Abstract
Phytophthora megakarya, the causative agent of cacao black pod disease in West African countries causes an extensive loss of yield. In this study we have analyzed 4 libraries of ESTs derived from Phytophthora megakarya infected cocoa leaf and pod tissues. Totally 6379 redundant sequences were retrieved from ESTtik database and EST processing was performed using seqclean tool. Clustering and assembling using CAP3 generated 3333 non-redundant (907 contigs and 2426 singletons) sequences. The primary sequence analysis of 3333 non-redundant sequences showed that the GC percentage was 42.7 and the sequence length ranged from 101 – 2576 nucleotides. Further, functional analysis (Blast, Interproscan, Gene ontology and KEGG search) were executed and 1230 orthologous genes were annotated. Totally 272 enzymes corresponding to 114 metabolic pathways were identified. Functional annotation revealed that most of the sequences are related to molecular function, stress response and biological processes. The annotated enzymes are aldehyde dehydrogenase (E.C: 1.2.1.3), catalase (E.C: 1.11.1.6), acetyl-CoA C-acetyltransferase (E.C: 2.3.1.9), threonine ammonia-lyase (E.C: 4.3.1.19), acetolactate synthase (E.C: 2.2.1.6), O-methyltransferase (E.C: 2.1.1.68) which play an important role in amino acid biosynthesis and phenyl propanoid biosynthesis. All this information was stored in MySQL database management system to be used in future for reconstruction of biotic stress response pathway in cocoa.
Keywords: cocoa, EST, annotation, cDNA
Background
Theobroma cacao (cocoa) is a diploid tree grown in tropical countries [1]. Worldwide many people depend on cocoa for their income. Cocoa is grown in a range of conditions such as full sun, or more traditionally under shade. In India, cocoa has been grown as a mixed crop under arecanut, coconut and oil palm shades. Demand for cocoa has been increased tremendously not only as a raw material for chocolate industry, but also for its flavor and other properties which imparts several health benefits [2, 3]. Diseases are major problem for decline in cocoa production and causing annual crop loss of 20– 30 % [4]. The major diseases of cocoa include black pod (Phytophthora spp.), witches' broom (Crini pellis perniciosa), and frosty pod rot (Moniliophthora roreri) causing heavy loss in production worldwide. Phytophthora megakarya, causative agent for black pod disease in West African countries is the most damaging pathogen in cocoa industry. Although Phytophthora megakarya only exists in Africa, the species Phytophthora palmivora and Phytophthora capsici are responsible for the disease in South America and India. Fungicides are used to control the disease with varying success and at significant cost to small hold farmers [5, 6]. Genomic research provides new tools to study the genetic and molecular bases of different traits. Complete genome of the cocoa has been recently published [7]. Expressed Sequence Tags (ESTs) are sequenced regions of cDNA copies of mRNA that are expressed under different conditions and represents part of the transcribed portion of the genome [8]. ESTs can be used for gene annotation, gene discovery and sequence determination. Various cocoa EST sequencing projects have been done to understand the transcriptome of cocoa [9, 10]. The EST sequence information is essential for the molecular based assays leading to cocoa crop improvement. With the objective of identifying the functional genes expressed in diseased condition (black pod), we analyzed 4 libraries of ESTs derived from Phytophthora megakarya infected cocoa leaf and pod tissues. These studies would lead to the development of secondary cocoa EST database for specific stress conditions (biotic, abiotic) that will be helpful for the researchers of cocoa crop improvement.
Methodology
Primary sequence source:
Four libraries of EST sequences derived from Phytophthora megakarya infected cocoa leaf and pod tissues belonging to the genotypes PNG and UPA134 were used in this study. Totally 6379 redundant EST sequences were retrieved from ESTtik database [9] Table 1 (see supplementary material).
EST Analysis:
EST analysis includes the following steps: 1) EST preprocessing, 2) EST assembly, and 3) functional annotation. The implemented steps are illustrated in Figure 1.
Figure 1.
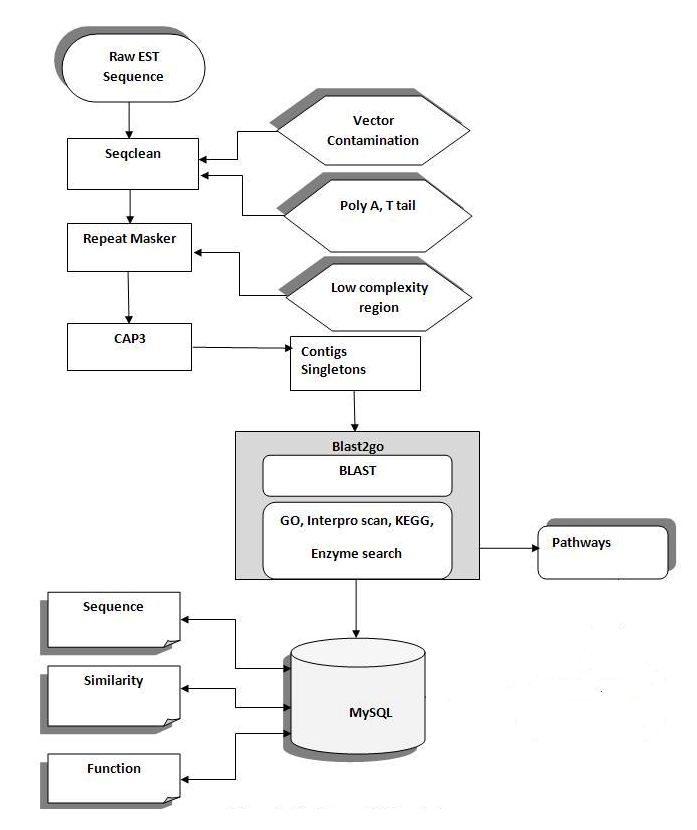
EST analysis work flow
EST pre -processing and assembly:
EST processing like removal of vector contamination, trimming poly A/ T tail and low complexity region, removal of linker and adaptor sequence were performed using SeqClean [11] tool. Vector contamination database UniVec was configured with local Blast [12] and used in SeqClean tool. Repeatmasker (http://www.repeatmasker.org) [13] tool was used to remove the low complexity regions from the EST sequences. Clustering and assembling were done by adopting CAP3 tool [14].
Primary sequence analysis:
The primary sequence analysis of GC percentage, average length of contigs and length range of contigs were processed using custom developed perl script (DSA.pl). For the detection of number of clustered sequences present in different contigs the CAP3 assembly files (.ace) were analyzed using a perl script (cap3_analyzer.pl).
Functional annotation:
Non redundant EST sequences were subjected to blastx [12] similarity search. Further, the homologous sequences were made stringent by selecting those having E-value below e-10. Gene Ontology [15] search, enzyme search, Interproscan and KEGG mapping were done using Blast2go (www.blast2go.org) tool [16].
Database design:
The information which was obtained from the processing and annotation of the EST sequences were deposited in a MySQL relational database. Three different tables were created using SQL for storing sequences, blast hit and functional annotation.
Results and Discussion
EST pre -processing and assembly:
The 6379 EST sequences retrieved from ESTtik database were processed using SeqClean tool resulting in 6349 good quality EST sequences which were used for further analysis. By the contig assembly using CAP3 tool 3333 non redundant EST (907 contigs and 2426 singletons) sequences were obtained Table 2 (see supplementary material).
Primary sequence analysis:
The primary sequence analysis showed that total GC content of non redundant EST collection as 42.7%, average length of the EST collections is 419 residues/ sequence and the sequence length ranged from 101 residues to 2576 residues Table 2 (see supplementary material). CAP3 clustered the total ESTs in to 907 contigs. Number of sequences in different contigs ranged from 2 to 141. Contig374 contained the maximum number of sequences i.e. 141 (Figure 2).
Figure 2.

Contigs vs number of sequences clustered in corresponding contigs.
Functional annotation:
Similarity search (blastx) was executed against the nonredundant database. Totally 1230 orthologous genes were annotated with a significant E-value of < e-10. The blast result showed that contig473 (2 sequence assembled) showed high similarity (95.75% with E-value 3.5E-138) to heat shock protein and contig396 (3 sequence assembled) showed high similarity (93% with E-value 1.9E-71) to low molecular weight heat shock protein. Contig338, contig345, contig668 and contig687 showed similarity in defense of related proteins.
Various biotic stress related proteins like GTP-binding protein (Contig366, Contig635), chitinase (contig568), beta-cyanoalanine synthase (Contig292), metallothionein (contig 542), thaumatin, trypsin inhibitor (contig810) heat shock protein, hydroxyproline-rich glycoprotein(Contig526), Omethyltransferase( Contig846), abc transporter family proteins, 12-oxophytodienoate reductase (Contig230, Contig379), carbonic anhydrase (Contig576), glutamine synthetase (Contig308), thioredoxin (Contig80, Contig280, Contig667), cyclophilin (Contig758), f-box family protein (Contig660), glutathione peroxidase (Contig184), ascorbate peroxidase (Contig573, Contig487), lipid transfer protein (Contig480, Contig342, ), cellulose synthase (Contig356), expansin, and pathogen related protein (contig 868) could be identified in the present study. Other major proteins involved in cell growth, cellular communication, cellular transport, transport mechanisms, energy pathway, protein destination and protein synthesis process can also be found in our EST collection (see supplementary material). Gesteira et al [17] identified pathogenesis related proteins, receptor kinase, MAP kinase and trypsin inhibitors as proteins related to Moniliophthora perniciosa infection in cocoa through comparative analysis of EST. In a similar work, Verica et al [18] identified proteins like chitinase, heat-shock proteins and beta-cyanoalanine synthase in cocoa were upregulated when treated with inducer of defense response. The cDNAs developed for the differently expressed genes in cocoa in response to witche's broom disease were putatively categorized as belonging to signal transduction, response to biotic and abiotic stress, metabolism, RNA and DNA metabolism, protein metabolism and cellular maintenance classes [19]. Gene Ontology classification (GO), HMMER search against Pfam database, Interproscan and Enzyme search were done using Blast2go tool. Gene ontology results revealed that most of the sequences were related to cellular function; stress response and biological process (see supplementary material). Enzyme search against KEGG, annotated 272 enzymes belonging to 114 metabolic pathways. The annotated enzymes were aldehyde dehydrogenase (E.C: 1.2.1.3), catalase (E.C: 1.11.1.6), acetyl-CoA C-acetyltransferase (E.C: 2.3.1.9), threonine ammonia-lyase (E.C: 4.3.1.19), acetolactate synthase (E.C: 2.2.1.6), dihydroxy-acid dehydratase (E.C: 4.2.1.9), Omethyltransferase (E.C: 2.1.1.68) and cinnamoyl-CoA reductase (E.C: 1.2.1.44) and most of them play an important role in amino acid biosynthesis. Many other enzymes involved in biosynthesis of secondary metabolites, fatty acid metabolism, and fructose and mannose metabolism were annotated. Three different tables were created using SQL commands in MySQL relational database management system. The results obtained in EST processing and primary sequence analysis were organized in the first table. The second table possessed the information obtained in similarity search and further functional annotation results were saved in the third table. These three tables were logically linked. Each row in the table was assigned a unique serial number. All the information was deposited in 3333 rows in each table that can be retrieved by either logical or key word search.
Conclusion
Four libraries of EST sequences derived from Phytophthora megakarya infected cocoa tissues have been analysed. Functional annotation resulted in 1230 orthologous genes, which included 272 enzymes and others were defense related and cellular functional genes. The annotated information was organized in a MySQL database. This information will be useful for the reconstruction of biotic stress response pathways in cocoa.
Supplementary material
Acknowledgments
The work was supported by a grant from Department of Information Technology (DIT), Government of India. We are thankful to Dr. George V. Thomas, Director, CPCRI, Kasaragod for the encouragement, guidance and facilities.
Footnotes
Citation:Naganeeswaran et al, Bioinformation 8(2): 065-069 (2012)
References
- 1.GAR Wood, RAA Lass., editors. Cacao, 4th edn. Oxford: Blackwell; 2001. p. 620. [Google Scholar]
- 2.PM Eris-Etherton, CL Keen. Curr Opin Lipidol. 2002;13:41. doi: 10.1097/00041433-200202000-00007. [DOI] [PubMed] [Google Scholar]
- 3.MC: EBRD Offices London; 2007. pp. 10–16. Cocoa Resources in consuming Countries–ICCO Market Committee, 10th meeting. [Google Scholar]
- 4.D Guest. Phytopathology. 2007;97:1650. doi: 10.1094/PHYTO-97-12-1650. [DOI] [PubMed] [Google Scholar]
- 5.LH Purdy, RA Schmidt. Annu Rev Phytopathol. 1996;34:573. doi: 10.1146/annurev.phyto.34.1.573. [DOI] [PubMed] [Google Scholar]
- 6.TO Adejumo, et al. Afr J Biotechnol. 2005;4:143. [Google Scholar]
- 7.X Argout, et al. Nat Genet. 2011;43:101. [Google Scholar]
- 8.V Poncet, et al. Mol Genet Genomics. 2006;276:436. doi: 10.1007/s00438-006-0153-5. [DOI] [PubMed] [Google Scholar]
- 9.X Argout, et al. BMC Genomics. 2008;9:512. doi: 10.1186/1471-2164-9-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.PG Jones, et al. Planta. 2006;224:1449. [Google Scholar]
- 11. http://compbio.dfci.harvard.edu/tgi/software/
- 12.SF Altschul, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. http://www.repeatmasker.org.
- 14.X Huang, A Madan. Genome Res. 1999;9:868. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.E Camon, et al. Genome Res. 2003;13:662. [Google Scholar]
- 16.A Conesa, et al. Bioinformatics. 2005;21:3674. [Google Scholar]
- 17.AS Gesteira, et al. Ann Botany. 2007;100:129. [Google Scholar]
- 18.JA Verica, et al. Plant Cell Rep. 2004;23:404. doi: 10.1007/s00299-004-0852-5. [DOI] [PubMed] [Google Scholar]
- 19.AL Gildemberg, et al. Molecular plant pathology. 2007;8:279. doi: 10.1111/j.1364-3703.2007.00393.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


