Abstract
Courtship behaviour involves a complex exchange of signals and responses. These are usually studied at the phenotypic level, and genetic or transcriptional responses to courtship are still poorly understood. Here, we examine the gene-expression changes in Drosophila melanogaster females in response to one of the key male courtship signals in mate recognition, song produced by male wing vibration. Using long oligonucleotide microarrays, we identified several genes that responded differentially to the presence or absence of acoustic courtship stimulus. These changes were modest in both the number of genes involved and fold-changes, but notably dominated by antennal signalling genes involved in olfaction as well as neuropeptides and immune response genes. Second, we compared the expression patterns of females stimulated with synthetic song typical of either conspecific or heterospecific (Drosophila simulans) males. In this case, antennal olfactory signalling and innate immunity genes were also enriched among the differentially expressed genes. We confirmed and investigated the time course of expression differences of two identified immunity genes using real-time quantitative PCR. Our results provide novel insight into specific molecular changes in females in response to courtship song stimulation. These may be involved in both signal perception and interpretation and some may anticipate molecular interactions that occur between the sexes after mating.
Keywords: Drosophila melanogaster, gene expression, microarray, courtship song, Turandot
1. Introduction
Sexual reproduction often involves complex interactions between males and females extending from pre-mating courtship signalling to post-mating molecular interactions. Recently, some of the physiological and neuronal changes associated with the reception of sexually important signals have been identified in both vertebrates and invertebrates, including Drosophila [1–4]. Progress has also been made in identifying transcriptional changes associated with social interactions in Drosophila, particularly in males [5–7]. However, the genes involved in female responses to male signals still remain largely unknown, and very few studies have attempted to identify transcriptional changes involved in pre-mating responses to stimulation using a genome-wide analysis of gene expression. One such study [8] assessed gene expression of Drosophila melanogaster females 24 h after they had been courted by and rejected males, while another [9] focused on expression changes in females in response to visual cues of attractive males in swordtail fish (Xiphophorus nigrensis).
Although the molecular responses to courtship signals are a priori expected to include genes involved in mating preference, an intriguing and previously unexplored possibility is that courtship may also induce molecular and physiological changes in females in anticipation of mating. Recent studies of female Drosophila have identified male-induced molecular changes associated with a response to sperm and accessory gland proteins, many of which appear to be under antagonistic sexual selection [10,11]. Do changes start to occur during courtship, in anticipation of mating?
Drosophila melanogaster is an ideal species for studying genes involved in behaviour, because it has a long history as a model organism in genetic studies and a well-annotated genome, and also its courtship behaviour is well understood. Courtship in D. melanogaster involves visual, acoustic, olfactory and tactile signals [12]. Courtship song, produced by male wing vibration, is perhaps the most important courtship signal influencing male mating success [13]. Song is detected with a modified antennal receiver, which transfers air vibrations to the hearing neurons [14,15]. In many Drosophila species, song consists of two main components; pulse song and sine song [16]. Pulse song includes repetitive trains of pulses and their inter-pulse intervals (IPIs) as well as a distinctive rhythm in IPI [17–20] and contributes to interspecific mate discrimination [18,19,21,22]. However, apart from general hearing genes [23–25], very little is known about the genetic basis of female response to song, especially in comparison to song production.
In this study, we used D. melanogaster to trace transcriptomic changes that occur in females upon hearing male song in order to identify the molecular components involved in female response to acoustic stimulation as well as to study the species-specificity in this response. Gene-expression changes were studied in response to the presence and attractiveness of an acoustic courtship stimulus without exposing the females to courting males, i.e. by excluding the confounding effects of other male traits upon female gene expression. One study on the swordtail fish [9] is the only similar attempt to identify gene-expression responses to a sexually important courtship signal in isolation.
We combined a playback experimental approach with transcriptome profiling using two-colour microarrays and real-time quantitative PCR (qPCR). First, we confirmed song discrimination behaviour in our laboratory strain of D. melanogaster flies using synthetic song. Second, using long oligonucleotide microarrays, we identified several loci that responded differentially to the presence or the absence of the acoustic courtship stimuli. Although modest overall, many of the identified changes in gene expression were concentrated in antennal signalling genes (mainly known previously to be involved in olfactory reception) as well as in neuropeptides and immune response genes. Further, we compared the gene-expression patterns of females stimulated with synthetic songs typical of either conspecific or heterospecific (Drosophila simulans) males and found similar differential expression in several antennal olfactory signalling genes, and also in innate immunity peptides. In a third experiment, we used qPCR to confirm and explore the time frame of expression patterns of two identified immunity genes in response to stimulation with conspecific and heterospecific songs.
2. Material and methods
(a). Mating discrimination experiment
An isogenic wild-type D. melanogaster strain (Oregon-K) was used for all experiments. We used synthetic song playback to confirm the presence of song discrimination also in this strain. Five-day old virgin females were grouped with wingless 5-day old conspecific virgin males, and stimulated with either D. melanogaster- or D. simulans-like synthetic songs, with the mean IPI and pulse train cycle of approximately 35 ms and 55 s, and approximately 45 ms and 40 s, respectively (song synthesis is described in Ritchie et al. [22]). The number of copulating pairs was counted every 2 min over a period of 20 mins. We carried out 10 trials per song with 20 pairs of flies in each and analysed the mating data with a generalized linear mixed model with binomial error distribution, with the cumulative proportion of mated females (out of 200 per song) as a response variable, song type as fixed and trial as a random factor, and the observation time as a covariate, using R v. 2.9.1 [26].
(b). Microarrays
Behavioural playback experiments to obtain transcriptome profiles were carried out by aspirating 40 five-day old virgin females at a time into a chamber mounted on a loudspeaker. No males were introduced in order to isolate the effect of auditory signal perception from other male signals. Conspecific and heterospecific songs (described above), as well as white noise as a control, were played back to females for 15 min during peak mating activity time (ZT5-7, E. Immonen 2009, personal observation). After the trials, females were removed from the chamber by anesthetizing with CO2, snap-frozen with liquid nitrogen and stored in −70°C.
One hundred and twenty female heads from three playback-trials were randomly pooled to form each sample per treatment, with a total of four biological replicate samples prepared for the control and heterospecific song stimulation treatments and eight for the conspecific song stimulation treatment. Heads were removed individually from the frozen flies to minimize the loss of antennal segments and with the aid of liquid nitrogen to prevent thawing. Samples were hybridized into two-channel long oligonucleotide microarrays (FL003-INDAC; see electronic supplementary material, material and methods and www.flychip.org.uk for protocols and further information). Four arrays were probed with control and conspecific song stimulus groups, three with conspecific and heterospecific song groups. Three of the arrays had reverse labelling to account for dye-bias in hybridization efficiency. FlyChip at the University of Cambridge performed sample processing, array hybridization, image scanning and quality controls. Data are deposited in NCBI (reference no. GEO GSE31190).
We used expression information from FlyAtlas [27] to eliminate probes not expressed in the head. Several packages within Bioconductor in R were used for the data pre-processing and analysis (see electronic supplementary material, material and methods) [26,28]. Differential expression was tested using limma, with Bayesian approximation of standard errors [29,30]. False discovery rate (FDR) was estimated to account for multiple testing [31].
One of the main aims of this study was to explore whether any a priori defined functional sets of genes are over-represented among the genes showing differences in expression owing to song treatments. For this, we took advantage of predefined gene sets from several databases, including the Gene Ontology database, the Kyoto Encyclopedia of Genes and Genomes and the Integrated Documentation Resource for Protein Families, Domains, Regions and Sites, and tested overrepresentation of functional terms by calculating a moderated Fisher Exact p-value (FDR estimated to control for multiple testing), and subsequently clustering genes into groups that share a high degree of similar significantly enriched biological functions, as implemented in The Database for Annotation, Visualization and Integrated Discovery (DAVID) [32,33]. This approach not only reveals the major biological themes associated with the genes under study, but also the groups of genes that are likely to be co-regulated based on their functional similarity. Focusing only on the genes showing largest expression differences with a stringent FDR may fail to capture the biological mechanisms involved in expression changes, especially when the changes are small [34]. We therefore defined the lists of genes included in the gene functional enrichment analyses using a gene-specific p-value cut-off of 0.05 [35].
(c). Real-time qPCR
We chose two candidate genes, TotM and TotC, from the contrast of conspecific and heterospecific songs for validation owing their statistical significance and effect size. Six biological replicates per treatment were obtained by independent sample collections following the same experimental procedures as before. However, in addition to exposing the females to the song treatments and control for 15 min, another set of flies were exposed for only 5 min in order to test whether expression differences occur quickly after stimulation. One-step qPCR was carried out to test for expression differences between the treatment groups at each time point (see the electronic supplementary material and methods for further details of all the methods used).
3. Results
(a). Mate choice based on song
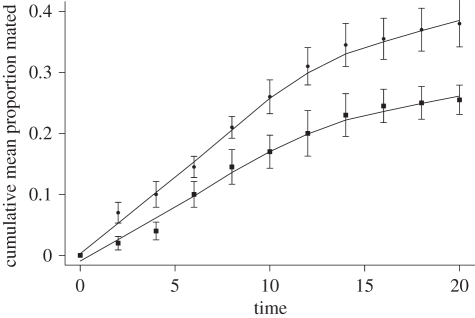
Females mated significantly more often when stimulated with a song model representing D. melanogaster pulse song when compared with that of D. simulans throughout the observation period (χ2 = 96.7, p < 0.001; figure 1), though both songs were stimulatory to females. Thus, the pulse song variation acts as a species-specific stimulatory signal [22] and is a target of female mating preferences in the Oregon-K isogenic line used in the present study.
Figure 1.
Cumulative mean proportion of mated females, with standard errors. Circles, conspecific song; squares, heterospecific song.
(b). The transcriptome response to courtship song
In order to detect differences in gene expression associated with hearing a song, we compared conspecific song-stimulated and control females. There were 412 differentially expressed genes, of which 41 were significant with a 5 per cent FDR (table 1). To identify gene-expression changes associated with song discrimination for attractive versus non-attractive songs, we compared the expression profile of conspecific song-stimulated females to that of heterospecific song-stimulated females. This contrast revealed 222 differentially expressed genes, of which two, TotM and TotC, were significant after correction for multiple testing with a strict 5 per cent FDR (table 1). That more differences in gene expression were detected between song and no-song than between the two songs is consistent with the results of behavioural experiments, which show that heterospecific song is still stimulatory to females, but less so.
Table 1.
Differentially expressed genes from the two sets of microarrays, with FDR < 5%. (FC (fold change) > 0 indicates upregulation with conspecific song.)
| contrast | gene | FC | p-value | FDR adj. p-value |
|---|---|---|---|---|
| conspecific–heterospecific song | TotM | 2.44 | 3.62E-08 | 1.7E-05 |
| TotC | 1.88 | 1.85E-06 | 4.0E-04 | |
| conspecific song–control | CG12726 | 2.13 | 6.48E-35 | 2.9E-31 |
| CG14645 | 1.70 | 5.75E-18 | 1.3E-14 | |
| Cdep | 1.57 | 1.92E-13 | 2.8E-10 | |
| CG10332 | −1.45 | 1.2E-09 | 1.3E-06 | |
| CG6188 | −1.44 | 3.6E-09 | 3.2E-06 | |
| ems | 1.42 | 1.3E-08 | 9.4E-06 | |
| Or83b | 1.37 | 2.7E-08 | 1.7E-05 | |
| CG31678 | 1.37 | 2.6E-07 | 1.4E-04 | |
| mRpS26 | 1.36 | 2.9E-07 | 1.4E-04 | |
| CG4230 | 1.36 | 6.0E-07 | 2.7E-04 | |
| DptB | −1.35 | 9.8E-07 | 3.9E-04 | |
| CG18542 | 1.34 | 1.8E-06 | 6.6E-04 | |
| CG32533 | 1.32 | 6.4E-06 | 2.0E-03 | |
| CG8600 | 1.32 | 7.3E-06 | 2.0E-03 | |
| CG13607 | 1.32 | 7.3E-06 | 2.0E-03 | |
| trn | 1.32 | 7.5E-06 | 2.0E-03 | |
| qkr58E-3 | 1.32 | 7.9E-06 | 2.0E-03 | |
| Att-A | −1.31 | 1.0E-05 | 2.4E-03 | |
| Gld | 1.30 | 1.7E-05 | 3.9E-03 | |
| X11Lbeta | 1.30 | 1.8E-05 | 3.9E-03 | |
| skf | 1.29 | 2.9E-05 | 6.2E-03 | |
| CG10635 | 1.29 | 3.2E-05 | 6.3E-03 | |
| Thd1 | 1.29 | 3.3E-05 | 6.3E-03 | |
| CG10889 | 1.28 | 4.8E-05 | 8.5E-03 | |
| CG14630 | −1.28 | 4.7E-05 | 8.5E-03 | |
| mthl8 | 1.27 | 9.6E-05 | 1.6E-02 | |
| CG7861 | 1.27 | 1.1E-05 | 1.7E-02 | |
| Cys | 1.27 | 1.1E-05 | 1.7E-02 | |
| Oseg1 | 1.27 | 1.1E-05 | 1.7E-02 | |
| CG10962 | −1.27 | 1.2E-05 | 1.7E-02 | |
| Pink1 | 1.26 | 1.3E-05 | 1.8E-02 | |
| Cyp6a14 | 1.26 | 1.3E-05 | 1.8E-02 | |
| CG31708 | 1.26 | 1.4E-05 | 1.9E-02 | |
| mbl | 1.26 | 1.7E-05 | 2.2E-02 | |
| CG11093 | 1.25 | 2.3E-05 | 2.9E-02 | |
| GstE9 | −1.25 | 2.5E-05 | 3.0E-02 | |
| CG5966 | 1.25 | 2.9E-05 | 3.5E-02 | |
| Nkd | 1.25 | 3.3E-05 | 3.9E-02 | |
| Atg5 | 1.24 | 3.7E-05 | 4.1E-02 | |
| CG4678 | 1.24 | 4.0E-05 | 4.4E-02 | |
| DIP1 | 1.24 | 4.4E-05 | 4.7E-02 |
To assess which biological processes are associated with the song responses, we performed functional enrichment analysis [32,33] on the genes that differed in expression in each of the stimulus comparisons (with gene-specific p-value < 0.05), and identified several clusters of genes that share similar, significantly enriched annotation terms (electronic supplementary material, table S1). Hearing conspecific song was significantly associated with a cluster of six genes that all share functions in signalling, odorant/pheromone binding and cognition (enrichment score = 2.39). A second significant gene cluster for this contrast contained four genes, all involved in signalling, hormone and neuropeptide activity (enrichment score = 2.11), and a third cluster five immune response genes (enrichment score = 1.69). Preference for species-specific song was significantly associated with two clusters of genes with shared annotations. The first cluster included five genes (four from the Turandot gene family), which are involved in humoral immunity and stress response as secreted signal peptides (enrichment score = 2.59). The second group was similar to the first cluster identified in comparison between song and control: here four genes shared functions in signalling, cognition and odorant/pheromone binding (enrichment score = 2.32). It is worth noting that both of the comparisons involved significant enrichment of seven antennal genes, the first comparison Os-C, Os-E, Pbprp3, a5, Or83b, Pbprp1 and Pbprp5 (fold enrichment = 8.0, FDR = 7.4E-04), and the second one Os-C, Os-E, Pbprp3, a5, Or43a, Pbprp4 and a10 (fold enrichment = 15.3, FDR = 1.1E-05). See the electronic supplementary material, table S1 for the genes and all of the functional annotations included in the clusters.
(c). Effect of song stimulation duration upon Turandot gene expression
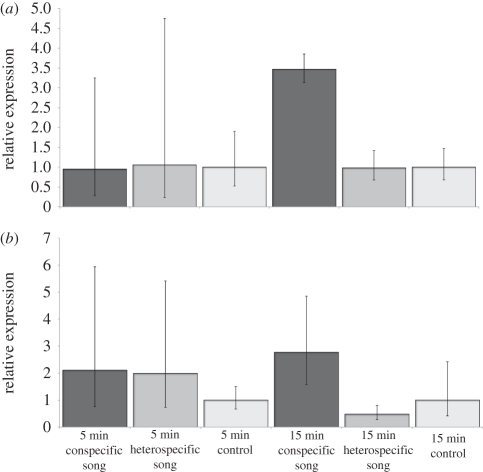
We chose to test the expression of two genes, TotM and TotC, with qPCR, based on their fold change and significant expression changes in the experiments with conspecific and heterospecific song. The expression of these two genes that belong to the family of Turandot genes was tested after stimulating the females with either of the two songs or white noise for 5 and 15 min in order to examine the time course of expression variation.
Five minutes stimulation did not induce significant changes in expression levels between any of the acoustic treatments for either of the genes (figure 2; TotM: χ22 = 2.9, p-value = 0.2; TotC: χ22 = 0.01, p-value = 0.9). However, after 15 min of song stimulation, both TotM and TotC were significantly upregulated with conspecific song when compared with heterospecific song, and TotC compared with the control (figure 2; TotM: χ22 = 9.6, p-value = 0.008; TotC: χ22 = 11.38, p-value = 0.003).
Figure 2.
Transcript abundance after 5 and 15 min song stimulation for (a) TotC and (b) TotM. 95% CI shown for mean relative expression values.
4. Discussion
We have examined gene-expression changes associated with the presence and the attractiveness of male courtship song in D. melanogaster. Song plays a key role in stimulating female mating and females are more stimulated by homospecific song, which we confirmed with Oregon-K females. Genes involved in this species-specific response may be under sexual selection and contribute to sexual isolation.
(a). Signalling and olfactory genes respond to song
Hearing a song in isolation from any other male traits resulted in relatively modest differences in gene expression, however, responses in distinct groups of genes were identified. Song induced changes in genes that function in signalling, and interestingly, many of them are expressed in the antennae, which are the Drosophila hearing organs [36]. Antennal genes were enriched seven and 15 times more among those differentially expressed between song and control and between conspecific and heterospecific songs, respectively. Because changes were more pronounced between the song types this cannot simply be owing to a general response to acoustic stimulation. Recently, it has been shown that antennae are actively tuned to the frequencies within homospecific song [37]. Four of the antennal genes are shared between the two comparisons, including Os-C, Os-E, Pbprp3 and a5. Although effect sizes are small, the significant enrichment of the antennal genes suggests that modest changes can be biologically meaningful. It should also be appreciated that our RNA preparations were from whole heads, so tissue-specific expression changes in antennal neurons are probably considerably greater. We did not detect any previously identified genes involved in hearing in either of the comparisons [23–25].
The signalling genes responding to song include a significant enrichment of genes (Pdf, crz, hug and tk) with functions in neuropeptide signalling pathways and hormone activity (electronic supplementary material, table S1). The neuropeptide Pdf regulates signalling in neurons involved in a variety of circadian rhythmic behaviours, in a species-specific way [38], and its regulation responds to selection for increased or decreased mating latency [39]. Crz has recently been implicated in sex-specific stress-related behaviours [40], and is linked with the regulation of dopamine [40], which modulates female sexual receptivity [41]. These neuropeptides may therefore participate in the perception of the rhythmic conspecific pulse song and downstream signalling modulating arousal [42].
Interestingly, song stimulation in both experiments evoked expression changes in genes involved in chemical communication (nearly all of the antennal genes, see electronic supplementary material, table S1), which cannot be induced by olfaction differences in our study as females only heard song. These include odorant receptor gene Or49a and the co-receptor gene Or83b (both differentially expressed between song and control) as well as several odorant-binding protein-coding genes, most of which are also involved in binding pheromones (for example, Pbprp3 and Os-E). Olfactory genes found in our study, including Pbprp3, Os-C, Pbprp5 and Obp99c, also respond to mating [43]. Why do we see subtle changes in so many genes involved in olfactory signalling? The simultaneous activation of the olfactory system when hearing conspecific song could enhance the sensitivity of pheromone detection during courtship. Drosophila sensory neurons responsible for odorant detection cover the surface of the third antennal segment (funiculus), including trichoid sensillae implicated in the recognition of the pheromone 11-cis-vaccenyl acetate [44]. Perhaps, mechanical vibrations of the antennal arista connected to the funiculus during song stimulation also influences the expression of other loci expressed in the antennae. Alternatively, antennal genes may have pleiotropic effects involved in other kinds of signal transmission.
(b). Immune response to song
Hearing an attractive, conspecific song induced expression changes in genes involved in immunity and stress response. While some were downregulated (Attacin-A (Att-A) and -C (Att-C), Diptericin B (DptB), Drosomycin (Drs) and Immune-induced molecule 18 (IM18)) compared with the control, four out of eight members from Turandot family (TotA, TotC, TotX and TotM) as well as Immune-induced molecule 4 (IM4) were upregulated compared with the heterospecific song. Two Turandot genes, TotC and TotM, were examined more closely with real-time qPCR. Significant changes in gene expression were only detected after 15 min of stimulation with attractive, conspecific song compared with the heterospecific song, and Tot-C also compared with the control. Upregulation within 15 min of the start of courtship may be sufficient to have Turandot genes expressed prior to mating. However, the difference between a biologically meaningful level of expression and what can be significantly detected by qPCR, is unknown.
Many immunity genes are involved in female reproduction in D. melanogaster. Also D. melanogaster males show expression differences in immunity-related genes when courting females [7]; however, the function of these changes is not yet known. Interestingly, long-term exposure to acoustic signals has been linked with increased immunity in field crickets (Teleogryllus oceanicus) [45], thus an increased probability of mating influences immune functions in a variety of organisms. Two of the Turandot genes, TotA and TotC, show sex-specific expression: they are upregulated in female heads relative to males, and are regulated downstream from the sex-specific pre-mRNA splicing factor transformer (tra) [46] which, together with doublesex [47], controls the sex determination cascade. All the Turandot genes observed in this study (TotM, -C, -X and -A) show similar expression patterns across tissues: they are enriched in the female spermatheca, head, heart, adult carcass and fat body (FlyAtlas, [27]), and are all probably secreted into the haemolymph [48]. Drs also shows sex-biased fat body-specific expression [49]. Fat body in the head is involved in expression of many genes that mediate sexual differentiation [50], as well as male responses to mating [51]. Immunity genes are differentially expressed in females when mating [8,10,43,52,53] and nearly all the immunity genes identified in our study are upregulated in mated females [10]. McGraw et al. [43] demonstrated induced expression of TotM and Att-C by male sperm and Att-A by Acps and in particular sex peptide (see also [52,54]). Interestingly, a recent study identified increased expression in TotC and -A in the brains of mated females [55]. Our results suggest that some of these changes, including increased expression of many Turandot genes, begin before copulation. However, other mating-induced immunity genes show decreased expression in response to courtship stimulation. Att-A and -C, as well as DptB are upregulated in the female abdomen after mating but not in the head tissues [54]. It is therefore possible that their downregulation in the head prior to mating represents a location shift in transcriptional activity and resource allocation. Several non-exclusive explanations have been suggested for the mating-related immune response in females [8,43,52,53], including protection from septic injury [56] and antagonistic male molecules [10,54]. Both TotM and TotC are among the fastest evolving immunity genes between D. melanogaster and D. simulans [57], and also their regulation has diverged between females of the two species [58]. Perhaps, the asymmetrical selection that arises from sexual conflict over components of female fitness may have contributed to the sexual dimorphism in the expression patterns of these genes, as well as their divergence between the species.
Previous studies on mated females have suggested that many proteins required for reproduction may be produced during pre-mating reproductive maturation [43,59]. Here, we have identified similar transcriptional changes in response to song as are seen in post-mated females, including Turandot and other immunity and olfactory genes. Another intriguing gene showing increased expression in response to song stimulation is Glucose dehydrogenase (Gld; table 1), which codes for a protein that facilitates sperm storage in mated females [60]. Our findings thus suggest that the transcription changes thought to occur in response to mating may begin during courtship and may represent an adaptive preparation for mating, including anticipation of sexually antagonistic post-mating interactions with male molecules or increased risk of pathogen infection. Indeed, our findings support a recent suggestion that increased immunity prior to mating may be a common female strategy in insects [56]. That some expression changes depend upon the species-specific nature of song could result from a more stimulatory effect of conspecific song or, perhaps more intriguingly, an influence of a female ‘decision’ to mate during courtship. The connection between mate recognition and the downstream effects makes the molecules involved a powerful target for studies of evolutionary divergence, and provide a starting point for characterizing the genetic pathways activated during courtship stimulation and how they are linked with the adaptive responses to mating, which will provide insights into key evolutionary processes ranging from species recognition, sexual selection and conflict to speciation.
Acknowledgements
We thank Anneli Hoikkala, Rhonda Snook, Arild Husby and two anonymous reviewers for their helpful comments on the manuscript, Lisa Olohan (FlyChip) for carrying out the microarray hybridization, Jeff Graves, Amit Arora and Stephen Goodwin for advice. This work was funded by the Marie Curie Initial Training Network, ‘Understanding the evolutionary origin of biological diversity' (ITN-2008-213780 SPECIATION). We thank all participants in the network for the inspiring and helpful discussions.
References
- 1.Gentner T. Q., Hulse S. H., Duffy D., Ball G. F. 2001. Response biases in auditory forebrain regions of female songbirds following exposure to sexually relevant variation in male song. J. Neurobiol. 46, 48–58 (doi:10.1002/1097-4695(200101)46:1<48::AID-NEU5>3.0.CO;2-3) [DOI] [PubMed] [Google Scholar]
- 2.Hoke K. L., Ryan M. J., Wilczynski W. 2005. Social cues shift functional connectivity in the hypothalamus. Proc. Natl Acad. Sci. USA 102, 10 712–10 717 10.1073/pnas.0502361102 (doi:10.1073/pnas.0502361102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sockman K. W., Gentner T. Q., Ball G. F. 2002. Recent experience modulates forebrain gene-expression in response to mate-choice cues in European starlings. Proc. R. Soc. Lond. B 269, 2479–2485 10.1098/rspb.2002.2180 (doi:10.1098/rspb.2002.2180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murthy M. 2010. Unraveling the auditory system of Drosophila. Curr. Opin. Neurobiol. 20, 281–287 10.1016/j.conb.2010.02.016 (doi:10.1016/j.conb.2010.02.016) [DOI] [PubMed] [Google Scholar]
- 5.Ellis L. L., Carney G. E. 2011. Socially-responsive gene expression in male Drosophila melanogaster is influenced by the sex of the interacting partner. Genetics 187, 157–169 10.1534/genetics.110.122754 (doi:10.1534/genetics.110.122754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carney G. E. 2007. A rapid genome-wide response to Drosophila melanogaster social interactions. BMC Genomics 8, 288. 10.1186/1471-2164-8-288 (doi:10.1186/1471-2164-8-288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis L. L., Carney G. E. 2009. Drosophila melanogaster males respond differently at the behavioural and genome-wide levels to Drosophila melanogaster and Drosophila simulans females. J. Evol. Biol. 22, 2183–2191 10.1111/j.1420-9101.2009.01834.x (doi:10.1111/j.1420-9101.2009.01834.x) [DOI] [PubMed] [Google Scholar]
- 8.Lawniczak M. K. N., Begun D. J. 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47, 900–910 10.1139/g04-050 (doi:10.1139/g04-050) [DOI] [PubMed] [Google Scholar]
- 9.Cummings M. E., Larkins-Ford J., Reilly C. R. L., Wong R. Y., Ramsey M., Hofmann H. A. 2008. Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc. R. Soc. B 275, 393–402 10.1098/rspb.2007.1454 (doi:10.1098/rspb.2007.1454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Innocenti P., Morrow E. H. 2009. Immunogenic males: a genome-wide analysis of reproduction and the cost of mating in Drosophila melanogaster females. J. Evol. Biol. 22, 964–973 10.1111/j.1420-9101.2009.01708.x (doi:10.1111/j.1420-9101.2009.01708.x) [DOI] [PubMed] [Google Scholar]
- 11.Wolfner M. F. 2009. Battle and ballet: molecular interactions between the sexes in Drosophila. J. Hered. 100, 399–410 10.1093/jhered/esp013 (doi:10.1093/jhered/esp013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenspan R. J., Ferveur J. F. 2000. Courtship in Drosophila. Annu. Rev. Genet. 34, 205–232 10.1146/annurev.genet.34.1.205 (doi:10.1146/annurev.genet.34.1.205) [DOI] [PubMed] [Google Scholar]
- 13.Rybak F., Sureau G., Aubin T. 2002. Functional coupling of acoustic and chemical signals in the courtship behaviour of the male Drosophila melanogaster. Proc. R. Soc. Lond. B 269, 695–701 10.1098/rspb.2001.1919 (doi:10.1098/rspb.2001.1919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gopfert M. C., Robert D. 2002. The mechanical basis of Drosophila audition. J. Exp. Biol. 205, 1199–1208 [DOI] [PubMed] [Google Scholar]
- 15.Gopfert M. C., Robert D. 2003. Motion generation by Drosophila mechanosensory neurons. Proc. Natl Acad. Sci. USA 100, 5514–5519 10.1073/pnas.0737564100 (doi:10.1073/pnas.0737564100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing A. W., Bennett-Clark H. C. 1968. The courtship songs of Drosophila. Behavior 31, 288–301 10.1163/156853968X00298 (doi:10.1163/156853968X00298) [DOI] [Google Scholar]
- 17.Kyriacou C. P., Hall J. C. 1980. Circadian-rhythm mutations in Drosophila melanogaster affect short-term fluctuations in the males courtship song. Proc. Natl Acad. Sci. USA 77, 6729–6733 10.1073/pnas.77.11.6729 (doi:10.1073/pnas.77.11.6729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyriacou C. P., Hall J. C. 1982. The function of courtship song rhythms in Drosophila. Anim. Behav. 30, 794–801 10.1016/S0003-3472(82)80152-8 (doi:10.1016/S0003-3472(82)80152-8) [DOI] [PubMed] [Google Scholar]
- 19.Kyriacou C. P., Hall J. C. 1986. Interspecific genetic control of courtship song production and reception in Drosophila. Science 232, 494–497 10.1126/science.3083506 (doi:10.1126/science.3083506) [DOI] [PubMed] [Google Scholar]
- 20.Kyriacou C. P., Vandenberg M. J., Hall J. C. 1990. Drosophila courtship song cycles in normal and period mutant males revisited. Behav. Genet. 20, 617–644 10.1007/BF01065875 (doi:10.1007/BF01065875) [DOI] [PubMed] [Google Scholar]
- 21.Bennet-Clark H. C., Ewing A. 1969. Pulse interval as a critical parameter in the courtship song of Drosophila melanogaster. Anim. Behav. 17, 755–759 10.1016/S0003-3472(69)80023-0 (doi:10.1016/S0003-3472(69)80023-0) [DOI] [Google Scholar]
- 22.Ritchie M. G., Halsey E. J., Gleason J. M. 1999. Drosophila song as a species-specific mating signal and the behavioural importance of Kyriacou & Hall cycles in D. melanogaster song. Anim. Behav. 58, 649–657 10.1006/anbe.1999.1167 (doi:10.1006/anbe.1999.1167) [DOI] [PubMed] [Google Scholar]
- 23.Kamikouchi A., Inagaki H. K., Effertz T., Hendrich O., Fiala A., Göpfert M. C., Ito K. 2009. The neural basis of Drosophila gravity-sensing and hearing. Nature 458, 165–172 10.1038/nature07810 (doi:10.1038/nature07810) [DOI] [PubMed] [Google Scholar]
- 24.Eberl D. F., Duyk G. M., Perrimon N. 1997. A genetic screen for mutations that disrupt an auditory response in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 94, 14 837–14 842 10.1073/pnas.94.26.14837 (doi:10.1073/pnas.94.26.14837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Z., et al. 2004. Two interdependent TRPV channel subunits, inactive and Nanchung, mediate hearing in Drosophila. J. Neurosci. 24, 9059–9066 10.1523/JNEUROSCI.1645-04.2004 (doi:10.1523/JNEUROSCI.1645-04.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Development Core Team 2007. R: a language and environment for statistical computing, 2.7.2 edn Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 27.Chintapalli V. R., Wang J., Dow J. A. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 10.1038/ng2049 (doi:10.1038/ng2049) [DOI] [PubMed] [Google Scholar]
- 28.Gentleman R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. 10.1186/gb-2004-5-10-r80 (doi:10.1186/gb-2004-5-10-r80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1–26 10.2202/1544-6115.1027 (doi:10.2202/1544-6115.1027) [DOI] [PubMed] [Google Scholar]
- 30.Smyth G. K. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (eds Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W.), pp. 397–420 New York, NY: Springer [Google Scholar]
- 31.Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 32.Dennis G. J., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. 10.1186/gb-2003-4-5-p3 (doi:10.1186/gb-2003-4-5-p3) [DOI] [PubMed] [Google Scholar]
- 33.Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 (doi:10.1038/nprot.2008.211) [DOI] [PubMed] [Google Scholar]
- 34.Mootha V. K., et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 10.1038/ng1180 (doi:10.1038/ng1180) [DOI] [PubMed] [Google Scholar]
- 35.Falcon S., Gentleman R. 2008. Hypergeometric testing used for gene set enrichment analysis. In Bioconducter case studies (eds Hahne F., Huber W., Gentleman R., Falcon S.), pp. 207–220 New York, NY: Springer Science+Business Media [Google Scholar]
- 36.Eberl D. F. 1999. Feeling the vibes: chordotonal mechanisms in insect hearing. Curr. Opin. Neurobiol. 9, 389–393 10.1016/S0959-4388(99)80058-0 (doi:10.1016/S0959-4388(99)80058-0) [DOI] [PubMed] [Google Scholar]
- 37.Riabinina O., Dai M., Duke T., Albert J. T. 2011. Active process mediates species-specific tuning of Drosophila ears. Curr. Biol. 21, 658–664 10.1016/j.cub.2011.03.001 (doi:10.1016/j.cub.2011.03.001) [DOI] [PubMed] [Google Scholar]
- 38.Bahn J. H., Lee G., Park J. H. 2009. Comparative analysis of Pdf-mediated circadian behaviors between Drosophila melanogaster and D. virilis. Genetics 181, 965–975 10.1534/genetics.108.099069 (doi:10.1534/genetics.108.099069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackay T. F., Heinsohn S. L., Lyman R. F., Moehring A. J., Morgan T. J., Rollmann S. M. 2005. Genetics and genomics of Drosophila mating behavior. Proc. Natl Acad. Sci. USA 102(Suppl. 1), 6622–6629 10.1073/pnas.0501986102 (doi:10.1073/pnas.0501986102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Bretz C. A., Hawksworth S. A., Hirsh J., Johnson E. C. 2010. Corazonin neurons function in sexually dimorphic circuitry that shape behavioral responses to stress in Drosophila. PLoS ONE 5, e9141. 10.1371/journal.pone.0009141 (doi:10.1371/journal.pone.0009141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neckameyer W. S. 1998. Dopamine modulates female sexual receptivity in Drosophila melanogaster. J. Neurogenet. 12, 101–114 10.3109/01677069809167259 (doi:10.3109/01677069809167259) [DOI] [PubMed] [Google Scholar]
- 42.Andretic R., Van Swinderen B., Greenspan R. J. 2005. Dopaminergic modulation of arousal in Drosophila. Curr. Biol. 15, 1165–1175 10.1016/j.cub.2005.05.025 (doi:10.1016/j.cub.2005.05.025) [DOI] [PubMed] [Google Scholar]
- 43.McGraw L. A., Gibson G., Clark A. G., Wolfner M. F. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14, 1509–1514 10.1016/j.cub.2004.08.028 (doi:10.1016/j.cub.2004.08.028) [DOI] [PubMed] [Google Scholar]
- 44.Kaupp U. B. 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat. Rev. Neurosci. 11, 188–200 10.1038/nrn2789 (doi:10.1038/nrn2789) [DOI] [PubMed] [Google Scholar]
- 45.Bailey N. W., Gray B., Zuk M. 2011. Exposure to sexual signals during rearing increases immune defence in adult field crickets. Biol. Lett. 7, 217–220 10.1098/rsbl.2010.0659 (doi:10.1098/rsbl.2010.0659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldman T. D., Arbeitman M. N. 2007. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3, 2278–2295 10.1371/journal.pgen.0030216 (doi:10.1371/journal.pgen.0030216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rideout E. J., Dornan A. J., Neville M. C., Eadie S., Goodwin S. F. 2010. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13, 458–466 10.1038/nn.2515 (doi:10.1038/nn.2515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ekengren S., Hultmark D. 2001. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem. Biophys. Res. Commun. 284, 998–1003 10.1006/bbrc.2001.5067 (doi:10.1006/bbrc.2001.5067) [DOI] [PubMed] [Google Scholar]
- 49.Parisi M., et al. 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5, R40. 10.1186/gb-2004-5-6-r40 (doi:10.1186/gb-2004-5-6-r40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujii S., Amrein H. 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO. J. 21, 5353–5363 10.1093/emboj/cdf556 (doi:10.1093/emboj/cdf556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellis L. L., Carney G. E. 2010. Mating alters gene expression patterns in Drosophila melanogaster male heads. BMC Genomics 11, 558. 10.1186/1471-2164-11-558 (doi:10.1186/1471-2164-11-558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng J., Zipperlen P., Kubli E. 2005. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 15, 1690–1694 10.1016/j.cub.2005.08.048 (doi:10.1016/j.cub.2005.08.048) [DOI] [PubMed] [Google Scholar]
- 53.McGraw L. A., Clark A. G., Wolfner M. F. 2008. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics 179, 1395–1408 10.1534/genetics.108.086934 (doi:10.1534/genetics.108.086934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Domanitskaya E. V., Liu H., Chen S., Kubli E. 2007. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 274, 5659–5668 10.1111/j.1742-4658.2007.06088.x (doi:10.1111/j.1742-4658.2007.06088.x) [DOI] [PubMed] [Google Scholar]
- 55.Dalton J. E., Kacheria T. S., Knott S. R., Lebo M. S., Nishitani A., Sanders L. E., Stirling E. J., Winbush A., Arbeitman M. N. 2010. Dynamic, mating-induced gene expression changes in female head and brain tissues of Drosophila melanogaster. BMC Genomics 11(Suppl. 1), 541. 10.1186/1471-2164-11-541 (doi:10.1186/1471-2164-11-541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siva-Jothy M. T. 2009. Reproductive immunity. In Insect infection and immunity: evolution, ecology and mechanisms (eds Rolff J., Reynolds S.), pp. 241–250 Oxford, UK: Oxford University Press [Google Scholar]
- 57.Obbard D. J., Welch J. J., Kim K. W., Jiggins F. M. 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5, e1000698. 10.1371/journal.pgen.1000698 (doi:10.1371/journal.pgen.1000698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graze R. M., McIntyre L. M., Main B. J., Wayne M. L., Nuzhdin S. V. 2009. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics 183, 547–561 10.1534/genetics.109.105957 (doi:10.1534/genetics.109.105957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mack P. D., Kapelnikov A., Heifetz Y., Bender M. 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 10 358–10 363 10.1073/pnas.0604046103 (doi:10.1073/pnas.0604046103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iida K., Cavener D. R. 2004. Glucose dehydrogenase is required for normal sperm storage and utilization in female Drosophila melanogaster. J. Exp. Biol. 207, 675–681 10.1242/jeb.00816 (doi:10.1242/jeb.00816) [DOI] [PubMed] [Google Scholar]