Abstract
Disintegrins are low molecular weight peptides isolated from viper venom. These peptides bind to integrin receptors using a conserved binding motif sequence containing an RGD or similar motif. As a consequence, disintegrins can inhibit platelet aggregation and inhibit cell migration, proliferation, and initiate apoptosis in cancer cell lines. Rubistatin is a MVD disintegrin cloned from a Crotalus ruber ruber venom gland. The biological activity of MVD disintegrins is poorly understood. Recombinant rubistatin (r-Rub) was cloned into a pET32b plasmid and expressed in reductase deficient E. coli. Expression was induced with IPTG and the resulting fusion peptide was affinity purified, followed by thrombin cleavage, and removal of vector coded sequences. r-Rub peptide inhibited ADP-induced platelet aggregation by 54% +/− 6.38 in whole blood. We assessed the ability of r-Rub to initiate apoptosis in three human cancer cell lines. Cultures of SK-Mel-28, HeLA, and T24 cells were grown for 24 h with 2.5µM r-Rub followed by Hoechst staining. Chromatin fragmentation was observed in treated SK-Mel-28, but not in T24 or HeLA cells. A TUNEL assay revealed that 51.55% +/− 5.28 of SK-Mel-28 cells were apoptotic after 18 h of treatment with 3.5µM of r-Rub. Cell migration and proliferation assays were performed in order to further characterize the biological effects of r-Rub on SK-Mel-28 cells. At 3µM, r-Rub inhibited cell migration by 44.4% +/− 0.5, while at 3.5µM it was able to inhibit cell proliferation by 83% +/− 6.0.
Keywords: melanoma cell line, recombinant disintegrin, MVD disintegrin, cell migration, cell proliferation, apoptosis induction
1. Introduction
Vital to the organization of metazoan tissue is the ability of individual cells to attach to an extracellular matrix (ECM). Critical to this process are the members of the integrin receptor family. Integrins are heterodimeric transmembrane proteins consisting of an α and β subunit. These subunits form a receptor that facilitates an anchoring attachment between the cell and ECM proteins such as fibronectin or collagen (Wegener and Campbell, 2008). Integrins also have the ability to transduce signals depending on the extracellular environment (Hynes, 2002). Conformational changes caused by the binding of a ligand allow the integrin to transduce a signal for the cell to migrate, proliferate, or undergo apoptosis (Ridley et al., 2003; Mousa, 2008).
Integrin-mediated apoptosis, or anoikis, is an important process that keeps a regulatory check on cancer development. When a cell loses its attachment to the ECM, integrins can begin a signal cascade that leads to the release of pro-apoptotic proteins from the mitochondria (Frisch and Ruoslathi, 1997). This in turn leads to the release of caspases through the intrinsic apoptotic pathway and ultimately leads to cell death (Lockshin and Zakeri, 2004). It is clear that avoidance of anoikis is a key step toward achieving metastasis during cancer development (Reed, 1999). It has been shown that integrin antagonists, such as disintegrins, can counteract this avoidance, causing cancer cells to undergo apoptosis after treatment (Taga et al., 2002).
Disintegrins are a well-studied class of integrin antagonists isolated from viper venom. Disintegrins are non-enzymatic, low molecular weight peptides synthesized as mRNAs lacking a metalloprotease domain (Okuda et al., 2002; Vija et al., 2009) or released from proteolytic cleavage of a larger snake venom metalloprotease (Kini and Evans, 1992). They contain an integrin-binding loop with a variable amino acid binding motif (McLane et al., 2004). An RGD motif is found in many disintegrins and is a classical characteristic of integrin antagonists (Rouslahti, 1996). However, many disintegrins contain other binding motifs such as KGD, MLD, VGD, MDG, and MVD (McLane et al., 2004; Calvete et al., 2005). Additionally the amino acids flanking this binding motif, have been shown to play a role in the biological activity of disintegrins (Kini and Evans, 1995a,b; Seoane et al., 2010).
Here we describe functional characterization of an MVD, medium-sized disintegrin, termed rubistatin, cloned from the venom gland of the red diamond rattlesnake (Crotalus ruber ruber). Atrolysin E/D, isolated from Crotalus atrox and a potent inhibitor of platelet aggregation, is the only other studied MVD disintegrin (Shimokawa et al., 1998). However, no additional functional studies were reported on Atrolysin E/D. Our present study presents data indicating that r-Rub is a strong apoptotic inducer, and a cell migration and proliferation inhibitor of SK-Mel-28 cells.
2. Materials and methods
2.1. Isolation of Rubistatin cDNA
A venom gland was extracted from a red diamond rattlesnake (Crotalus ruber ruber; Avid # 058-522-010) and stored at −72°C prior to mRNA isolation. The Invitrogen Fast Track 2.0 mRNA Isolation Kit was used to obtain mRNA from the venom gland. RT-PCR was used to generate double stranded cDNA (Promega Access RT-PCR), with the following disintegrin-specific primers: forward primer: 5' AATCCGTGCTGCGATGCTG 3', and reverse primer: 5' CTGCCTGTTGCTGCAGAC 3'. The reverse transcription reaction was performed at 45°C, for 45 min. PCR amplification consisted of 40 cycles of 94°C (30 sec), 60°C (1 min), and 68°C (2 min). A final extension step was performed for 7 min, at 68°C. RT-PCR cDNA products were separated by 1% agarose gel electrophoresis, and purified using the E gel system (Invitrogen).
2.2. Subcloning, expression, and purification of r-Rub
Rubistatin cDNA was amplified using the forward 5' CCGGGTACCAAT CCGTGCTGCGATGCTG 3' (Kpn I linker underlined), and the reverse 5' ACGCAAGCTTCTGCCTGTTGCTGCAGAC 3' (Hind III linker underlined) primers. PCR was performed using 32 cycles of 94°C (30 sec), 60°C (1 min), and 68°C (1 min). The 224 bp band containing Rubistatin cDNA was separated in a 1% agarose gel, digested with Kpn I and Hind III restriction enzymes and cloned into the pET 32b expression vector (Novagen). The recombinant pET 32b/Rubistatin construct was transformed into competent reductase-deficient, origami2 E. coli cells (Novagen).
Cultures were grown to an A600 of 0.6–0.8 in 2xYTA broth. After 4 h induction with 0.5 mM IPTG, cells were centrifuged at 6000 g for 15 min at 4 °C. Cell pellets were resuspended in a total of 12.5 mL of His binding/wash buffer (50mM NaPO4 pH 8, 300mM NaCl, 0.01% Tween-20). Cells were then lysed using 10 cycles of sonication for 20 sec each. The thioredoxin (Trx)-r-Rub fusion protein was purified from the cell lysate using His tag isolation Dynabeads (Invitrogen) according to the manufacture’s protocol. A thrombin cleavage reaction was performed using a thrombin cleavage/capture kit (Novagen) with thrombin optimized to 0.036 units/10µg to separate the Trx fusion partner from the r-Rub peptide. To remove the Trx tag after thrombin cleavage, dynabead purification was performed a second time. The thrombin cleavage reaction resuspended in 1× His binding/wash buffer was added to dynabeads in a 14:1 ratio. The mixture was allowed to incubate for 10 min with agitation before removing the supernatant from the beads. The concentration of purified peptide was determined with a Bradford assay using bovine serum albumin as a standard. Two micrograms of r-Rub peptide were mixed with 5 µL of 4×NuPAGE LDS sample buffer (Invitrogen) and 2 µL of 10× reducing agent in final volume of 20 µL. The samples were boiled at 70 °C for 10 min, loaded on a NuPAGE 4–12% Bis-Tris gel (Invitrogen), and separated at a constant 200 V for 40 min in NuPAGE 1X MES SDS running buffer (Invitrogen).
2.3. Cell culture conditions
SK-Mel-28 cells were grown in Eagle’s minimum essential medium (ATCC). HeLA cells were grown in Dulbecco's modified eagle medium (ATCC). T24 cells were grown in McCoy’s 5A medium (ATCC). All media was supplemented with 10% fetal bovine serum (FBS) and penicillin-100 (IU/mL), streptomycin (0.1 mg/mL), and amphotericin B (0.25µg/mL). All cells were grown at 37°C in a humidified incubator with 5% CO2.
2.4. Inhibition of platelet aggregation using whole blood
The inhibition of platelet aggregation study was done according to the Sánchez et al. (2010) method using a dual-channel Chrono-Log Whole-Blood Aggregometer [Ca+2] model 560 (Havertown, USA). Briefly, different concentrations of r-Rub were added to 10% citrated whole human blood, and pre-incubated at 37 °C for 2 and 4 min, respectively. Platelet aggregation was initiated by 10 µM ADP. Percentage of impedance was measured using whole blood. The maximal aggregation in the absence of r-Rub was given as 100% aggregation.
2.5. Chromatin fragmentation assay
Two well chamber slides were seeded with 106 cells in 1 mL of growth medium and were allowed to settle for 24 h. Cells were treated with 2.5µM r-Rub in one chamber and with an equal volume of His binding/wash buffer in the other chamber. After treatment for 24 h, cells were washed in 1× PBS and fixed in 3% formaldehyde for 1 h, followed by two additional PBS washes. Hoechst stain (10µg/mL) was added to each well and allowed to incubate for 20 min in the dark, followed by two PBS washes. Coverslips were mounted using 30% glycerol and the slides analyzed using 1000× magnification on a Zeiss AXIO fluorescent microscope.
2.6. Apoptotic assay
Five hundred thousand SK-Mel-28 cells were seeded in six wells of a 12 well plate and allowed to grow for 24 h. Cells were treated for 18 h with 3.5µM r-Rub, equal volume of His binding/wash buffer, or no treatment. Then, non-adherent cells were transferred to a flow cytometry tube, followed by the remaining cells detached with 0.05% trypsin EDTA. Cells were pelleted by centrifugation at 300×G for 5 min and fixed in 1% paraformaldehyde for 1 h. Cells were washed twice with 1× PBS and resuspended in 70% ethanol. Cell suspensions were stored at −20°C for 24 h. Cells were processed using an APO-BRDU™ Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s protocol. Cell suspensions were analyzed using a Becton Dickinson FACSCalibur flow cytometer with CellQuest software. Results were normalized to take into account the effect of the His binding/wash buffer. Experiments were performed in triplicate.
2.7. Wound Healing Assay
A Cell BioLab Wound Healing Assay was used to measure cell migration inhibition. A cell suspension of 5×105 SK-Mel-28 cells was seeded onto the 24 well-plate according the manufacturer’s protocol. After 24 h incubation, the insert was removed and the cells were washed twice with culture media. A picture was taken from each well before the addition of 500 µL of media containing 3µM of r-Rub peptide. Positive controls consisted of cells treated with 3µM echistatin (Sigma). Negative controls consisted of cells treated with equal volume of His binding/wash buffer. Cells were incubated for 24 h with their respective treatments. Then pictures were taken of each well and the percentage closure was calculated following the manufacturer’s instructions. Experiments were performed in triplicate.
2.8. Cell Proliferation Assay
A WST-1 assay (GenDEPOT) was used to determine cell proliferation inhibition. Fifty thousand SK-Mel-28 cells/well were added to a 96-well plate grown for 24 h at 37°C in a 5% CO2 environment. After 24 h, growth media was aspirated. Then, 100 µL of culture media containing 3.5 µM of r-Rub peptide were added to their respective wells. Positive controls consisted of cells treated with 3.5µM echistatin (Sigma). Negative controls consisted of cells treated with equal volume of His binding/wash buffer or PBS. After 24 h all wells were then treated with 10 µL of WST-1 cell proliferation assay reagent for 1 h. The same volume of culture medium and WST-1 reagent used to treat the cells was used as the control blank for the microplate reader. The formation of the formazan product was measured at 450 nm. The reference wavelength was 630 nm. A dual wavelength reading was used to obtain the results, where the reference wavelength was subtracted from the reading wavelength, 450–630 nm. Results were normalized to take into account the effect of the His binding/wash buffer. Percentage of cell proliferation inhibition was calculated using the following formula: (Abs450nm–600nm of treated ÷ Abs450nm–600nm of untreated) × 100. Experiments were performed in triplicate.
3. Results
3.1. An MVD disintegrin from C. ruber ruber
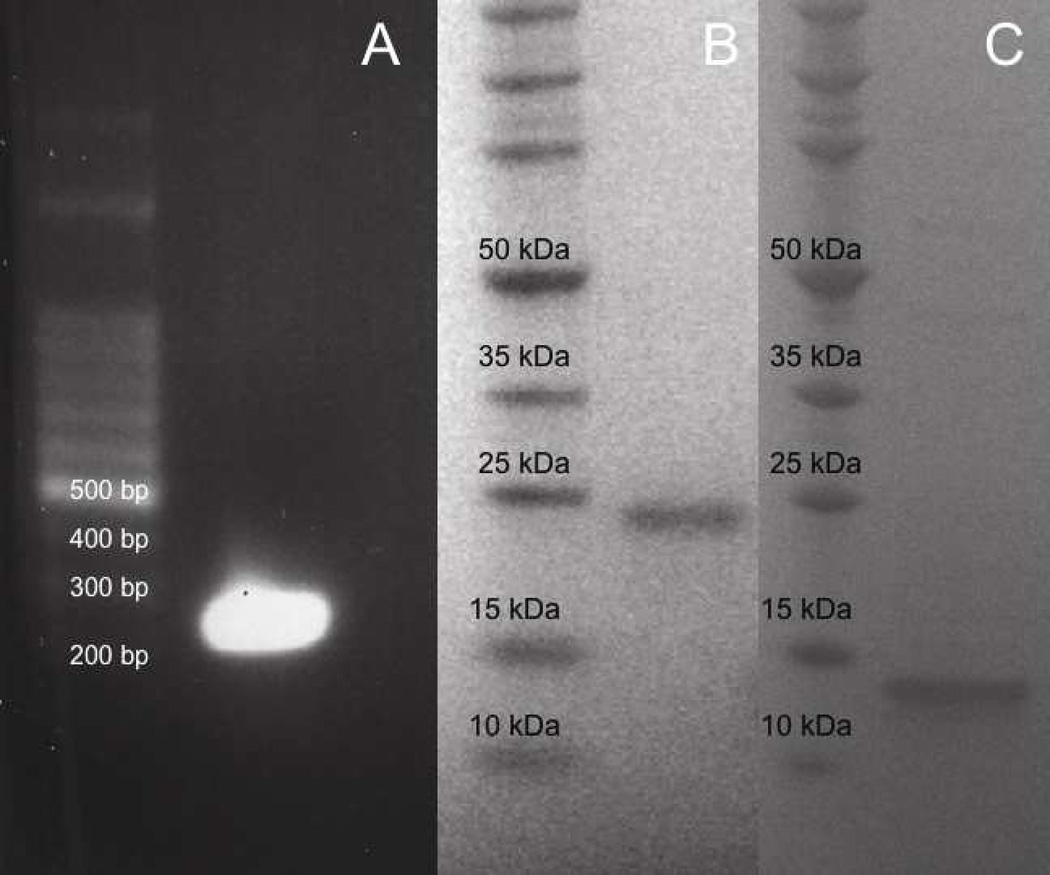
A 224 bp cDNA fragment was obtained from RT-PCR amplification of a venom gland from a red rattlesnake (Fig. 1A). The small cDNAs isolated in this study coded for a truncated 61 amino acid disintegrin containing an MVD in its binding motif. The disintegrin termed rubistatin begins with the amino acid Asn, includes a stop codon, and 20 nucleotides of its 3' UTR (Fig. 2).
Fig. 1.
Cloning, expression, and purification of r-Rub. (A) 100 bp DNA marker (left lane). Rubistatin cDNA (right lane). SDS-PAGE of expressed protein resolved in a 4–12 % NuPAGE Bis-Tris gel under reducing condition (B–C). (B) Molecular weight marker (left lane). Thrx/r-Rub fusion protein (right lane). (C) Molecular weight marker (left lane). Purified r-Rub peptide (right lane).
Fig. 2.
Partial cDNA sequences and deduced amino acid sequences of rubistatin (GenBank accession number: JN22479). The cDNA sequences are shown on the upper line. The deduced amino acid sequence (one-letter abbreviation) is on the lower line. The stop codon is indicated with an asterisk (*). The Cys residues are shaded in gray. The MVD motif is shown in a black box.
3.2. Expression and purification of recombinant rubistatin
After inducing expression of recombinant rubistatin (r-Rub) in reductase-deficient E. coli, the recombinant peptide was isolated by His tag affinity purification. An SDS-PAGE gel showed a distinct band at approximately 24 kDa, corresponding to the TRX-r-Rub fusion protein (Fig. 1B). After cleavage with thrombin and removal of the TRX tag, SDS PAGE analysis showed a single band (Fig. 1C).
3.3. Inhibition of platelet aggregation of r-Rub
r-Rub peptide inhibited ADP-induced platelet aggregation by 54% +/− 6.38 in whole blood (Table 1).
Table 1.
Inhibition of platelet aggregation with whole blood in the presence of r-Rub.
| Sample | Whole blood |
|---|---|
| Control (PBS) | 0 |
| r-Rub | 54 +/− 6.38 |
The experiments were repeated thrice. The results are expressed in % of inhibition. The final concentration of r-Rub used with blood was 4.2 mg/mL.
3.4. r-Rub induced apoptosis of SK-Mel-28 cells
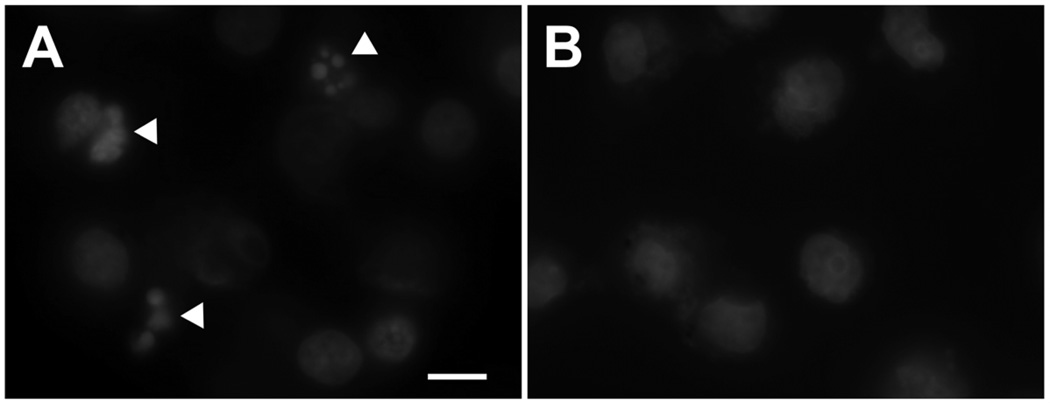
The r-Rub peptide’s ability to induce apoptosis on SK-Mel-28, T24, and HeLa cells was determined using an APO-BRDU TUNEL and chromatin fragmentation assays. Cells were incubated with 2.5 µM r-Rub peptide for 24 h. The r-Rub peptide was able to induce chromatin fragmentation of SK-Mel-28 cells as determined by fluorescent microscopy and Hoechst staining (Fig. 3A). Chromatin fragmentation was not detected in untreated cells (Fig. 3B), T24, or HeLa cells treated with r-Rub peptide (data not shown).
Fig. 3.
r-Rub induced nuclear fragmentation of SK-Mel-28 cells. (A) Hoechst-stained cells after treatment with 2.5 µM r-Rub for 24 h (chromatin fragmentation, white arrowheads). (B) Untreated cells stained with Hoechst.
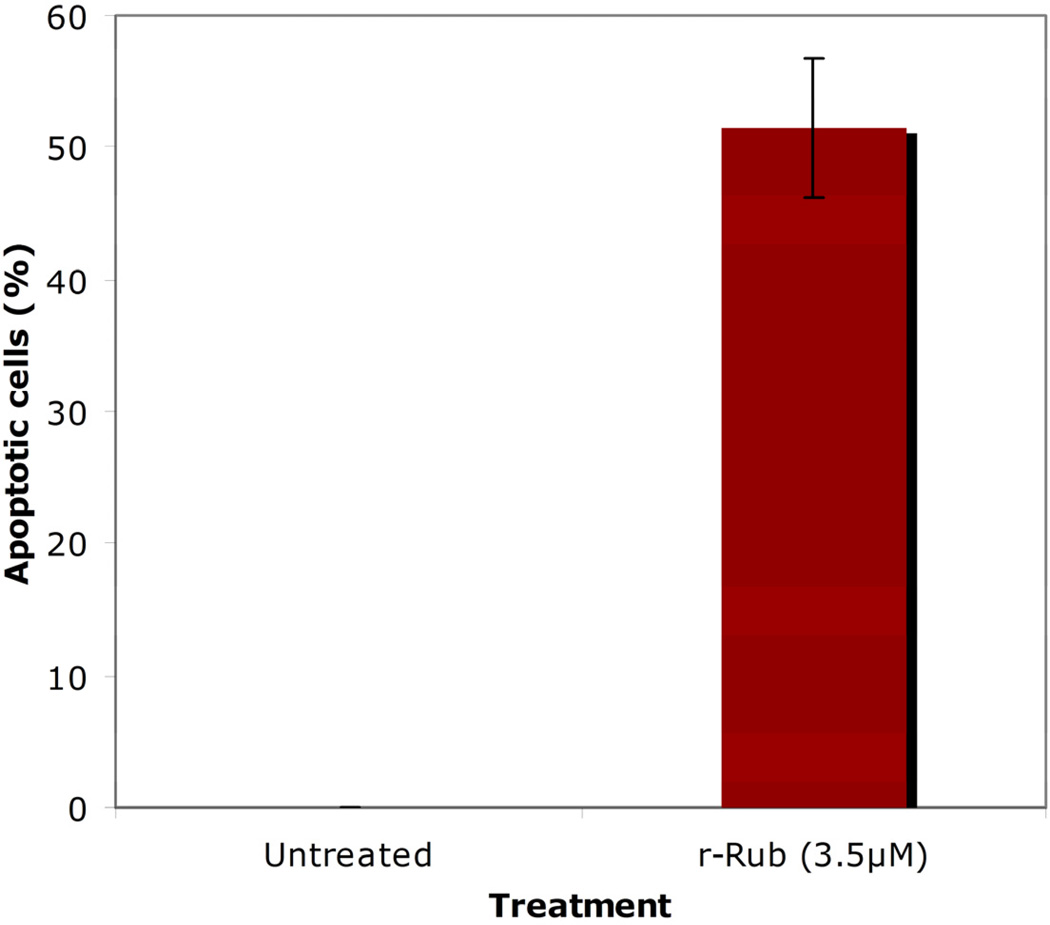
A TUNEL assay was performed to quantify the population of apoptotic SK-Mel-28 cells after treatment with r-Rub peptide for 18 h. At 3.5µM r-Rub peptide, 51.55% +/− 5.28 of treated SK-Mel-28 cells were undergoing apoptosis (Fig. 4). The apoptotic activity of the r-Rub peptide was statistically significant (p <0.005).
Fig. 4.
r-Rub induced apoptosis of SK-Mel-28 cells. Cells were incubated with 3.5 µM of r-Rub peptides for 18 h. Untreated cells were used as a negative control. Experiments were done in triplicate. Standard deviation data are included as bars.
3.5. r-Rub inhibits cell migration and proliferation of SK-Mel-28
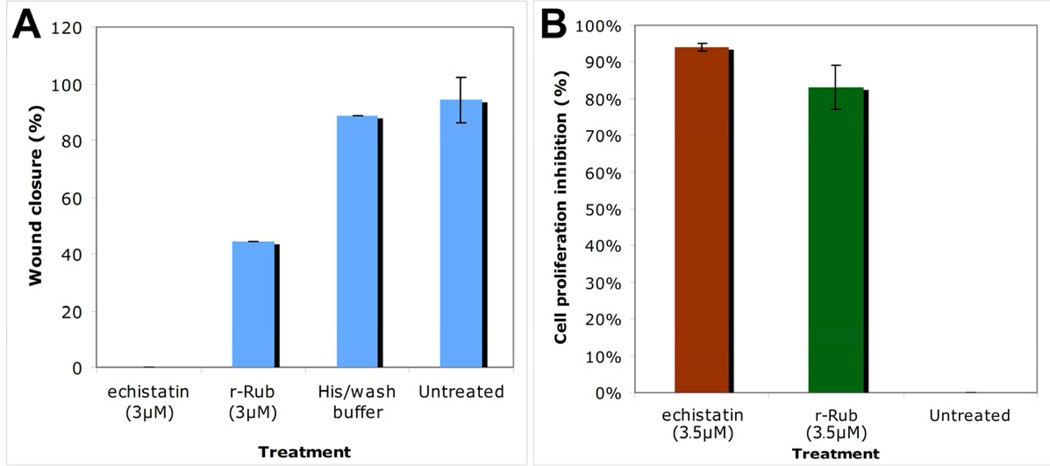
Cell migration and proliferation assays were performed in order to further characterize the biological effects of r-Rub on SK-Mel-28 cells. At 3µM, r-Rub inhibited cell migration by 44.4% +/− 0.5 (Fig. 5A), while at 3.5µM it was able to inhibit cell proliferation by 83% +/− 6.0 (Fig. 5B).
Fig. 5.
r-Rub inhibited migration and proliferation of SK-Mel-28 cells. (A) At 3µM r-Rub inhibited Sk-Mel-28 cell migration. (B) At 3.5µM r-Rub inhibited proliferation of SK-Mel-28 cells. In both assays, cells were incubated with echistatin (positive control) or buffer (negative control). Experiments were performed in triplicate.
4. Discussion
4.1. rubistatin, an MVD disintegrin
In this study, we cloned and expressed a 224 bp cDNA (designated as Rubistatin). Analysis of the predicted amino acid sequence revealed that rubistatin is a medium-sized disintegrin with an MVD motif. Figure 6 shows an amino acid sequence comparison of rubistatin with other closely related medium sized disintegrins. Rubistatin shares 95% amino acid identity with atrolysin E/D (accession # AAB23201.1), 87% amino acid identity with barbourin (accession # P22827.1), tergeminin (accession # P22828.1), cerastin (accession # AAB24805.1), lutosin (accession # AAB24809.1), and 84% identity with mojastin (accession # ABG77588.1). Cysteine residues are conserved in all of these sequences.
Fig. 6.
Amino acid comparison between rubistatin and other medium-sized disintegrins. Dashes indicate amino acid identities as compared to the rubistatin sequence. The tripeptide binding motif is shaded in a black box.
Disintegrins are characterized by their size and the number of disulfide bonds (4–7). The disintegrin family includes: short (41–51 amino acids), medium (70 amino acids), long (84 amino acids), dimeric, or as disintegrin-like domains of PIII snake venom metalloproteases (100 amino acids) (Calvete et al., 2005). Disintegrins’ binding strength to the integrin receptor is determined by the conformation of the binding loop containing a tripeptide motif (Juarez et al., 2008). This conformation is dependent on the formation of the appropriate disulfide bonds in the disintegrin peptide (Calvete et al., 2005).
Most of the disintegrins contain an RGD tripeptide motif in the binding loop (McLane et al., 2004). However, the small disintegrins, eristocophin II (accession # AAB2266), jerdostatin (accession # AAP20878), and obustatin (accession # P83569) contain a MGD, RTS, and KTS, respectively. The medium-sized disintegrins barbourin, tergeminin, cerastin, lutosin, MT-C (accession # AF051790), and salmosin 2 (accession # AAC42597) have a KGD instead. Atrolysin E/D has an MVD motif. The long disintegrin bilitoxin-1 has an MGD motif (Nikai et al., 2000). Thirteen of the dimeric disintegrins contain non-RGD tripeptide motifs. Five of these contain an MLD motif: EC3B (accession # P81630), EC6A (accession # P82465), EMS11A (reference), EO5A (Bazan-Socha et al., 2004), and VLO5B (Bazan-Socha et al., 2004). Four dimeric disintegrins contain a VGD motif, EC3A (accession # P81630), EO5B (Bazan-Socha et al., 2004), lebetase (accession # CAA66471), and VLO5A (Bazan-Socha et al., 2004). Three contain a KGD, ussuristatin-2 (accession # A59412), VB7B (Calvete et al., 2003), and piscivostatin 2B (accession #BAC55946). One contains an MGD motif, EMF10B (accession # P81743).
Different disintegrin tripeptide motifs bind to different integrin receptors (Marcinkiewicz, 2005). For instance, KTS disintegrins bind to αlβ1 integrins, while RGD disintegrins bind to αIIbβ3, αvβ3, and αvβ5. MLD disintegrins bind to α4β1, α4β7, and α9β1 integrin receptors (Marcinkiewicz et al., 1999; Marcinkiewicz et al., 2000; Bazan-Socha et al., 2004). It is also clear that for three MLD-dimeric disintegrins (VLO5, EO5, and EC3) the amino acid N-terminal to the MLD motif affects potency and selectivity to the integrin receptor (Bazan-Socha et al., 2004; Walsh and Marcinkiewicz, 2011). Furthermore, the KGD-containing medium-sized disintegrins salmosin 2, tergeminin, cerastin, and lutosin bind to αIIbβ3 and are strong platelet aggregation inhibitors. However, the KGD-containing dimeric VB7B has low platelet aggregation activity and can inhibit cell adhesion of cells containing α5β1 integrin receptors to fibronectin (Calvete et al., 2003).
4.2. Recombinant disintegrins
Heterologous expression of disintegrins in bacterial hosts provides many advantages. Among these are the ability to rapidly express and purify high amounts of recombinant disintegrins, and the ability to induce targeted mutations. Previous studies have shown that GST-disintegrin fusion proteins expressed in E. coli display biological activity (Rahman et al., 1998; Chang et al., 1993; Mahimkar et al., 2005; Seoane et al., 2010; Taklemeriam et al., 2011). In the present study, we used a pET32 expression vector, transformed into a bacterial strain that allows formation of disulfide bonds.
4.3. Apoptotic-inducing disintegrins
Several disintegrins and disintegrin-like peptides induce apoptosis (Wu et al., 2003). These include: accutin (Yeh et al., 1998), agkistin-s (Ren et al., 2006) echistatin (Brassard et al., 1999; Alimenti et al., 2004), rhodostomin (Yeh et al., 2001), salmosin (Hong et al., 2003), the disintegrin portion of the recombinant (r) metargidin (Trochon-Joseph et al., 2004), the mutant mojastin r-Moj-DM (Seoane et al., 2010), and the recombinant partial PIII-SVMP, GST-acocostatin (Taklemeriam et al., 2011).
r-Rub’s apoptotic activity was cell-specific, as it induced apoptosis of SK-Mel-28, but not of T24 or HeLa cell lines. Cell-specific apoptotic-inducing activity was also shown with GST-acocostatin (Taklemeriam et al., 2011). This type of specificity can be attributed to differences in integrin expression. We have previously characterized the integrin expression on SK-Mel-28, T24, and HeLa cell lines. The closest homolog to r-Rub, atrolysin E/D inhibited ADP and collagen-activated platelet aggregation (Shimokawa et al., 1998). This inhibitory activity suggests the potential binding of atrolysin E/D to integrin receptors αIIbβ3, α2β1, or α1β1. It is possible that r-Rub binds to the same receptors, since r-Rub also inhibits ADP-activated platelet aggregation. However, we have shown that SK-Mel-28 cells lack α1β1 (Seoane et al., 2010). Both HeLa and SK-Mel-28 cell lines express α2β1 (Taklemariam et al., 2011), but r-Rub failed to induce apoptosis of HeLa cells. We also hypothesize that it is unlikely that r-Rub binds to αvβ3, an integrin known to bind to apoptotic-inducing disintegrins (Yeh et al., 1998; Wierzbicka-Patynowski et al., 1999), as this receptor is found in both SK-Mel-28 and T24 cell lines (Seoane et al., 2010; Taklemeriam et al., 2011). Therefore at this time, it is difficult to predict the receptor that is targeted by r-Rub to induce apoptosis.
4.4. Apoptosis induction, proliferation and cell migration inhibition
r-Rub inhibited cell migration and proliferation and induced apoptosis of SK-Mel-28 cells. Anti-proliferative and apoptotic-inducing effects on cancer cells due to a single agent have been reported. For instance, three carbonic anhydrase IX inhibitors induced both effects on HeLa cells (Cianchi et al., 2010). Furthermore, Gullo et al. (2010) demonstrated that CD137 (a member of the TNF family) agonists both inhibited cell proliferation and induced apoptosis on multiple myeloma cell lines. In addition, rapamycin inhibited proliferation and the invasive ability of two gastric cancer cell lines and induced their apoptosis (Yao et al., 2011).
A connection between cell proliferation, migration, and apoptotic inhibition is also emerging. For instance, endothelin-1 promotes cell proliferation, cell invasion, and apoptotic inhibition of osteosarcomas (Zhao et al., 2011). López and Alvarez-Salas (2011) demonstrated that microRNA (miR) 34c-3p inhibited cell proliferation and migration and induced apoptosis of the cervical carcinoma cell line SiHa.
The mechanistic relationship between the activation of cell proliferation and inhibition of apoptosis is clear for some tumor-promoting factors. For instance, Ammoun et al. (2011) demonstrated that IGFBP-1 (insulin-like growth factor-binding protein-1) signaling results in schwannoma cell proliferation via β1 integrin-driven activation of the Src/FAK pathway. This in turn leads to the increased activation of AKT, an anti-apoptotic protein. The FAK pathway is also known to inhibit and promote degradation of p53, an apoptotic inducer (Ilic et al., 1998; Lim et a., 2008).
However, there is evidence demonstrating that different pathways regulate cell proliferation and apoptosis for some potential anti-tumor agents. Immobilized isthmin (ISM) activates the FAK survival pathway, but soluble ISM induces apoptosis by recruiting pro-caspase 8 to the cell membrane (Zhang et al., 2011). Both of these pathways are activated by ISM binding to αvβ5. Finally, MUC1 knockdown in MKN45, a human gastric carcinoma cell line, resulted in the activation of both cell proliferation and apoptotic pathways (Costa et al., 2011).
4.5. Conclusions
Disintegrins perform a number of useful biological functions and are a source of potential cancer treatments. Based on the amino acid alignment of rubistatin and its closest relative, it is likely that atrolysin E/D shares similar activities. r-Rub peptide blocked platelet aggregation of whole blood. Our data demonstrate that r-Rub with its unusual MVDRN motif is a potent apoptotic inducer of SK-Mel-28, but not T24 or HeLA cells. r-Rub also inhibits cell migration and proliferation of SK-Mel-28. It is possible that some of the cell proliferation inhibition may be due to apoptosis, as r-Rub induced apoptosis of treated SK-Mel-28 cells. Although the signal transduction pathways activated by r-Rub remained to be elucidated, the end result is the apoptotic activation, resulting in the down-regulation of survival and cell migration of treated SK-Mel-28 cells.
Highlights.
> r-Rub is a potent apoptotic inducer of SK-Mel-28, but not T24 or HeLA cells. >r-Rub also inhibits cell migration and proliferation of SK-Mel-28. >It is possible that some of the cell proliferation inhibition may be due to apoptosis. >It is likely that atrolysin E, the closest homolog to rubistatin, shares similar activities.
Acknowledgements
We thank Cleber Ouverney and Lucy Arispe for technical assistance. We also thank Stephanie Mandal for the preparation and labeling of composite figures. Funding for this project was provided by NSF-REU #DBI 1004350, NIH/SCORE # 2SO6 GM008192, NIH Grant # R25GM071381, and NCRR/Viper # 2P40RR018300.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there is no conflict of interest.
References
- Alimenti E, Tafuri S, Scibelli A, D'Angelo D, Manna L, Pavone LM, Belisario MA, Staiano N. Pro-apoptotic signaling pathway activated by echistatin in GD25 cells. Biochim. Biophys. Acta. 2004;1693:73–80. doi: 10.1016/j.bbamcr.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Ammoun S, Schmidt MC, Zhou L, Ristic N, Ercolano E, Hilton DA, Perks CM, Hanemann CO. Insulin-like growth factor-binding protein 1 (IGFBP-1) regulates human schwannoma proliferation, adhesion, and survival. Oncogene. 2011 doi: 10.1038/onc.2011.357. [DOI] [PubMed] [Google Scholar]
- Bazan-Socha S, Kisiel DG, Young B, Theakston DG, Calvete JJ, Sheppard D, Marcinkiewicz C. Structural requirements of MLD-containing disintegrins for functional interaction with α4β1 and α9 β1 integrins. Biochemistry. 2004;43:1639–1647. doi: 10.1021/bi035853t. [DOI] [PubMed] [Google Scholar]
- Brassard DL, Maxwell E, Malkowski M, Nagabhushan TL, Kumar CC, Armstrong L. Integrin alpha(v)beta(3)-mediated activation of apoptosis. Exp. Cell. Res. 1999;251:33–45. doi: 10.1006/excr.1999.4559. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Moreno-Murciano MP, Theakston RDG, Kisiel DG, Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem. J. 2003;372:725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez PP, Sanz PL. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Chang HH, Hu ST, Huang TF, Chen SH, Lee YH, Lo SJ. Rhodostomin, an RGD-containing peptide expressed from a synthetic gene in Escherichia coli, facilitates the attachment of human hepatoma cells. Biochem. Biophys. Res. Commun. 1993;190:242–249. doi: 10.1006/bbrc.1993.1037. [DOI] [PubMed] [Google Scholar]
- Cianchi F, Vinci MC, Supuran CT, Peruzzi B, De Giuli P, Fasolis G, Pastorekova S, Papucci L, Masini F, Puccetti L. Selective inhibition of carbonic anhydrase IX decreases cell proliferation and induces ceramide-mediated apoptosis in human cancer cells. J Pharmacol Exp Ther. 2010;334:710–719. doi: 10.1124/jpet.110.167270. [DOI] [PubMed] [Google Scholar]
- Costa NR, Paulo P, Caffrey T, Hollingsworth MA, Santos-Silva F. Impact of MUC1 Mucin downregulation in the phenotypic characteristics of MKN45 gastric carcinoma cell line. PLos One. 2011;6:e26970. doi: 10.1371/journal.pone.0026970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Ruoslahti E. Integrins and anoikis. Current Opinion in Cell Biology. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- Gullo C, Koh LK, Pang WL, Ho KT, Tan SH, Schwarz H. Inhibition of Proliferation and induction of apoptosis in multiple myeloma cell lines by CD137 ligand signaling. PLoS ONE. 2010;5:e10845. doi: 10.1371/journal.pone.0010845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Lee H, You W, Chung K, Kim D, Song K. The snake venom disintegrin salmosin induces apoptosis by disassembly of focal adhesions in bovine capillary endothelial cells. Biochem. Biophys. Res. Commun. 2003;302:502–508. doi: 10.1016/s0006-291x(03)00213-4. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Illic D, Almeida EA, Schlaepfer DD, Dazin P, Aizawa S, Damsky CH. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J Cell Biol. 1998;143:547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez P, Comas I, Gonzalez-Candelas F, Calvete JJ. Evolution of snake venom disintegrins by positive Darwinian selection. J Mol Biol and Evolution. 2008;25:2391–2407. doi: 10.1093/molbev/msn179. [DOI] [PubMed] [Google Scholar]
- Kini RM, Evans HJ. Structural domains in venom proteins: evidence that metalloproteinases and nonenzymatic platelet aggregation inhibitors (disintegrins) from snake venoms are derived by proteolysis from a common precursor. Toxicon. 1992;30:265–293. doi: 10.1016/0041-0101(92)90869-7. [DOI] [PubMed] [Google Scholar]
- Kini RM, Evans HJ. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem Biophys Res Commun. 1995a;212:1115–1124. doi: 10.1006/bbrc.1995.2084. [DOI] [PubMed] [Google Scholar]
- Kini RM, Evans HJ. A novel approach to the design of potent bioactive peptides by incorporation of proline brackets: antiplatelet effects of Arg-Gly-Asp peptides. FEBS Lett. 1995b;375:15–27. doi: 10.1016/0014-5793(95)01144-4. [DOI] [PubMed] [Google Scholar]
- Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Illic D. Nuclear FAK promotes cell proliferation and survival through FERM-induced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin RA, Zakeri Z. Apoptosis, autophagy, and more. Int. J. Biochem. Cell Biol. 2004;36:2405–2419. doi: 10.1016/j.biocel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- López JA, Alvarez-Salas M. Differential effects of miR-34c-3p and miR-34c-5p on SiHa cells proliferation, apoptosis, migration, and invasion. Biochem Byophys Res Commun. 2011;409:513–519. doi: 10.1016/j.bbrc.2011.05.036. [DOI] [PubMed] [Google Scholar]
- Mahimkar RM, Visaya O, Pollock AS, Lovett DH. The disintegrin domain of ADAM9: a ligand for multiple beta1 renal integrins. Biochem. J. 2005;385:461–468. doi: 10.1042/BJ20041133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz C. Functional characteristic of snake venom disintegrins: potential therapeutic implication. Current Pharm Des. 2005;11:815–827. doi: 10.2174/1381612053381765. [DOI] [PubMed] [Google Scholar]
- Marcinkiewicz C, Taooka Y, Yokosaki Y, Calvete JJ, Marcinkiewicz MM, Lobb RR, Niewiarowski S, Sheppard D. Inhibitory effects of MLDG-containing heterodimeric disintegrins reveal distinct structural requirements for interaction of the integrin alpha 9beta 1 with VCAM-1, tenascin-C, and osteopontin. J Biol Chem. 2000;275:31930–31937. doi: 10.1074/jbc.M003209200. [DOI] [PubMed] [Google Scholar]
- Marcinkiewicz C, Calvete JJ, Vijay-Kumar S, Marcinkiewicz MM, Raida M, Schick P, Lobb RR, Niewiarowski S. Structural and functional characterization of EMF10, a heterodimeric disintegrin from Eristocophis macmahoni venom that selectively inhibits alpha 5 beta 1 integrin. Biochemistry. 1999;38:13302–13309. doi: 10.1021/bi9906930. [DOI] [PubMed] [Google Scholar]
- McLane MA, Sanchez EE, Wong A, Paquette-Straub C, Perez JC. Disintegrins. Curr Drug Targets Cardiovasc Haematol Disord. 2004;4:327–355. doi: 10.2174/1568006043335880. [DOI] [PubMed] [Google Scholar]
- Mousa SA. Cell adhesion molecules: potential therapeutics & diagnostic implications. Mol Biotechnol. 2008;38:33–40. doi: 10.1007/s12033-007-0072-7. [DOI] [PubMed] [Google Scholar]
- Nikai T, Taniguchi K, Komon Y, Masuda K, Fox JW, Sugihara H. Primary structure and functional characterization of bilitoxin-1, a novel dimeric P-II snake venom metalloproteinase from Agkistrodon bilineatus venom. Arch Biochem Biophys. 2000;378:6–15. doi: 10.1006/abbi.2000.1795. [DOI] [PubMed] [Google Scholar]
- Okuda D, Koike H, Morita T. A new gene structure of the disintegrin family: a subunit of dimeric disintegrin has a short coding region. Biochemistry. 2002;41:14248–14254. doi: 10.1021/bi025876s. [DOI] [PubMed] [Google Scholar]
- Rahman S, Aitken A, Flynn G, Formstone C, Savidge GF. Modulation of RGD sequence motifs regulates disintegrin recogni-tion of αIIbβ3 and α5β1 integrin complexes. Replacement of elegantin alanine-50 with proline, N-terminal to the RGD sequence, diminishes recognition of the α5β1 complex with restoration induced by Mn2þ cation. Biochem. J. 1998;335:247–257. doi: 10.1042/bj3350247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC. Mechanisms of apoptosis avoidance in cancer. Curr Opin Oncol. 1999;11:68–75. doi: 10.1097/00001622-199901000-00014. [DOI] [PubMed] [Google Scholar]
- Ren A, Wang S, Cai W, Yang G, Zhu Y, Wu X, Zhang Y. Agkistin-s, a disintegrin domain, inhibits angiogenesis and induces BAECs apoptosis. J Cell Biochem. 2006;99:1517–1523. doi: 10.1002/jcb.20859. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Sánchez EE, Lucena SE, Reyes S, Soto JG, Cantu E, Lopez-Johnston JC, Guerrero B, Salazar AM, Rodríguez-Acosta A, Galán JA, Tao WA, Pérez JC. Cloning, expression, and hemostatic activities of a disintegrin, r-mojastin 1, from the Mohave rattlesnake (Crotalus scutulatus scutulatus) Thromb Res. 2010;126:211–219. doi: 10.1016/j.thromres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane AI, Tran VL, Sanchez EE, White SA, Choi JL, Gaytán B, Chavez N, Reyes SR, Ramos CJ, Tran LH, Lucena SE, Sugarrek M, Perez JC, Mandal SA, Ghorab S, Rodriguez-Acosta A, Fung BK, Soto JG. The mojastin mutant Moj-DM induces apoptosis of the human melanoma Sk-Mel-28, but not the mutant Moj-NN nor the non-mutated recombinant Moj-WN. Toxicon. 2010;56:391–401. doi: 10.1016/j.toxicon.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa K, Jia L, Shannon JD, Fox JW. Isolation, Sequence Analysis, and Biological Activity of Atrolysin E/D/D, the Non-RGD Disintegrin Domain from Crotalus atroxVenom. Archives of Biochemistry and Biophysics. 1998;354:239–246. doi: 10.1006/abbi.1998.0698. [DOI] [PubMed] [Google Scholar]
- Taga T, Suzuki A, Gonzalez-Gomez I, Gilles FH, Stins M, Shimada H, Barsky L, Weinberg KI, Laug WE. αv-Integrin antagonist EMD 121974 induces apoptosis in brain tumor cells growing on vitronectin and tenascin. Int. J. Cancer. 2002;98:690–697. doi: 10.1002/ijc.10265. [DOI] [PubMed] [Google Scholar]
- Taklemariam T, Seoane AI, Ramos CJ, Sanchez EE, Lucena SE, Perez JC, Mandal SA, Soto JG. Functional analysis of a recombinant PIII-SVMP, GST-acocostatin: an apoptotic inducer of HUVEC, HeLa, but not SK-Mel-28 cells. Toxicon. 2011;57:646–656. doi: 10.1016/j.toxicon.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trochon-Joseph V, Martel-Renoir D, Mir LM, Thomaïdis A, Opolon P, Connault E, Li H, Grenet C, Fauvel-Lafeve F, Soria J, Legrand C, Soria C, Perricaudet M, Lu H. Evidence of antiangiogenic and antimetastatic activities of the recombinant disintegrin domain of metargidin. Cancer Res. 2004;64:2063–2069. doi: 10.1158/0008-5472.can-03-3272. [DOI] [PubMed] [Google Scholar]
- Vija H, Samuel M, Siigur E, Tonismägi, Trummal K, Sibbi J, Siigur J. VGD and MLD-motifs containing heterodimeric disintegrin viplebedin-2 from Vipera lebetina snake venom: purification and cDNA cloning. Comp Biochem Physiol Part B. 2009;153:253–260. doi: 10.1016/j.cbpb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Walsh EM, Marcinkiewicz C. Non-RGD-containing snake venom disintegrins, functional and structural relations. Toxicon. 2011 doi: 10.1016/j.toxicon.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Wegener KL, Campbell ID. Transmembrane and cytoplasmic domains in integrin activation and protein-protein interactions (review) Mol. Membr. Biol. 2008;25:376–387. doi: 10.1080/09687680802269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I, Niewiarowski S, Marcinkiewicz C, Calvete JJ, Marcinkiewicz MM, McLane MA. Structural requirements of echistatin for the recognition of alpha(v)beta(3) and alpha(5)beta(1) integrins. J Biol Chem. 1999;274:37809–37814. doi: 10.1074/jbc.274.53.37809. [DOI] [PubMed] [Google Scholar]
- Wu WB, Peng HC, Huang TF. Disintegrin causes proteolysis of beta-catenin and apoptosis of endothelial cells. Involvement of cell- cell and cell-ECM interactions in regulating cell viability. Exp. Cell Res. 2003;286:115–127. doi: 10.1016/s0014-4827(03)00105-8. [DOI] [PubMed] [Google Scholar]
- Yao C, Liu J, Shao L. Rapamycin inhibits the proliferation and apoptosis of gastric cancer cells by downregulating the expression of surviving. Hepatogastroenterology. 2011;58:1075–1080. [PubMed] [Google Scholar]
- Yeh CH, Peng HC, Huang TF. Accutin, a new disintegrin, inhibits angiogenesis in vitro and in vivo by acting as integrin alphavbeta3 antagonist and inducing apoptosis. Blood. 1998;92:3268–3276. [PubMed] [Google Scholar]
- Yeh C, Peng H, Yang R, Huang T. Rhodostomin, A Snake Venom Disintegrin, Inhibits Angiogenesis Elicited by Basic Fibroblast Growth Factor and Suppresses Tumor Growth by A Selective αvβ3 Blockade of Endothelial Cells. Molecular Pharmacology. 2001;59:1333–1342. doi: 10.1124/mol.59.5.1333. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen M, Venugopal S, Zhou Y, Ziang W, Li Y-H, Lin Q, Kini RM, Chong Y-S, Ge R. Isthmin exerts pro-survival and death-promoting effect on endothelial cells through alphavbeta5 integrin depending on its physical state. Cell Death and Disease. 2011;2 doi: 10.1038/cddis.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liao Q, Long H. Endothelin-1 promotes osteosarcoma cell invasion and survival against cisplatin-induced apoptosis. Clin Orthop Res. 2011 doi: 10.1007/s11999-011-1939-2. [DOI] [PMC free article] [PubMed] [Google Scholar]