Rpn10 K to R mutants including all Rpn10 lysines (oligos are listed in Table S3) were expressed under galactose induction. Pulled down GST-Rpn10 forms were analyzed by Rpn10 western blotting.
(E) Monoubiquitination reactions of Rpn10K84only mutant using wild-type and methylated ubiquitin. Asterisk, electrophoretic artifact shown by the mutant, observed also in non-incubated samples. Short and long exposures of the film are shown.
(F) Ability of a set of Rpn10 mutants to rescue growth defect exhibited by rpn10Δrad23Δ yeast strain (see supplemental data). Rpn10 forms were expressed at 22 °C using a galactose inducible vector, selected with a URA3 marker. Cells (3×104) were spotted in the first column and 3/7 serial dilutions were made for the successive columns.
To further characterize Rpn10 modification, lysine residues of Rpn10 were mutated (see supplemental data). In GST pulldowns, forms involving Rpn10K84R mutation showed a strong decrease in monoubiquitination (Figure 5B, lanes 4, 7 and 8), while mutations in other lysines produced only mild or undetectable effects (Figure 5B). Nonetheless, our results show that additional lysines may be modified. Thus, to analyze Rpn10 forms defective in ubiquitination we prepared Rpn10K71,84,99,268R and Rpn10NO–K mutants, and the control mutant Rpn10K104,130,133,134R. Analysis of GST pulldowns of these forms showed that the Rpn10K104,130,133,134R mutant exhibited monoubiquitination to wild-type levels (Figure 5C, lanes 1 and 4), whereas Rpn10K71,84,99,268R (lane 3) and Rpn10NO–K (lane 2) mutants showed very low levels of modification. In a parallel assay, Rpn10K84R mutant showed a decrease in monoubiquitination (lane 5) similar to that observed in the Rpn10K71,84,99,268R mutant. K84 monoubiquitination was also analyzed with the Rpn10K84only mutant, in which we mutated all lysine residues to arginine except for K84. This form was expressed in yeast and showed monoubiquitination nearly to physiological levels (Figure 5D), suggesting a dominant role of K84 in the process of Rpn10 monoubiquitination. We performed reactions in vitro using recombinant Rpn10K84only as a substrate. We observed that, while methylated ubiquitin produced one band of monoubiquitination, wild-type ubiquitin produced several bands (Figure 5E), suggesting that, secondarily (see Figures 4D and E), Rsp5 is able to build short ubiquitin chains, maybe due to the absence of additional lysine residues in the mutant Rpn10 sequence.
To assess the functional relevance of Rpn10 monoubiquitination, a complementation test was performed. Strains carrying a double deletion of RPN10 and RAD23 genes are strongly deficient in recruiting substrates to the proteasome and exhibit slow growth (Figure S4C; Chen and Madura, 2002). We tested the capacity of plasmid-borne Rpn10 to rescue the absence of the RPN10 gene. We observed that wild-type form of Rpn10 efficiently rescued growth of a rpn10Δrad23Δ strain (Figure 5F). Similarly, the rpn10K104,130,133,134R mutant, which shows monoubiquitination at wild-type levels, fully complemented Rpn10 function. However, the rpn10K71,84,99,268R and the rpn10NO–K mutants, which show impaired monoubiquitination, did not rescue Rpn10 function to WT levels (Figure 5F). To our knowledge, this is the first genetic evidence linking proteasome function with monoubiquitination of a protein.
Monoubiquitinated Rpn10 Shows Low Affinity to Ubiquitin Conjugates
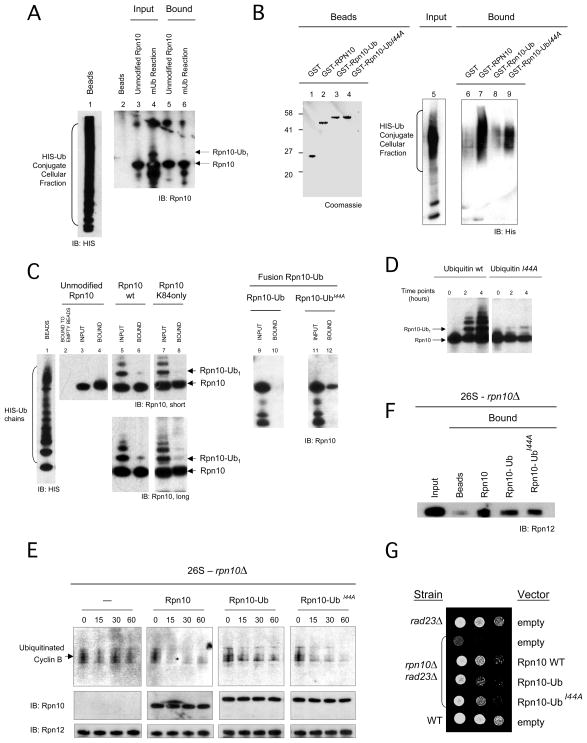
We asked what was the role of monoubiquitination in the context of proteasome function. The UIM of Rpn10 is involved in the recruitment of substrates to the proteasome (Fu et al., 1998; Elsasser et al., 2004, Verma et al., 2004). The interaction between ubiquitin and Rpn10 UIM relies on a hydrophobic patch on the surface of ubiquitin, composed of Leucine 8, Isoleucine 44, and Valine 70, which define a pocket for the methyl group of a strictly conserved alanine within the UIM, alanine 231 in the case of Rpn10 (Wang et al., 2005). We tested whether in mUb-Rpn10, the covalently linked ubiquitin group could impose a functional restriction to Rpn10 UIM by means of a ‘fold-back’ interaction (Di Fiore, 2003; Woelk et al., 2006). We obtained a cellular fraction containing 6xHIS tagged polyubiquitin conjugates (see supplemental data and Figure S5A, lane 10) and immobilized it to Ni-NTA resin (Figure 6A, lane 1). The binding capacity of Rpn10 and mUb-Rpn10 (synthesized in vitro, as in Figure 4A) to immobilized conjugates was challenged. We observed that unmodified Rpn10 could bind very efficiently to conjugates, but mUb-Rpn10 did not (Figure 6A, lanes 5 and 6), suggesting that monoubiquitination of Rpn10 inhibits the activity of the UIMs. In the conditions of the binding assay no Rpn10 deubiquitination was observed (Figure S5B). To further characterize this interaction, we generated permanently monoubiquitinated forms by appending a ubiquitin group to Rpn10. We included the Rpn10-UbI44A mutant, which shows a decreased affinity of linked ubiquitin to the UIM (Hoeller et al., 2006). Thus, different forms were expressed and bound to GSH-beads (Figure 6B, lanes 1–4) and their affinity to the cellular fraction of polyubiquitin-conjugates, as the liquid phase, was tested (lane 5). As in previous assay, conjugates bound strongly to Rpn10 (lane 7) but not to Rpn10 linked to Ub (lane 8). Interestingly, the Rpn10-UbI44A mutant partially re-established the capacity of Rpn10 to bind conjugates, showing that the non-covalent ubiquitin-UIM interaction is involved in the inhibition of the UIM.
Figure 6. Monoubiquitinated Rpn10 shows an inactive UIM and decreases the proteolytic activity of the proteasome.
(A) Binding assay of Rpn10 and mUb-Rpn10 to a fraction of endogenous HIS-ubiquitin conjugates (see Figure S5A and supplemental data) immobilized on Ni-NTA beads (lane 1). Liquid phase inputs include unmodified Rpn10, as a control (lane 3), and a monoubiquitination reaction sample (mUb reaction), which contains both unmodified Rpn10 and mUb-Rpn10 (Rpn10-Ub1) (lane 4). Bound material is shown in lanes 5 and 6.
(B) Binding assay of Rpn10 and C-terminal ubiquitin fusions (Rpn10-Ub and Rpn10-UbI44A), immobilized in beads (lanes 1 to 4), to a fraction of endogenous HIS-ubiquitin conjugates (input, lane 5). Bound material was eluted and analyzed by HIS western blotting (lanes 6 to 9).
(C) Binding assay of unmodified and mUb-Rpn10 (wild-type and K84only mutant), Rpn10-Ub and Rpn10-UbI44A to unanchored polyubiquitin chains immobilized in beads (lane 1). Input and bound material are shown for each sample (lanes 3 to 12). A control of Rpn10 binding to empty beads is included in lane 2. A longer film exposure is shown for lanes 5 to 8.
(D) Reaction of Rpn10 monoubiquitination in vitro using wild-type ubiquitin and I44A mutant.
(E) Time-course degradation assays in vitro with proteasomes deficient in Rpn10, equimolar amounts of Rpn10 forms and ubiquitinated cyclin B. Points at indicated times were taken and analyzed by anti cyclin B, Rpn10 and Rpn12 western blotting.
(F) Rpn10-Ub binding to rpn10Δ proteasomes. GST-Rpn10, GST-Rpn10-Ub and GST-Rpn10-UbI44A were immobilized to beads and proteasomes were applied as the soluble phase.
(G) Rescue of Rpn10 function by Rpn10-Ub forms, including Rpn10, Rpn10-Ub and Rpn10-UbI44A expressed from a vector. The assay was performed as in figure 5F. Levels of protein expression are shown in Figure S5D.
In the previous assay we used polyubiquitin conjugates isolated from cells, therefore we could not exclude the interference of cellular factors associated to ubiquitin conjugates in the binding assay. Thus, we challenged the affinity of Rpn10 and Rpn10 modified forms towards pure unanchored polyubiquitin chains (see supplemental data). In this assay the Rpn10K84only mutant was included to observe the behaviour of Rpn10 modified uniquely at K84. Again, binding assays showed that immobilized polyubiquitin chains (Figure 6C, lane 1) bound efficiently to unmodified Rpn10 (wild-type and Rpn10K84only mutant) but not their respective monoubiquitinated forms (Figure 6C, lanes 3–8). We also tested the affinity of the Rpn10-Ub fusion towards unanchored polyubiquitin chains. Consistently with the assay using cellular polyubiquitin conjugates (Figure 6B), Rpn10-Ub did not bind to ubiquitin chains (Figure 6C, lanes 9–10), whereas the Rpn10-UbI44A mutant partially re-established binding (lanes 11–12). Therefore, mUb-Rpn10 synthesized by Rsp5 and the Rpn10-Ub chimera exhibit the same feature: a dramatic decrease in their affinity to ligands, suggesting that Rpn10-Ub fusion is a good approach to study the effect of monoubiquitination in Rpn10 UIM function. Moreover, the partial rescue of polyubiquitin binding capacity of the Rpn10-UbI44A mutant showed that the decreased affinity to polyubiquitin is due to an intramolecular UIM-monoubiquitin hydrophobic interaction.
According to the presented data, the interaction UIM-ubiquitin appears to be essential for both catalysis (Figure 4B and C) and function (Figure 6A–C) of Rpn10 monoubiquitination. The capacity of ubiquitinI44A to promote Rpn10 monoubiquitination was tested in vitro and very poor activity was observed, as compared to the reaction using wild-type ubiquitin (Figure 6D). These results complement the data obtained using Rpn10UIM mutant (Figure 4B), and, altogether, show that Rsp5 requires the Rpn10 UIM-ubiquitin hydrophobic interaction to be active.
Monoubiquitination of Rpn10 Reduces the Proteolytic Activity of the Proteasome
In Figures 6A–C we show that mUb-Rpn10 exhibits low affinity to polyubiquitin and polyubiquitinated substrates but we do not show the effect in active proteasomes. To address this question, we performed cyclin B degradation tests in a time course fashion, using equimolar amounts of Rpn10 and Rpn10-Ub forms added to rpn10Δ proteasomes (Figure S5C). We observed that degradation rates of cyclin B were strongly accelerated when Rpn10 was added, slightly accelerated when Rpn10-UbI44A was added, and substantially inhibited in absence of Rpn10 or adding Rpn10-Ub (Figure 6E). However, these results could also be explained by a decreased proteasome interaction of the Rpn10-Ub form. To rule out this possibility, we compared the affinity of Rpn10, Rpn10-Ub and Rpn10-UbI44A to rpn10Δ proteasomes by binding assays, and observed that all Rpn10 forms bound identically to the proteasome (Figure 6F), showing that degradation of cyclin B is promoted by a proteasome-bound Rpn10.
If mUb- Rpn10 shows an inactive UIM, one could expect a strong decrease of functional complementation in vivo of Rpn10-Ub compared to wild-type Rpn10, and an intermediate complementation of Rpn10-UbI44A (see Figures 6B, C and E). To check this hypothesis, we expressed these proteins in rpn10Δ rad23Δ cells. We observed that Rpn10-Ub could not complement rpn10 null mutants efficiently, showing the strong effect of permanent monoubiquitination in Rpn10 (Figure 6G). This effect was dependent on the UIM-ubiquitin interaction of Rpn10-Ub, because the rpn10-UbI44A mutation, consistently with binding assays, partially re-established complementation (Figure 6G). The levels of expression of plasmid-borne RPN10 variants were similar to that of endogenous Rpn10 in all cases (Figure S5D).
Rpn10 Monoubiquitination is Reduced Under Conditions of Proteolytic Stress
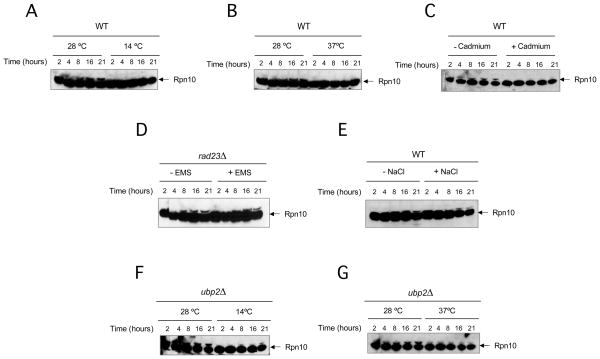
Our observations suggested that monoubiquitination of Rpn10 could regulate the capacity of this substrate receptor to bind ubiquitinated substrates. We asked whether the levels of mUb-Rpn10 could be influenced by conditions that require functional Rpn10, such as proteolytic stress (Medicherla et al., 2008). We analyzed the status of Rpn10 in cultures growing at 14, 28 or 37 °C (Figure 7A, B, F and G). We observed that Rpn10 monoubiquitination was strongly decreased in cells subjected to either cold or heat shock. We asked whether a similar response to stress could also be observed at 28 °C in the presence of cadmium, a compound that promotes proteolytic stress (Jungmann et al., 1993; Medicherla et al., 2008). We observed that, in the presence of cadmium, Rpn10 monoubiquitination was not induced (Figure 7C). We then asked whether the tight control of mUb-Rpn10 levels was a specific response to proteolytic stress or could be promoted by other kind of perturbations. To address this question we supplemented cultures with ethyl methyl sulfoxide (EMS) to cause DNA damage, or NaCl to induce osmotic stress in the same manner than in previous assays. We observed that Rpn10 monoubiquitination persisted under these conditions (Figure 7D and E), showing a specific correlation of elevated protein degradation with low levels of mUb-Rpn10.
Figure 7. Monoubiquitination of Rpn10 is decreased under certain stress conditions.
(A–G) Yeast cells (wild-type, rad23Δ and ubp2Δ) were grown in YPD liquid media at the described conditions. Cadmium: 200 μM, Ethyl methyl sulfoxide (EMS): 0.08%, NaCl: 500 mM. Samples were taken at indicated times, number of cells normalized and analyzed by western blotting with Rpn10 antibody.
Discussion
Monoubiquitination Of A Polyubiquitin Receptor
Monoubiquitination is conserved from yeast to mammals and involved in important cellular processes such as endocytosis and regulation of nuclear functions (Hicke, 2001; Dupré et al., 2004; Kirkin and Dikic, 2007). Monoubiquitinated proteins are usually recognized by specific receptors that contain UBDs. For example, in the endocytic pathway, Vps9, Sts1, Sts2, Eps15 or Hrs are UBD-containing protein adaptors that trigger protein internalization by binding to monoubiquitinated cargo. In addition, these factors are all monoubiquitinated in vivo (Di Fiore et al., 2003; Hicke and Dunn, 2003; Hoeller et al., 2006; Woelk et al., 2006). In the context of the proteasome, Rpn10 binds polyubiquitinated substrates by means of one or two UIMs (Van Nocker et al., 1996; Elsasser et al., 2004; Verma et al., 2004; Wang et al., 2005). In this work we show that Rpn10 is regulated by ‘coupled’ monoubiquitination, constituting to our knowledge the first link between regulation of proteasome function and monoubiquitin signal. In vivo, Rpn10 shows multi-monoubiquitination and polyubiquitination. The formation of polyubiquitinated forms of Rpn10 depends on the presence of the ubiquitin chain elongating factor, Hul5, and correlates with Rpn10 turnover (Crosas et al., 2006). We could uncouple Rpn10 monoubiquitination from polyubiquitination by deleting the HUL5 gene. Moreover, in vivo levels of mUb-Rpn10 are not affected in ubp6Δ cells, suggesting that the machinery involved in the process of Rpn10 monoubiquitination is essentially independent of ubiquitin chain formation and remodeling.
Rsp5/Nedd4-like Ubiquitin-Protein Ligase and Ubp2 Deubiquitinating Enzyme Regulate Rpn10 Monoubiquitination
The ubiquitin-protein ligase involved in Rpn10 monoubiquitination is Rsp5, a member of the Nedd4 HECT ligase family. Rsp5 and its mammalian orthologue Nedd4.2 have emerged as multitasking enzymes with conserved functions, being involved in endocytosis and nuclear roles (Hicke and Dunn, 2003; Kirkin and Dikic, 2007). It has been proposed that Rsp5-Ubp2 association provides a mechanism of monoubiquitination based on the capacity of Ubp2 to disassemble K63 ubiquitin chains synthesized by Rsp5 (Kee et al., 2006). Our data shows that Rsp5 and Ubp2 exert opposed driving forces in the control of mUb-Rpn10 homeostasis in vivo, however, Rsp5 is sufficient for the production of mUb-Rpn10. Ubp2 is highly active towards Rpn10-monoubiquitin isopeptide bonds and exerts a homeostatic control of mUb-Rpn10 (Figure 2A, 2B and 3A). It should be noted that, in the absence of Ubp2, a significant fraction of Rpn10 remains unmodified suggesting that other DUBs may be involved in deubiquitination of mUb-Rpn10 in vivo.
We have reconstituted the reaction of Rpn10 monoubiquitination using recombinant proteins and observed that monoubiquitination can be efficiently catalyzed with either wild-type or methylated ubiquitin, showing a pattern of multi-monoubiquitination. MS analysis confirmed modification at four distinct lysine residues: preferentially to K84, placed within the VWA domain, and to K268, in the C-terminus of the sequence, and secondarily to K71 and K99.
Monoubiquitination, a Mechanism Controlling Substrate-Proteasome Interaction and Proteasome Catalytic Rates
We have found that the UIM-ubiquitin interaction is essential in the monoubiquitination reaction, since mutations at Rpn10 UIM or at ubiquitin I44 abrogate catalysis (Figures 4 and 6). This feature usually implies that the reaction product establishes an intramolecular interaction between the UIM and the linked monoubiquitin moiety, thus impairing further ubiquitination events and favoring monoubiquitination rather than polyubiquitination (Di Fiore, 2003; Woelk et al., 2006). Indeed, our data supports that Rpn10 and mUb-Rpn10 are two functionally distinct molecules: mUb-Rpn10 has lost the capacity exhibited by unmodified Rpn10 to bind ubiquitin conjugates or unanchored polyubiquitin chains (Figure 6). This impairment is due to a UIM-ubiquitin interaction in cis, because the RPN10-UbI44A mutant partially recovers the capacity to bind polyubiquitin. In addition, Rpn10 linked to monoubiquitin imposes a dramatic inhibition of cyclin B degradation, and in the cell, impairs full rescue of Rpn10 function, but simply restoring the UIM availability with the Rpn10-UbI44A mutant results in a strong rescue of Rpn10 function (Figure 6). An additional aspect that should be considered is the putative effect of K84 monoubiquitination, not only on UIM, but also on the VWA domain, involved in protein degradation in a UIM-independent manner (Verma et al., 2004). The fact that K84 is located within the VWA domain suggests that this domain could be affected by K84 monoubiquitination. An interesting hypothesis is that modifications at K268 and at K84 inhibit protein degradation with different efficiency.
With these observations, it could be predicted that when mUb-Rpn10 levels are increased in the cell, protein targeting and degradation to the proteasome would be decreased. Cells carrying a deletion of the UBP2 gene show substantial stabilization of proteasomal mUb-Rpn10 (Figure 3C), and consistently, these cells accumulate K48 ubiquitin linkages, as established in a recent study (Xu et al., 2009). This result would not be expected considering only the activity of Ubp2 on K63 ubiquitin chains (Kee et al., 2006). However, the accumulation of K48 chains fits in our model of attenuated proteasomal activity when mUb-Rpn10 levels are increased due to deletion of UBP2. The relative low abundance mUb-Rpn10 in growing cultures at standard conditions could be explained by the importance of the homeostasis of proteasome activity in the cell. Indeed, high levels of Rpn10-Ub cause a strong phenotype (Figure 6G). An additional explanation could be that Rpn10 is targeted for monoubiquitination only in a subpopulation of proteasomes, but further work is required to test this hypothesis.
We have observed that mUb-Rpn10 is dramatically decreased in cultures grown at low and high temperatures, and in the presence of cadmium. According to our results, suppression of Rpn10 monoubiquitination would increase the availability of Rpn10 UIM, promoting activation of this ubiquitin receptor. Rpn10 is essential for the degradation of damaged newly synthesized proteins, which are strongly increased at low and high temperatures and in the presence of cadmium (Medicherla et al., 2008), and in this scenario, Rpn10 function requires an active UIM (Elsasser et al., 2004; Verma et al., 2004). We have observed that Rpn10 monoubiquitination is dramatically decreased in these conditions, suggesting a mechanism to increase the availability of Rpn10 UIM.
Regulation of the Proteasome by Associated Ubiquitin Conjugating and Deconjugating Activities
Rsp5-dependent monoubiquitination of Rpn10, by impairing substrate binding, constitutes an efficient control of proteasome activity. In absence of monoubiquitination, substrate recognition by Rpn10 would promote the engagement of proteasomal ubiquitin hydrolases, such as Ubp6/Usp14 or Rpn11, and chain elongating factor Hul5 (Yao and Cohen, 2002; Verma et al., 2004; Crosas, et al., 2006; Hanna et al., 2006), which define a second level of proteasome regulation. Thus, two evolutionarily conserved ubiquitin ligases, Hul5/KIAA10 and Rsp5/Nedd4.2, and their related deubiquitinating enzymes, Ubp6/Usp14 and Ubp2, respectively, regulate early steps of proteasomal mediated degradation, underscoring the relevance of associated enzymatic factors in proteasome function. It has been recently published that p54, the Drosophila orthologue of Rpn10, is modified by up to four ubiquitin groups in vivo (Lipinszki et al., 2009), resembling multiple monoubiquitination of yeast Rpn10, characterized herein. Further work will be required to establish the physiological role of Rpn10 monoubiquitination in higher eukariotes.
Methods
Yeast methods, proteasome purification, ubiquitin conjugating and deconjugating reactions and binding assays are described elsewhere (Rose et al., 1990; Leggett et al., 2002; Crosas et al., 2006; Lu et al; 2008) or in the supplemental data. Cloning, expression and purification of the proteins are described in the supplemental data. In mass spectrometry analysis, ubiquitination sites were identified by excising gel bands containing Rpn10 and in-gel digested with trypsin. Peptide were separated by nanoscale reversed phase liquid chromatography coupled to hybrid tandem mass spectrometer (LTQ Orbitrap; ThermoElectron). MS/MS spectra were matched to Rpn10 sequence using sequest algorithm (Yates et al., 1995) with a mass increment of 114.0429 (signature diglycine generated by trypsin digest of conjugated ubiquitin) on lysine residues.
Supplementary Material
Acknowledgments
This work was performed in the IBMB, supported by Spanish Government (MICINN) grant BFU2006-02928. MS analysis was performed in HMS. We acknowledge NIH grants GM065592 (D.F.) and GM67945 (S.G). We thank F. Hanaoka, J.Y. Lu, H. Zhu., R. Vierstra, D. Kornitzer, J. Huibregtse, K. Madura, S. Elsasser, I. Dikic and S. Jentsch for plasmids and yeast strains. We also thank N.A. Hathaway and R.W. King for ubiquitinated cyclin B, and A. Sànchez (CRAG) for images.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Dupré S, Urban-Grimal D, Haguenauer-Tsapis R. Ubiquitin and endocytic internalization in yeast and animal cells. Biochim Biophys Acta. 2004;1695:89–111. doi: 10.1016/j.bbamcr.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Mueller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Sadis S, Rubin DM, Glickman M, van Nocker S, Finley D, Vierstra RD. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Hamazaki J, Sasaki K, Kawahara H, Hisanaga S, Tanaka K, Murata S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol Cell Biol. 2007;27:6629–6638. doi: 10.1128/MCB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985;128:1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dötsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Reins HA, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Muñoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 2009;28:1867–1877. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Lipinszki Z, Kiss P, Pál M, Deák P, Szabó A, Hunyadi-Gulyas E, Klement E, Medzihradszky KF, Udvardy A. Developmental-stage-specific regulation of the polyubiquitin receptors in Drosophila melanogaster. J Cell Sci. 2009;122:3083–3092. doi: 10.1242/jcs.049049. [DOI] [PubMed] [Google Scholar]
- Lu JY, Lin YY, Qian J, Tao SC, Zhu J, Pickart C, Zhu H. Functional dissection of a HECT ubiquitin E3 ligase. Mol Cell Proteomics. 2008;7:35–45. doi: 10.1074/mcp.M700353-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1990. [Google Scholar]
- Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong KM, Baek JH, Ahn BY, Yu MH, Kim J. Rpn10p is a receptor for ubiquitinated Gcn4p in proteasomal proteolysis. J Mol Cells. 2007;24:194–199. [PubMed] [Google Scholar]
- Van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8:1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–33. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.