Abstract
Estimates of hybrid fitness have been used as either a platform for testing the potential role of natural hybridization in the evolution of species and species complexes or, alternatively, as a rationale for dismissing hybridization events as being of any evolutionary significance. From the time of Darwin's publication of The Origin, through the neo-Darwinian synthesis, to the present day, the observation of variability in hybrid fitness has remained a challenge for some models of speciation. Yet, Darwin and others have reported the elevated fitness of hybrid genotypes under certain environmental conditions. In modern scientific terminology, this observation reflects the fact that hybrid genotypes can demonstrate genotype × environment interactions. In the current review, we illustrate the development of one plant species complex, namely the Louisiana Irises, into a ‘model system' for investigating hybrid fitness and the role of genetic exchange in adaptive evolution and diversification. In particular, we will argue that a multitude of approaches, involving both experimental and natural environments, and incorporating both manipulative analyses and surveys of natural populations, are necessary to adequately test for the evolutionary significance of introgressive hybridization. An appreciation of the variability of hybrid fitness leads to the conclusion that certain genetic signatures reflect adaptive evolution. Furthermore, tests of the frequency of allopatric versus sympatric/parapatric divergence (that is, divergence with ongoing gene flow) support hybrid genotypes as a mechanism of evolutionary diversification in numerous species complexes.
Keywords: natural hybridization, habitat selection, hybrid fitness
Introduction
‘Considering the several rules now given, which govern the fertility of first crosses and of hybrids, we see that when forms, which must be considered as good and distinct species, are united, their fertility graduates from zero to perfect fertility, or even to fertility under certain conditions to excess. That their fertility, besides being eminently susceptible to favorable and unfavorable conditions, is innately variable…it is by no means always the same in degree in the first cross and in the hybrids produced from this cross' (Darwin, 1859).
The above quote demonstrates that Darwin observed variation in fertility within and among species and considered its importance for understanding evolutionary processes. Notwithstanding this clarity, Darwin indicated throughout The Origin his concept that reticulate evolution had a relatively unimportant role during descent with modification (for example, see the only figure included in this book). Yet, his description of a wide range of hybrid fitness, and the influence of interactions between individuals in response to a given environment in determining this range, begs an alternative conclusion. Thus, relatively fit hybrids, like other ‘more fit' genotypes might be expected to contribute significantly to adaptation and diversification.
Since Darwin's time, many researchers have reported on the relative fitness of hybrids and have found that in many cases, hybrids are as fit, or fitter than their pure species counterparts (see Arnold and Hodges (1995); Arnold (2006, 2008) and Arnold and Martin (2010) for reviews). Many have debated what these estimates might mean with regard to the impact of genetic exchange on evolutionary pattern and process (Anderson, 1949; Anderson and Stebbins, 1954; Mayr, 1963; Lewontin and Birch, 1966; Grant, 1981; Barton and Hewitt, 1985; Arnold, 1992, 1997, 2006, 2008; Grant and Grant, 1992, 2010; Dowling and DeMarais, 1993; Rieseberg, 1997; Rieseberg et al., 2003; Seehausen, 2004; Seehausen et al., 2008; Anderson et al., 2009). Significantly, there are now numerous studies that have inferred a causal link between natural hybridization and adaptive evolution and/or diversification (Grant and Grant, 2002; Salzburger et al., 2002; Rieseberg et al., 2003; Seehausen, 2004; Schwarz et al., 2005; Martin et al., 2006; Anderson et al., 2009; Johnson et al., 2010).
Many examples of diversification and adaptation as the result of reticulate evolution have come to light, leading to the formation of several related hypotheses. These include: (1) hybrid genotypes with high relative fitness are produced more often than previously assumed (Arnold and Hodges, 1995); (2) even hybrids with low relative fitness may act as a conduit for genetic exchange, given that they have some level of fertility (Arnold and Hodges, 1995; Arnold, 1997, 2006); and (3) for many taxonomic groups, allopatric divergence is not as frequent as divergence accompanied by some level of at least intermittent gene flow (that is, parapatric or sympatric divergence; Pinho and Hey (2010)). Numerous authors including Darwin himself—see above quote—contributed to the formulation of the first hypothesis (relating to the production of relatively fit hybrid genotypes). Since Darwin, numerous reviews and books have addressed the concept of hybrid fitness, with some addressing the relative importance or unimportance of hybrid genotypes in the evolutionary and ecological trajectories of plant and animal lineages (Dobzhansky, 1937; Mayr, 1963; Endler, 1977; Moore, 1977; Barton and Hewitt, 1985; Howard, 1993; Arnold and Hodges, 1995; Arnold, 1997, 2006, 2008; Arnold and Martin, 2010; Grant and Grant, 2010). The data available for these reviews (particularly those deriving from genomic information) have multiplied exponentially over the past decade due largely to the advances in methodologies for collecting and analyzing DNA sequences (Green et al., 2010) and for deciphering the function of the underlying genes (Rosas et al., 2010). However, the species complexes that have provided the most rigorous and detailed descriptions of hybrid fitness have done so because of the availability of both genetic and ecological information (Grant and Grant, 1992, 2010; Rieseberg et al., 2003; Martin et al., 2005, 2006; Arnold et al., 2010; and see Arnold and Martin (2010) for a review). These various catalogs of hybrid fitness estimates have demonstrated that ‘all hybrids are not created equal' in terms of their fitness. As Darwin recognized, hybrid genotypes can demonstrate lower, equivalent or higher fitness relative to progenitor and other hybrid genotypes, and this variation can reflect genotype × environment interactions.
The observation of relatively high fitness of some hybrid genotypes suggests one causal factor by which both parapatric and sympatric divergence are accompanied by introgression. In particular, as lineages diverge, some genetic exchange can be the result of selection favoring the incorporation of foreign alleles, and thus the formation of admixed genotypes (Grant and Grant, 2002; Rieseberg et al., 2003; Martin et al., 2006). However, as suggested by hypothesis 2, this selective incorporation can be accomplished through relatively unfit hybrid genotypes as well. This is possible because a hybrid with lower overall fitness (for example, one that has low, but not zero, fertility) by definition produces gametes that are viable and thus able to contribute to the next hybrid generation (Arnold and Hodges, 1995). This subsequent generation may or may not have elevated fitness relative to the previous, low-fitness genotype, but it too can contribute to further generations if its fitness is above zero. Examples of the action of this process, by which hybrids with extremely low fitness have a significant role in adaptive evolution and lineage formation, include such diverse clades as sunflowers, Australian grasshoppers and Drosophila (see Arnold (1997, 2006) for reviews of the studies of these and additional organisms).
The third hypothesis derives from the recognition that a majority of plant lineages have allopolyploid ancestry (that is, polyploid lineages formed through hybridization; Arnold (1997, 2006)), as well as from the proliferation of studies (mainly involving animal complexes) that have detected past gene flow during speciation through the application of various algorithms (Wakeley and Hey, 1997; Templeton, 2001; Geneva and Garrigan, 2010; Wang and Hey, 2010) to genomic data sets (Salzburger et al., 2002; Seehausen, 2004; Won and Hey, 2005; Anderson et al., 2009; Green et al., 2010; Pinho and Hey, 2010; see also Arnold (2006, 2008) for a review of additional examples). Less frequently referred to are the literally thousands of studies documenting genetic exchange within groups of organisms as diverse as viruses, prokaryotes, plants and animals (see Grant and Grant (1992); Arnold (1997, 2006, 2008); Rieseberg (1997) and Seehausen (2004) for references to some of these studies). In the case of plants and animals, this exchange often takes the form of introgressive hybridization (or introgression—that is, the exchange of genes between lineages through the formation of an initial F1 hybrid generation, followed by backcrossing of F1 to individuals belonging to one or both of the parental lineages; Anderson and Hubricht (1938)).
In the current review, we will highlight how each of the above hypotheses has been tested during our own work on the plant group known as the Louisiana Irises. In particular, we wish to emphasize the development of this plant species complex into a model for identifying some of the genetic and ecological components that affect hybrid fitness. This definition of components affecting the fitness of Louisiana Iris hybrid genotypes has allowed us to make comparisons with other reticulate complexes and thus to test whether different hybrid genotypes vary in fitness across environments and whether hybrid genotypes with either elevated or lowered fitness have contributed to genetic exchange during the diversification of lineages and species complexes.
Hybrid fitness varies: testing a truism?
One of the hallmarks of research into genetic exchange during evolutionary diversification has been the assumption of either creative or destructive consequences. Within the creative paradigm, hybridization (or lateral exchange or viral recombination) is ascribed the potential role of a catalyst for the formation of novel lineages and/or adaptations. For example, Rieseberg et al. (2003) and Seehausen (2004) inferred a primary causality for hybridization between divergent lineages of sunflowers and cichlid fish, respectively, to the origin of novel adaptations and adaptive radiations. Similarly, Grant and Grant (2008, 2010) have documented the adaptive evolutionary impact of low levels of gene flow between various Darwin finch species.
In contrast to the concept of genetic exchange as a novelty-generating process, is the perception of hybridization and lateral exchange as disruptive of the natural order of diversification. Classically, Mayr (1963) stated, ‘The total weight of the available evidence contradicts the assumption that hybridization plays a major evolutionary role among higher animals.' Similarly, Excoffier et al. (2009) have posited the overwhelming influence of neutral (that is, demographic) processes for most introgression events associated with range expansion. In this regard, they concluded, ‘Hence, when massive introgression of genes into an invading species is observed, it appears neither necessary nor parsimonious to invoke mechanisms such as selection.' Thus, they argued for their conceptual framework to be considered a null model for testing hypotheses regarding other factors (Petit and Excoffier, 2009). Importantly for the current review, these authors argued that their approach would allow an estimate of which (if any) genetic elements were introgressing, or were prevented from introgressing, because of natural selection (Excoffier et al., 2009; Petit and Excoffier, 2009).
Louisiana Irises: defining hybrid fitness
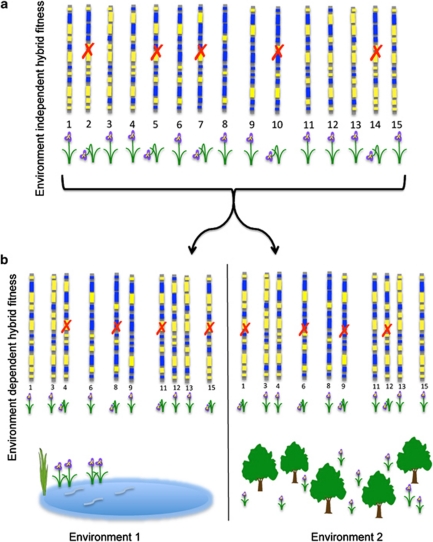
If research on Louisiana Irises has revealed anything about hybrid fitness, it is that fitness varies among genotypes and across habitats (Figure 1). In particular, some hybrids will suffer from reduced fitness regardless of the environment (Figure 1a), whereas other individuals may thrive in one particular habitat but not in another (Figure 1b). For this reason, it is important to evaluate hybrid fitness in the context of well-defined hybrid genotypes or genotypic classes. For example, Cruzan and Arnold (1994) detected differential selection at early life-history stages (that is, seed viability) that contributed to the presence/absence of different hybrid genotypes in hybrid zones. Similarly, Johnston et al. (2001a, 2001b, 2003, 2004) and Burke et al. (1998a, 1998b) , in a series of greenhouse and field studies, provided evidence that different environmental components were causal in the varying fitness of parental and hybrid genotypic classes (for example, F1, B1, F2 and natural hybrids. Similarly, Emms and Arnold (1997) detected both consistently high F1 hybrid fitness and varying fitness of parental genotypes across natural habitats. Finally, Cornman et al. (2004) detected patterns indicating that natural selection favored the production of hybrid seeds produced by mating between similar genotypes.
Figure 1.
Hybrid fitness varies by genotype and by environment. (a) Individuals (represented by linkage groups; the different colored regions reflect genetic material from hybridizing species) may suffer from reduced fitness as the result of genetic incompatibilities regardless of the environment. These hybrid genotypes are then lost from the population (indicated by a red ‘X'). (b) The fitness of the remaining individuals can then be tested in different environmental conditions (such as wet and sunny versus dry and shady).
The studies of fitness described above provided tests of the general hypothesis that hybrid fitness was uniformly low (Mayr, 1963; Barton and Hewitt, 1985), but also found that individual fitness varied by genotype and environment. As with our own earlier reviews (Arnold, 1992; Arnold and Hodges, 1995) and likewise some recent reviews of the literature (Coyne and Orr, 2004), reported estimates of hybrid fitness generally reflect mean values across classes rather than for individual genotypes. Although such data are useful for testing some hypotheses, for example, the ‘strength' of certain types of barriers to reproduction (Sobel et al., 2009), they are limited in their descriptive or predictive value for estimates of hybrid fitness or, for that matter, the extent to which speciation occurs with intermittent gene flow (see below).
The previous conclusion can be exemplified by comparison with the method for inferring the fitness of ‘non-hybrids'. In particular, the fitness of the latter is always placed in the context of individual genotypes rather than classes. The history of estimates of hybrid fitness being given in the context of pooled genotypes seems to reflect an underlying assumption that all hybrids are less fit and thus their fitness can be reflected accurately by a mean value. Therefore, tests of hypotheses regarding the fitness of hybrid genotypes require both the definition of specific genotypes and the incorporation of environmental contributions to fitness. Furthermore, for such tests to be most effective, the genotypes must be defined with sufficient detail to allow the description of different genomic regions associated with differential fitness estimates (Figure 1b). This is required if we are to be able to make and test predictions regarding which genomic elements are likely to contribute to reproductive isolation, adaptive trait introgression and/or hybrid speciation (Arnold and Hodges, 1995; Rieseberg, 1997; Rieseberg et al., 2003; Seehausen, 2004; Arnold, 2006). However, it is important to emphasize that hybrid fitness is almost certainly affected by genetic interactions. Furthermore, what is meant by a specific ‘hybrid genotype' will vary at different loci from a heterozygous condition to homozygosity of introgressed alleles. Once again, this reflects the need to derive fitness estimates from well-defined genotypes, rather than from genotypic classes.
With regard to Louisiana Irises, several studies that defined genotypic variation have paved the way for inferences regarding hybrid fitness and its possible relationship with adaptive evolution and lineage diversification (Bouck et al., 2005, 2007; Martin et al., 2005, 2006, 2008; Taylor et al., 2009; Tang et al., 2010). Each of these studies involved the application of linkage and/or quantitative trait loci (QTL) mapping methodologies and thereby allowed inferences regarding the number and position of genes contributing to the fitness of hybrids. Bouck et al. (2005, 2007) began these analyses, producing the first genetic maps for two members of this species complex, namely Iris fulva and Iris brevicaulis. From these analyses, the genetic architecture of both segregation distortion (reflecting the permeability of the two species' genomes to introgression) and floral phenotypes was determined. Inferences drawn from these studies suggested that genetic architecture and selection were likely to allow widespread introgression of genes between these two species, but at the same time impede the introgression of some phenotypic traits due to genetic linkage (Bouck et al., 2005, 2007). For example, in a backcross population of I. fulva and I. brevicaulis, alleles from I. fulva were significantly favored in 12 genomic regions compared with just 5 regions from I. brevicaulis (Tang et al., 2010). These findings reflect not only the detection of reduced fitness for some recombinant genotypes but also elevated fitness for others. These data were also transformative for understanding the potential evolutionary significance of natural hybridization for the Louisiana Iris species in that they opened a window from which to view microevolutionary processes at individual loci and QTL (Bouck et al., 2005, 2007). The power of such genomic information for deciphering the evolutionary role of admixture between divergent lineages has been well illustrated by studies of organisms as diverse as mice, influenza, sunflowers and yeast (Payseur et al., 2004; Yatabe et al., 2007; Novo et al., 2009; Vijaykrishna et al., 2010). Similarly, linkage and QTL mapping studies of Louisiana Irises provided the groundwork for making predictions regarding the role of selection and genetic architecture on introgression, adaptation and diversification.
Introgression and adaptation: transferring genetic material can provide adaptive benefits to the recipient
A longstanding hypothesis regarding introgression is that some genetic transfer events can provide the basis for adaptive evolution in the introgressed lineage. For example, Anderson (1949), in his book Introgressive Hybridization, reflected this hypothesis in the following passage: ‘There are some circumstantial data suggesting that introgression may be one of the main sources of that variability which provides the raw material for evolution'. Although there are still relatively few studies that have rigorously tested the hypothesis of adaptive trait transfer through introgression, we are past the stage of having only ‘circumstantial data'. In particular, Anderson and Hubricht (1938) and Anderson (1949) pointed to a lack of discrete genetic markers required to define specific hybrid genotypes as the starting point for testing for adaptive trait introgression.
We are no longer faced with a dearth of discrete genetic information. For example, the complete genome sequence for domestic dogs allowed a recent description of adaptive trait introgression involving genes that control coat color. Alleles from domestic dogs, which caused darker pelage, have apparently introgressed into North American wolf populations (Anderson et al., 2009). The pattern of genetic variation detected in wolf populations falsified the hypothesis of neutral diffusion of coat color alleles from dogs, and instead, supported the hypothesis that the introgression involved positive selection for the resulting darker coats in the hybrid wolves (Anderson et al., 2009). Similarly, Fitzpatrick et al. (2009) used single-nucleotide polymorphisms in a scan of the genomes of hybrid populations formed between native and introduced species of salamanders in North America (Ambystoma californiense and Ambystoma mavortium, respectively). From this survey, they detected the fixation of alleles from the introduced taxon in the native species at a handful of loci, but low levels of introgression at the majority of sampled markers. Like the canid data, this observation was used as the basis for falsifying a hypothesis that posits neutrality in the transfer of alleles between hybridizing lineages (Fitzpatrick et al., 2009). A final example of apparent adaptive trait introgression comes from work on wild species of annual sunflowers. Whitney et al. (2006, 2010) estimated the affects from both biotic and abiotic factors on the fitness of Helianthus annuus annuus, Helianthus debilis and the hybrid subspecies, Helianthus annuus texanus, formed by introgressive hybridization between H. a. annuus and H. debilis (Heiser, 1951). By placing both parental taxa, their natural introgressed hybrid and experimental first-generation backcross hybrids into natural settings, Whitney et al. (2006, 2010) demonstrated that introgression from H. debilis into H. a. annuus had resulted in the transfer of adaptations from the former into the latter, thus facilitating the spread of H. annuus (in the form of the new hybrid lineage, H. a. texanus) into novel habitats in eastern Texas. By combining an experimental ecological approach with genomic information, it was thus possible to not only test for adaptive affects from introgression but also to identify specific genotype × environment associations that were (at least partially) the foci of positive selection (Whitney et al., 2006, 2010).
Louisiana Irises and evidence for adaptive trait transfer
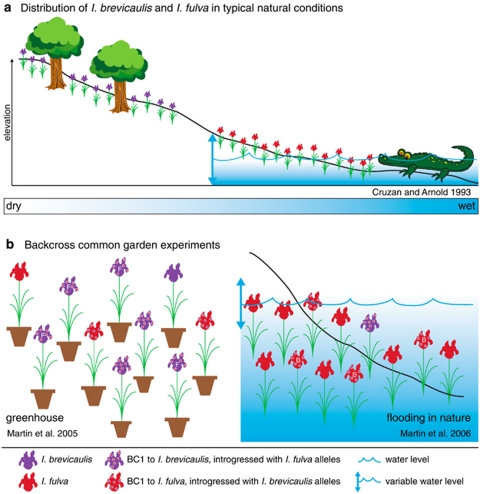
A series of recent genomic and ecological analyses of Louisiana Iris species have also tested the hypothesis that introgression in nature is due to neutral diffusion. Although often found in sympatry near bayous and waterways, I. fulva and I. brevicaulis nonetheless exhibit habitat partitioning. I. fulva typically grows at lower elevations along the bayou edges, where rhizomes are often submerged in water, whereas I. brevicaulis generally occurs at slightly higher elevations in mixed hardwood forest (Figure 2a) (Viosca, 1935; Cruzan and Arnold, 1993; Johnston et al., 2001b). Martin et al. (2005, 2007) have tested for QTL underlying survivorship in different habitats. These analyses involved I. fulva × I. brevicaulis first-generation backcross (that is, BC1) populations. Analyses across years and habitats were possible because Louisiana Irises are long lived and can be replicated clonally by subdividing rhizomes of a given genotype. The first two studies described below were ‘accidental' in that habitats into which the genotypes were transplanted were revealed subsequently to be highly selective, yet still within the range of those found in nature.
Figure 2.
(a) The typical natural distribution of Iris brevicaulis and Iris fulva. I. brevicaulis and I. fulva often occur in sympatry near bayous and waterways in southern Louisiana. Although plants from each species may grow within a meter of one another, elevational gradients along bayou edges and differences in microhabitats provide distinct growth environments for each species. I. brevicaulis is generally found in mixed hardwood forest at slightly higher elevations that do not experience flooding, whereas I. fulva is found at lower elevations in soils that are often submerged in water. (b) Results of experiments on I. brevicaulis and I. fulva backcross individuals. Genetic clones from two backcross populations, one toward I. brevicaulis and one toward I. fulva, were grown in both a greenhouse common garden and in a common garden in a natural Louisiana Iris setting. In the drier greenhouse common garden, backcross genotypes toward I. brevicaulis survived at a higher frequency than did those in the direction of I. fulva. Three QTLs introgressed from I. brevicaulis increased survivorship of I. fulva backcrosses. Interestingly, some alleles from wet-adapted I. fulva increased the fitness of I. brevicaulis backcross genotypes even in these dry conditions. The natural common garden experienced higher water levels than usual, resulting in prolonged flooding across the site. A cline in survivorship was seen with I. fulva parents showing the highest percentage survival (27%), followed by individuals backcrossed to I. fulva (9%), and then individuals backcrossed to I. brevicaulis (5.5%). I. brevicaulis parents had no survivors. Although individuals composed primarily of the dry-adapted I. brevicaulis alleles were generally less fit in the flooded environment, a QTL from I. brevicaulis was associated with higher survival of certain I. fulva BC1 individuals in the flooded conditions.
The first experiment examined the genetic architecture associated with the survivorship of hybrid genotypes in a greenhouse setting (Figure 2b) (Martin et al., 2005). Although well watered in this common garden experiment, BC1 iris genotypes were not placed in standing water as is typical for I. fulva populations. Inferences from this long-term survivorship analysis falsified a neutral model of introgression and strongly supported adaptive trait introgression between these two iris species. First, the backcross genotypes toward the dry-adapted I. brevicaulis survived at a significantly higher frequency than did those in the direction of the wet-adapted, I. fulva. Second, as expected from a model of adaptive trait introgression, introgression of three genomic regions (that is, QTLs) from I. brevicaulis was significantly associated with survivorship in the I. fulva BC1 genotypes.
A second analysis involved the same backcross populations, but in this case, the genotypes were placed into southern Louisiana in regions occupied by native I. fulva, I. brevicaulis and natural hybrids (Viosca, 1935; Arnold, 1993; Johnston et al., 2001b). The microhabitats selected for transplants spanned hardwood forests (I. brevicaulis-like habitats; Viosca, 1935; Cruzan and Arnold, 1993; Johnston et al., 2001b) and bayous (I. fulva-like habitats; Viosca, 1935; Cruzan and Arnold, 1993; Johnston et al., 2001b). Yet, within 1 month, the entire transplant area was inundated with >1m of water that remained for several months (Figure 2b) (Martin et al., 2006). The resulting percentage survivorship reflected a cline among four genotypic classes: I. brevicaulis=0%, backcrosses toward I. brevicaulis=5.5%, backcrosses toward I. fulva=9% and I. fulva=27%. Furthermore, the surviving genotypes of the BC1 generation toward I. brevicaulis contained significantly more alleles from I. fulva than those genotypes from this backcross population that did not survive the flooded conditions. It was concluded that trait introgression had occurred that provided some hybrid genotypes with adaptations to this extreme environment (Martin et al., 2006).
A third analysis by Martin et al., involving pollinator interactions with these same parental and BC1 genotypes, also uncovered evidence for adaptive trait introgression. In this instance, Martin et al. (2008) defined a complex genetic architecture of QTLs that caused certain genotypes to be either attractive or unattractive to three pollinator classes—bumblebees, butterflies and hummingbirds. The patterns suggesting selective consequences from recombination between the I. fulva and I. brevicaulis genomes were similar to those found in both of the above analyses. In particular, I. brevicaulis genotypes were avoided completely by butterflies, and almost completely by hummingbirds (Martin et al., 2008). In contrast, BC1 hybrids towards this species were significantly greater in attractiveness to both of these pollinator classes. It thus appears that alleles from I. fulva, introgressed onto the I. brevicaulis genomic background, provided a selective benefit to the resulting hybrid genotypes.
Overall, the patterns described above reflect apparent signatures of positive selection for certain introgressed alleles underlying adaptations to particular ecological settings. However, there were also data reflecting unpredicted complexity (Table 1). Grant and Grant (2002) reflected this class of inference (in the context of their findings from a 30-year study of Darwin's finches) in the following manner: ‘Hybridization occurred repeatedly though rarely, resulting in elevated phenotypic variances in G. scandens and a change in beak shape. The phenotypic states of both species at the end of the 30-year study could not have been predicted at the beginning.'
Table 1. Results of QTL mapping in Louisiana Irises summarizing expected and unexpected findings in terms of patterns of introgression in different habitats.
| Expected results | Unexpected results | |
|---|---|---|
| Drier greenhouse common garden | Iris brevicaulis BC1 individuals survived at a higher rate than did Iris fulva BC1 individuals. Introgression of three genomic loci from I. brevicaulis into I. fulva BC1 individuals is associated with increased survival | I. fulva alleles increased fitness of I. brevicaulis BC1 individuals. An I. fulva QTL was associated with higher survival in I. fulva BC1 individuals. I. fulva alleles introgressed into I. brevicaulis BC1 individuals at a higher frequency than expected under a neutral model |
| Flooded natural area common garden | I. fulva parents and BC1 individuals survived at a significantly higher rate than did I. brevicaulis parents and BC1 individuals. The I. brevicaulis BC1 individuals who survived had significantly more I. fulva alleles than did those that did not survive | A QTL associated with higher survivorship in I. fulva BC1 individuals is associated with alleles from dry-adapted I. brevicaulis |
Abbreviation: BC1, first-generation backcross.
Although data collected from Louisiana Irises are a mere fraction of that for Darwin's finches, they reflect similar unpredictability. For example, in the relatively dry greenhouse environment, (1) alleles from ‘wet-adapted' I. fulva increased the fitness of introgressed I. brevicaulis genotypes (Martin et al., 2005); (2) I. fulva alleles marked a QTL associated with higher survivorship of I. fulva BC1 genotypes (Martin et al., 2005); and (3) although I. fulva BC1 genotypes died at twice the frequency of BC1 hybrids toward I. brevicaulis, I. fulva alleles introgressed into I. brevicaulis at a significantly greater frequency than expected under a neutral model (Arnold et al., 2010). In the flooded environment (Martin et al., 2006), one of the two QTLs associated with increased survivorship of introgressed I. fulva (that is, BC1 genotypes toward I. fulva) was associated with an allele from ‘dry-adapted' I. brevicaulis. In addition, although varying fitness across environments has been a common observation in each of the studies designed to test for such effects, not all of the analyses have detected genotype × environment interactions (Taylor et al., 2009). Each of these findings suggest that, as for other well-studied species complexes, detecting both predicted and unpredicted phenomena often requires experimental analyses that span many years and numerous habitats, and that incorporate a wide array of approaches from many scientific sub-disciplines.
Divergence with gene flow: testing for non-allopatric diversification
Darwin (1859) summarized his conclusion that new species formed in sympatry or parapatry in the following manner: ‘…I believe that many perfectly defined species have been formed on strictly continuous areas…'. In the ensuing 150 years, this developed into a relatively controversial hypothesis (Mayr, 1963; Via, 2001; Bolnick and Fitzpatrick, 2007; de Aguiar et al., 2009; Cristescu et al., 2010). For example, Coyne and Orr (2004) state that ‘the resurgence of interest in sympatric speciation has produced a deluge of new information…these data have not supported the view that sympatric speciation is frequent in nature, either overall or in specific groups'.
Using genomic information, it is now possible to test for the geographic component of speciation and thus begin to estimate the frequency distribution of those lineages that diverged in complete isolation (that is, allopatry) versus those that shared space and time with related lineages, resulting in gene flow during divergence. For example, Pinho and Hey (2010) used a meta-analysis to estimate the frequency of related lineages that experienced introgressive hybridization during their divergence. Their meta-analysis included studies that used the isolation-with-migration (or IM) models as the basis for estimating population parameters, including gene flow. Although many of the 200+ studies inferred a lack of gene flow, over half of the cases examined detected 2NM estimates of >0.1 (Pinho and Hey, 2010); this result is not consistent with an hypothesis of purely allopatric divergence with no introgression.
Studies using genomic data, but applying methodologies other than IM, have also led to falsification of the allopatric model of divergence for various species complexes. Kulathinal et al. (2009) detected autosomal gene introgression between Drosophila pseudoobscura and Drosophila persimilis. Similarly, both Garrigan et al. (2005) and Green et al. (2010) examined genomic data sets (the latter study involving a comparison of the Homo sapiens and Homo neanderthalenis genome sequences), leading to the inference of introgression between different Homo species. One final exemplar of the process of divergence with gene flow comes from the work of Sturmbauer et al. In this case, genome scans (both nuclear and mitochondrial) and phylogenetic reconstructions inferred multiple bouts of introgression during the radiation of Lake Tanganyika cichlids (Sturmbauer et al., 2010).
Louisiana Irises diversify while introgressing
Two classes of observation reveal the extent to which introgression accompanied the evolutionary diversification of members of the Louisiana Iris species complex. The first has been discussed above in the context of hybrid fitness and adaptive trait introgression. In particular, natural hybrid populations formed between I. fulva, I. brevicaulis and Iris hexagona reflect not only hybridization but also gene transfer between species.
The second observation indicating the cumulative effects from introgression+natural selection is reflected by formation of the homoploid hybrid species, Iris nelsonii. On the basis of morphological and chromosomal characteristics, Randolph (1966) described this species as a three-way hybrid derivative from natural crosses between I. fulva, I. brevicaulis and I. hexagona. Genetic markers from the nuclear and chloroplast genomes supported this inference, with alleles from each of the three species detected in the genome of I. nelsonii (Arnold, 1993). Whether the divergent ecological adaptations or distinctive morphological traits demonstrated by this species, relative to the progenitor taxa, are consequences of its hybrid origin has not yet been tested. However, regardless of whether admixture has caused divergence in the various traits, this new lineage resulted from hybridization and introgression (Randolph, 1966; Arnold, 1993). Thus, as with cichlids, Drosophila, Homo and numerous other species clades, gene flow has continued during, and in the case of I. nelsonii, contributed to, the diversification and divergence of Louisiana Irises.
Conclusions
Gene flow during divergence has, and continues to, impact the Louisiana Iris species complex. Introgression among the various species has provided the material for further evolution in the form of adaptive trait transfer and homoploid hybrid speciation. The foundation of this reticulate evolution rests on hybrid genotypes with varying fitnesses across different ecological settings. Findings from evolutionary, genomic and ecological studies of numerous other species complexes lead to the same conclusions. Thus, it seems that Darwin's observation that different hybrid genotypes could vary in fitness across environments is correct, and that his model of speciation in the face of gene flow has also been supported by many recent findings. Such inferences challenge the utility and ubiquity of models of evolutionary diversification that assume the necessity of hybrid dysfunction or allopatric distributions. At the same time, they provide the impetus for evolutionary biologists interested in the process of speciation to continue to test for the occurrence of, and effects from, divergence with gene flow.
Acknowledgments
We thank Jürgen Gadau for the invitation to contribute this review. The Louisiana Iris research discussed in this review has been supported by numerous National Science Foundation grants. Current NSF funding for this work comes from DEB-0949479/0949424 (collaborative grant with the NH Martin of Texas State University) and DEB-1049757.
The authors declare no conflict of interest.
References
- Anderson E. Introgressive Hybridization. John Wiley and Sons: New York; 1949. [Google Scholar]
- Anderson E, Hubricht L. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am J Bot. 1938;25:396–402. [Google Scholar]
- Anderson E, Stebbins GL., Jr Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML. Natural hybridization as an evolutionary process. Annu Rev Ecol Syst. 1992;23:237–261. [Google Scholar]
- Arnold ML. Iris nelsonii: origin and genetic composition of a homoploid hybrid species. Am J Bot. 1993;80:577–583. doi: 10.1002/j.1537-2197.1993.tb13843.x. [DOI] [PubMed] [Google Scholar]
- Arnold ML. Natural Hybridization and Evolution. Oxford University Press: Oxford; 1997. [Google Scholar]
- Arnold ML. Evolution Through Genetic Exchange. Oxford University Press: Oxford; 2006. [Google Scholar]
- Arnold ML. Reticulate Evolution and Humans. Oxford University Press: Oxford; 2008. [Google Scholar]
- Arnold ML, Hodges SA. Are natural hybrids fit or unfit relative to their parents. Trends Ecol Evol. 1995;10:67–71. doi: 10.1016/S0169-5347(00)88979-X. [DOI] [PubMed] [Google Scholar]
- Arnold ML, Martin NH. Hybrid fitness across time and habitats. Trends Ecol Evol. 2010;25:530–536. doi: 10.1016/j.tree.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Arnold ML, Tang S, Knapp SJ, Martin NH. Asymmetric introgressive hybridization among Louisiana Iris species. Genes. 2010;1:9–22. doi: 10.3390/genes1010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- Bolnick DI, Fitzpatrick BM. Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst. 2007;38:459–487. [Google Scholar]
- Bouck AC, Peeler R, Arnold ML, Wessler SR. Genetic mapping of species boundaries in Louisiana Irises using IRRE retrotransposon display markers. Genetics. 2005;171:1289–1303. doi: 10.1534/genetics.105.044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck AC, Wessler SR, Arnold ML. QTL analysis of floral traits in Louisiana Iris hybrids. Evolution. 2007;61:2308–2319. doi: 10.1111/j.1558-5646.2007.00214.x. [DOI] [PubMed] [Google Scholar]
- Burke JM, Carney SE, Arnold ML. Hybrid fitness in the Louisiana Irises: analysis of parental and F1 performance. Evolution. 1998a;52:37–43. doi: 10.1111/j.1558-5646.1998.tb05136.x. [DOI] [PubMed] [Google Scholar]
- Burke JM, Voss TJ, Arnold ML. Genetic interactions and natural selection in Louisiana Iris hybrids. Evolution. 1998b;52:1304–1310. doi: 10.1111/j.1558-5646.1998.tb02012.x. [DOI] [PubMed] [Google Scholar]
- Cornman RS, Burke JM, Wesselingh RA, Arnold ML. Contrasting genetic structure of adults and progeny in a Louisiana Iris hybrid population. Evolution. 2004;58:2669–2681. doi: 10.1111/j.0014-3820.2004.tb01620.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates Inc.: Sunderland; 2004. [Google Scholar]
- Cristescu ME, Adamowicz SJ, Vaillant JJ, Haffner DG. Ancient lakes revisited: from the ecology to the genetics of speciation. Mol Ecol. 2010;19:4837–4851. doi: 10.1111/j.1365-294X.2010.04832.x. [DOI] [PubMed] [Google Scholar]
- Cruzan MB, Arnold ML. Ecological and genetic associations in an Iris hybrid zone. Evolution. 1993;47:1432–1445. doi: 10.1111/j.1558-5646.1993.tb02165.x. [DOI] [PubMed] [Google Scholar]
- Cruzan MB, Arnold ML. Assortative mating and natural selection in an Iris hybrid zone. Evolution. 1994;48:1946–1958. doi: 10.1111/j.1558-5646.1994.tb02225.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species by Means of Natural Selection or the Preservation of Favored Races in the Struggle for Life. Prometheus Books: Buffalo; 1859. [Google Scholar]
- de Aguiar MAM, Baranger M, Baptestini EM, Kaufman L, Bar-Yam Y. Global patterns of speciation and diversity. Nature. 2009;460:384–387. doi: 10.1038/nature08168. [DOI] [PubMed] [Google Scholar]
- Dobzhansky TH. Genetics and the Origin of Species. Columbia University Press: New York; 1937. [Google Scholar]
- Dowling TE, DeMarais BD. Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature. 1993;362:444–446. [Google Scholar]
- Emms SK, Arnold ML. The effect of habitat on parental and hybrid fitness: reciprocal transplant experiments with Louisiana Irises. Evolution. 1997;51:1112–1119. doi: 10.1111/j.1558-5646.1997.tb03958.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Geographic Variation, Speciation, and Clines. Princeton University Press: Princeton; 1977. [PubMed] [Google Scholar]
- Excoffier L, Foll M, Petit R. Genetic consequences of range expansions. Annu Rev Ecol Evol Syst. 2009;40:481–501. [Google Scholar]
- Fitzpatrick BM, Johnson JR, Kump DK, Shaffer HB, Smith JJ, Voss SR. Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid Tiger Salamanders. BMC Evol Biol. 2009;9:176. doi: 10.1186/1471-2148-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF. Evidence for archaic Asian ancestry on the human X chromosome. Mol Biol Evol. 2005;22:189–192. doi: 10.1093/molbev/msi013. [DOI] [PubMed] [Google Scholar]
- Geneva A, Garrigan D. Population genomics of secondary contact. Genes. 2010;1:124–142. doi: 10.3390/genes1010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BR, Grant PR. Fission and fusion of Darwin's finches populations. Phil Trans R Soc Lond B. 2008;363:2821–2829. doi: 10.1098/rstb.2008.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin's finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Natural selection, speciation and Darwin's finches. Proc Cal Acad Sci. 2010;61 (supp II:245–260. [Google Scholar]
- Grant V. Plant Speciation. Columbia University Press: New York; 1981. [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser CB., Jr Hybridization in the annual sunflowers: Helianthus annuus x H. debilis var Cucumerifolius. Evolution. 1951;5:42–51. [Google Scholar]
- Howard DJ.1993Reinforcement: origin, dynamics, and fate of an evolutionary hypothesisIn: Harrison RG (ed)Hybrid Zones and the Evolutionary Process Oxford University Press, Oxford; 46–69. [Google Scholar]
- Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden RC, et al. Genetic restoration of the Florida Panther. Science. 2010;329:1641–1645. doi: 10.1126/science.1192891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Arnold ML, Donovan LA. High hybrid fitness at seed and seedling life history stages in Louisiana Irises. J Ecol. 2003;91:438–446. [Google Scholar]
- Johnston JA, Donovan LA, Arnold ML. Novel phenotypes among early generation hybrids of two Louisiana Iris species: flooding experiments. J Ecol. 2004;92:967–976. [Google Scholar]
- Johnston JA, Grise DJ, Donovan LA, Arnold ML. Environment-dependent performance and fitness of Iris brevicaulis, I. fulva (Iridaceae) and hybrids. Am J Bot. 2001a;88:933–938. [PubMed] [Google Scholar]
- Johnston JA, Wesselingh RA, Bouck AC, Donovan LA, Arnold ML. Intimately linked or hardly speaking? The relationship between genotype and environmental gradients in a Louisiana Iris hybrid population. Mol Ecol. 2001b;10:673–681. doi: 10.1046/j.1365-294x.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Birch LC. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- Martin NH, Bouck AC, Arnold ML. Loci affecting long-term hybrid survivability in Louisiana Irises: implications for reproductive isolation and introgression. Evolution. 2005;59:2116–2124. [PubMed] [Google Scholar]
- Martin NH, Bouck AC, Arnold ML. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics. 2006;172:2481–2489. doi: 10.1534/genetics.105.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NH, Sapir Y, Arnold ML. The genetic architecture of reproductive isolation in Louisiana Irises: pollination syndromes and pollinator preferences. Evolution. 2008;62:740–752. doi: 10.1111/j.1558-5646.2008.00342.x. [DOI] [PubMed] [Google Scholar]
- Mayr E.1963Animal Species and Evolution Belknap Press: Cambridge; Mass. [Google Scholar]
- Moore WS. An evaluation of narrow hybrid zones in vertebrates. Q Rev Biol. 1977;52:263–277. [Google Scholar]
- Novo M, Bigey F, Beyne E, Galeote V, Gavory F, Mallet S, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci USA. 2009;106:16333–16338. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mouse. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Pinho C, Hey J. Divergence with gene flow: models and data. Annu Rev Ecol Evol Syst. 2010;41:215–230. [Google Scholar]
- Randolph LF. Iris nelsonii, a new species of Louisiana iris of hybrid origin. Baileya. 1966;14:143–169. [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Annu Rev Ecol Syst. 1997;28:359–389. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rosas U, Barton NH, Copsey L, Barbier de Reuille P, Coen E. Cryptic variation between species and the basis of hybrid performance. PLoS Biol. 2010;8:e1000429. doi: 10.1371/journal.pbio.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Baric S, Sturmbauer C. Speciation via introgressive hybridization in East African cichlids. Mol Ecol. 2002;11:619–625. doi: 10.1046/j.0962-1083.2001.01438.x. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Matta BM, Shakir-Botteri NL, McPheron BA. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436:546–549. doi: 10.1038/nature03800. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HDJ, Miyagi R, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–627. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW. The biology of speciation. Evolution. 2009;64:295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- Sturmbauer C, Salzburger W, Duftner N, Schelly R, Koblmüller S. Evolutionary history of the Lake Tanganyika cichlid tribe Lamprologini (Teleostei: Perciformes) derived from mitochondrial and nuclear DNA data. Mol Biol Evol. 2010;57:266–284. doi: 10.1016/j.ympev.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Okashah RA, Knapp SJ, Arnold ML, Martin NH. Reproductive isolation in Louisiana irises: transmission ratio distortion. BMC Plant Biol. 2010;10:48. doi: 10.1186/1471-2229-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Arnold ML, Martin NH. The genetic architecture of reproductive isolation in Louisiana Irises: hybrid fitness in nature. Evolution. 2009;63:2581–2594. doi: 10.1111/j.1558-5646.2009.00742.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Using phylogeographic analyses of gene trees to test species status and processes. Mol Ecol. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. [DOI] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna D, Poon LLM, Zhu HC, Ma SK, Li OTW, Cheung CL, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viosca P., Jr The irises of southeastern Louisiantaxonomic and ecological interpretation. Bull Am Iris Soc. 1935;57:3–56. [Google Scholar]
- Wakeley J, Hey J. Estimating ancestral population parameters. Genetics. 1997;145:847–855. doi: 10.1093/genetics/145.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hey J. Estimating divergence parameters with small samples from a large number of loci. Genetics. 2010;184:363–379. doi: 10.1534/genetics.109.110528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am Nat. 2006;167:794–807. doi: 10.1086/504606. [DOI] [PubMed] [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. New Phytol. 2010;187:230–239. doi: 10.1111/j.1469-8137.2010.03234.x. [DOI] [PubMed] [Google Scholar]
- Won Y-J, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H.petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]