Abstract
In cardiovascular system, activation of Endothelin receptors causes vasoconstriction which leads to Pulmonary Arterial Hypertension (PAH). Endothelin receptor antagonism has emerged as an important therapeutic strategy in pulmonary arterial hypertension. Bosentan is intended to affect vasoconstriction, hypertrophic and fibrotic effects by blocking the actions of receptors ETA and ETB. In this study we identified the action of Bosentan on endothelin B receptor using docking studies with homology modeled endothelin B receptor. Through the modeled protein, the flexible Docking study was performed with Bosentan and its derivatives with theoretically predicted active sites. The results indicated that amino acid ARG82, ARG84 and HIS197 present in endothelin B receptor are core important for binding activities and these residues are having strong hydrogen bond interactions with Bosentan. We have investigated the Bosentan and its derivatives interactions and scoring parameters using gold docking package. Among the docked compounds, one of the Bosentan derivatives BD6 shows better interaction than Bosentan with endothelin B receptor. Our results may be helpful for further investigations in both in vivo and in vitro conditions.
Keywords: Bosentan, Endothelin receptors, Modeller7v7, Molecular docking, Pulmonary Arterial Hypertension
Background
Pulmonary Arterial Hypertension (PAH) is a severe disease of Pulmonary arteries involving vasoconstriction, pulmonary vascular remodeling, and insitu thrombosis [1]. PAH develops the blood flow through the pulmonary arteries and the right side of the heart is under increasing strain to pump blood through to the lungs, leads to breathlessness, chest stiffness and fatigue [2]. Laboratory and clinical investigations have clearly shown that Endothelin receptors are over expressed in several forms of pulmonary vascular disease and play a significant pathogenetic role in the development and progression of pulmonary vasculopathy [3]. There are two distinct receptors for the endothelin family of peptides, endothelin receptor A (ETA) and endothelin receptor B (ETB) and these two receptors have unique locations and binding affinities for the endothelin peptides [4]. The ETA receptors are expressed on pulmonary vascular smooth muscle cells, whereas ETB receptors are located on both pulmonary vascular endothelial cells and smooth muscle cells. ETB receptors have an important role in the ETdependent protracted presser effects. Under some circumstances it actually contributes to pulmonary vasoconstriction, through a population of ETB receptors located on vascular smooth muscle cells [5]. Bosentan is the first oral endothelin receptor antagonist (ERA) that blocks the effect of endothelin receptors. It belongs to a class of highly substituted pyrimidine derivatives, with no chiral centers. It is designated chemically as 4-tert-butyl-N - [6-(2-hydroxy-ethoxy) -5- (2- methoxy-phenoxy) - [2, 2] - bipyrimidin-4-yl] - benzene sulfonamide monohydrate [6]. In vitro functional experiments performed on animal and human tissues showed that Bosentan behaves as a competitive antagonist on ET receptors. It is intended to inhibit vasoconstriction, hypertrophic and fibrotic effects by blocking the actions of receptors (ETA and ETB) which leads to reduction of blood pressure in lungs. Bosentan interact with the binding of ET-1 and other ET peptides to both ETA and ETB receptors [7]. The objective of this work is to identify the role the mechanism of Bosentan on ETB receptor and to model the protein, and to identify the interactions with Bosentan and their derivatives.
Methodology
The sequence information of Endothelin-B receptor- Homosapiens (Accession No: P24530) is retrieved from Swiss Prot (http://www.swissprot.org) and submitted to SBASE server (http:// hydra icgeb.trieste.it/sbase) for domain prediction. The predicted domain (118-386) is search for similarity (Protein blast) and the related protein structure was identified by the BLAST program against PDB. The macromolecular structure (PDB ID 3C9M [8]), which shows maximum identity with high score and less e-value designated as template. The 3D model was generated by using the academic version of MODELER9v7 (http//:www.salilab.org/modeler), based on the information obtained from sequence alignment [9]. The 3D structure obtained from modeler were further refined through molecular dynamics and equilibration methods using NAMD 2.5 with CHARMM27 force field and the system contains TIP3P water model. The energy of the structure was minimized with 10,000 steps with cutoff of 12 Å for vdW interactions. The equilibrated system was simulated with 1 picoseconds (PS); to hold back the original conformation, for that (NPT) constant pressure and 310K constant temperatures has been choosed [10]. Finally, the structure having the least energy with low RMSD was used for further studies. The final structure obtained, was analyzed by Ramachandran's map drawn using PROCHECK v.3.0, and environment profile using ERRAT graph (Structure Evaluation server). The model satisfying all the parameters after evaluation was considered for the further process [9]. Theoretical Active site prediction of Endothelin-B receptor was predicted using CASTP server. CASTP identifies and measures pockets and pocket mouth openings, as well as cavities. Here we input the modeled protein for predicting the ligand binding sites and the CASTP server predicts the amino acids crucial for binding interactions [11]. The Bosentan library was generated manually by addition of polar groups in the required position and 30 new compounds were generated as derivatives of Bosentan Table 1 (see supplementary material). The Docking interactions are done with GOLD (Genetic Optimization of Ligand Docking), based on genetic algorithm which is used for docking of Bosentan and their derivative compounds [12]. The interaction of these derivatives with the active site residues are thoroughly studied using molecular mechanics calculations. The parameters used for GA were population size 100, selection pressure 1.1, and operator parameters for crossover, mutation and migration were set to 100, 100 and 10 respectively. Default cutoff values of 3.0 X (dH-X) for hydrogen bonds and 6.0 X for vdW were employed. During docking, the default algorithm speed was selected and the ligand binding site in the Endothelial B receptor was defined within a 10 A° radius with the centroid as CE atom of ASP81. The number of poses for each inhibitor was set 100, and early termination was allowed if the top three bound conformations of a ligand were within 1.5X RMSD [13]. After docking, the individual binding poses of each ligand were observed and their interactions with the protein were studied.
Results
Homology Modeling of Endothelin – B receptor Domain:
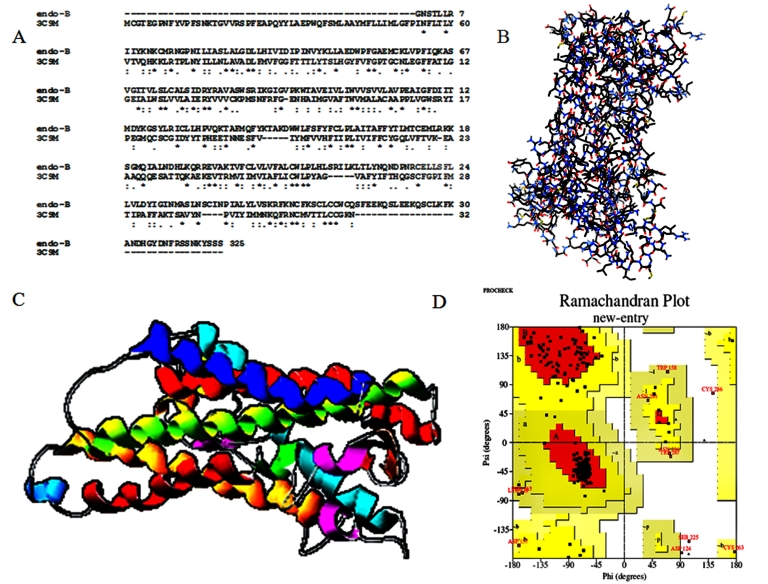
A high level of sequence identity should guarantee a more accurate alignment between the target sequence and template structure, and here 3C9M (Chain A, Structure of a mutant bovine Rhodopsin in hexagonal crystal form protein) with Endothelin-B receptor have high sequence identity and the identity of the reference protein with the Endothelin B receptor domain is 36%. Structurally conserved regions (SCRs) for the model and the template were determined by multiple sequence alignment (A) superimposition of template and model structure (B), 3D structure of modeled protein (C) and Ramachandran’s plot (D) are illustrated in Figure 1. In this study, the model is made up of residues 118 - 386 because the active domain region was identified in between these residues by the SBASE server. Coordinates from the reference protein (3C9M) to the SCRs, structurally variable regions (SVRs), N-termini and C-termini were assigned to the target sequence based on the satisfaction of spatial restraints (C).
Figure 1.
Modeling of Endothelin B receptor with sequence aligned (A) with 3C9M morphed (B) to build 3D model structure (C) and the model structure Ramachandran's plots (D)
Superimposition of 3C9M with Endothelin- B receptor domain:
This model was refined by molecular dynamics method and the final stable structure of the Endothelial B receptor is superimposed with Cα trace of template. The weighted root mean square deviation of Cα trace between the template and final refined models was 1.81 Å. This final refined model was used for the identification of active site and for docking of the substrate with the protein. This protein has the structural alignment with 13 helices and 2 sheets like a covered barrel shape. The overall scores indicates acceptable protein environment. After the refinement process, validation of the model was carried out using Ramachandran plot calculations computed with the PROCHECK program. Altogether 86.7% of the residues of Endothelin B receptor were in favored and allowed regions. The overall PROCHECK G-factor of Endothelin-B receptors was – 0.16 and VERIFY 3D environment profile was good. The compatibility score above zero in the VERIFY-3D program showed except ten, all residues are reasonable which makes us to believe that the structure of the Endothelin-B receptor protein is reliable.
Active site Identification of Endothelin B receptor:
The possible binding sites of Endothelin B receptor was searched based on the structural comparison of template and the model build with CASTP server. Since, Endothelin B and the 3C9M are well conserved in both sequence and structure; their biological function should be identical. The predicted results are interesting and its shows the amino acid position 17- 22, 78-98 and 197-208 are predicted to conserved with the active site. Here we consider the experimental binding sites of 3C9M include some of the residues as mentioned above. Thus in this study ARG 82, ARG 84 and HIS 197 are chosen as the more favorable sites to dock the substrate.
Docking of inhibitors within the active site of Endothelin B receptor:
In the presence of inhibitory results of Bosentan, new structural features and fictionalization requirements were proposed for the basic Bosentan scaffold that could increase affinity with Endothelin B receptor and consequently improves the anti hyper tense property [14]. Among these requirements, the modifications of the group in the structure by a more polar group were expected to increase the activity. These interesting results prompted us to prepare Bosentan analogues. Here we developed different analogues based on the structure of Bosentan by replacing with more polar groups than already existing groups, with little change in the properties of analogues represented in Table 1 (see supplementary material). These were used for docking studies to identify better drug derivative. Docking of inhibitors given in Table 1 (see supplementary material) with Endothelin B was performed using GOLD 3.0.1, which is based on genetic algorithm (CCDC). The structure of Bosentan used for designing derivatives. The docking procedure includes several steps. First, the protein-ligand complex is generated using the GOLD package without constraints between the ligand and the specific amino acids of the pocket. The algorithm exhaustively searches the entire rotational and translational space of the ligand with respect to the receptors. The flexibility of the ligand is given by dihedral angle variations. The various solutions evaluated by a score, which is equivalent to the absolute value of the total energy of the ligand in the protein environment. Thus docking with the program GOLD version 3.0.1 was employed to locate the appropriate binding orientation and conformation of compounds with Endothelin B receptor.
Gold Score fitness function:
Gold Score performs a force field based scoring function and is made up of four components namely, hydrogen bond energy (H-bond); vander Waals energy (External vdW); vander Waals energy (internal vdW) and Intermolecular hydrogen bond (internal H-bond) energy. The external vdw score is multiplied by a factor of 1.375 when the total fitness score is computed. This is an empirical correction to encourage protein-ligand hydrophobic contact. The fitness function has been optimized for the prediction of ligand binding positions [15].
GoldScore Fitness = S (hb_ext) + 1.3750*S (vdw_ext) + S (hb_int) + 1.0000*S (int) S (int) = S (vdw_int) + S (tors)
Where S (hb+ext) is the protein-ligand hydrogen bond score, S (vdw+ext) is the protein-ligand van der Waals score, S (hb+int) is the score from intramolecular hydrogen bond in the ligand and S (vdw+int) is the score from intramolecular strain in the ligand.
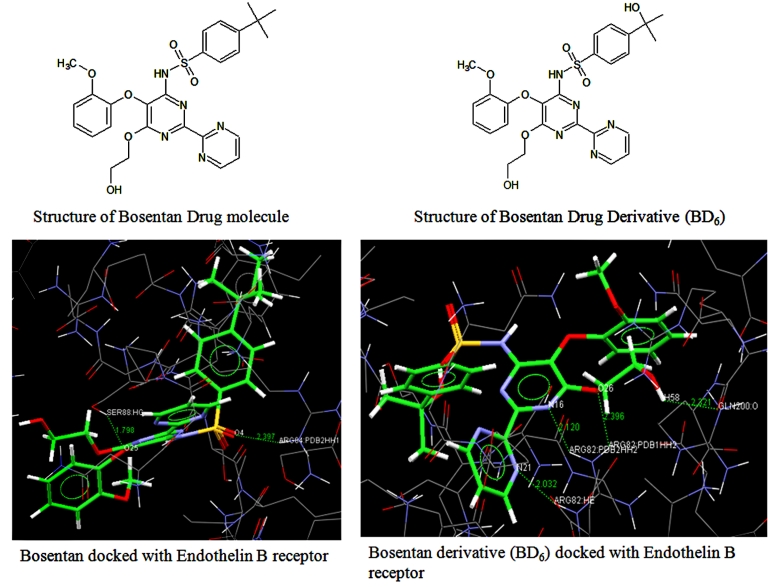
Among the 29 inhibitors BD6 derivate of Bosentan showed high affinity than others towards Endothelin B. The structure of the BD6 was shown in the Figure 2. Bosentan docked results shows interaction of Endothelin B receptor with 2 hydrogen bonds. The active residues in the Endothelin B receptor ARG84 binds to O4 of Bosentan with one hydrogen bond. The other hydrogen bond is formed between SER88 and O25 of Bosentan. BD6 derivative forms four hydrogen bonds with Endothelin B receptor with the active residues. It is observed that BD6 derivative was located at center of active site and is stabilized by hydrogen bonding interactions and these interactions are represented in Table 2. Bosentan total Gold Score fitness function is 30.03 Kcal/mol. This shows that Bosentan has good inhibitory activity on Endothelin B receptor. Gold Score fitness function of BD6: Total Gold Score fitness function is 45.21 Kcal/mol. Here we have chosen Gold Fitness function than Chem score fitness function as Gold score fitness function is marginally better than ChemScore fitness function. Even though, some other compounds in this library are having better scoring the BD6 shows better interactions and those interactions in Bosentan binding region Table 3 (see supplementary material). So, we have demonstrated that, among the 29 derivatives designed the binding capacity is highest for BD6 and can be further used for further investigations.
Figure 2.
Comparisons of interactions between Bosentan and its derivatives against Endothelin B receptor
Conclusion
In this work, we have constructed a 3D model of Endothelin B receptor of Homosapiens, and obtained a refined model after energy minimization. The validation of protein structure through SAVS server and NAMD validates the protein is stable enough. The stable structure is further used for docking with Bosentan and its derivatives. Docking results indicate that conserved amino-acid residues in Endothelin B receptor play an important role in maintaining a functional conformation and are directly involved in donor substrate binding. The interaction between the domain and the inhibitors proposed in this study are useful for understanding the potential mechanism of domain and the inhibitor binding. The hydrogen bonds play important role for the structure and function of biological molecule in this study, and we found that ARG 82, ARG 84 and HIS 197 of Endothelin B are important for strong hydrogen bonding interaction with these inhibitors. To the best of our knowledge ARG 82, ARG 84 is conserved in the domain and may be important for structural integrity or maintaining the hydrophobicity of the inhibitor-binding pocket. Among 29 derivatives used for docking, BD6 derivative showed best binding energy than other compounds. The total fitness function is high for BD6 when compared to other derivatives including Bosentan. According to our investigations from the docking results BD6 (C26H27N5O7S) can act as better Endothelin antagonist. Future investigation of other theoretical studies and experimental studies may confirm that BD6 as a potent inhibitor.
Supplementary material
Acknowledgments
One of the authors Mr. Chandrabose Selvaraj thanking Alagappa University for research grant and P. Seshapani Thanking Sri Venkateswara University for research support
Footnotes
Citation:Rayalu et al, Bioinformation 8(2): 081-086 (2012)
References
- 1.JC Honore, et al. Clin Sci. 2002;103:280S. doi: 10.1042/CS103S280S. [DOI] [PubMed] [Google Scholar]
- 2.B Mutlu, et al. Anadolu Kardiyol Derg. 2010;1:43. [PubMed] [Google Scholar]
- 3.T Raza, et al. Heart Reviews. 2007;8:90. [Google Scholar]
- 4.A Benigni, et al. Curr Opin Nephrol Hypertens. 1995;4:349. doi: 10.1097/00041552-199507000-00011. [DOI] [PubMed] [Google Scholar]
- 5.M Maegele, et al. Neurol Res. 2011;33:119. [Google Scholar]
- 6.G Strange, et al. Expert Rev Pharmacoecon Outcomes Res. 2011;11:253. doi: 10.1586/erp.11.26. [DOI] [PubMed] [Google Scholar]
- 7.S Boniface, et al. Rev Mal Respir. 2011;28:e94. doi: 10.1016/j.rmr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.RE Stenkamp, et al. Acta Crystallogr D Biol Crystallogr. 2008;64:902. [Google Scholar]
- 9.S Sundaram, et al. Bioinformation. 2010;5:177. [Google Scholar]
- 10.Y Wang, et al. Comput Sci Discov. 2011;4:015002. [Google Scholar]
- 11.SD Kaistha, R Sinha. Bioinformation. 2009;3:240. doi: 10.6026/97320630003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ML Verdonk, et al. Proteins. 2003;52:609. [Google Scholar]
- 13.MK Annamala, et al. Bioinformation. 2007;1:339. [Google Scholar]
- 14.F Goirand, et al. J Cardiovasc Pharmacol. 2003;41:117. doi: 10.1097/00005344-200301000-00015. [DOI] [PubMed] [Google Scholar]
- 15.S John, et al. BMC Bioinformatics. 2011:12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.