Abstract
Dissecting the genetic architecture of fitness-related traits in wild populations is key to understanding evolution and the mechanisms maintaining adaptive genetic variation. We took advantage of a recently developed genetic linkage map and phenotypic information from wild pedigreed individuals from Ram Mountain, Alberta, Canada, to study the genetic architecture of ecologically important traits (horn volume, length, base circumference and body mass) in bighorn sheep. In addition to estimating sex-specific and cross-sex quantitative genetic parameters, we tested for the presence of quantitative trait loci (QTLs), colocalization of QTLs between bighorn sheep and domestic sheep, and sex × QTL interactions. All traits showed significant additive genetic variance and genetic correlations tended to be positive. Linkage analysis based on 241 microsatellite loci typed in 310 pedigreed animals resulted in no significant and five suggestive QTLs (four for horn dimension on chromosomes 1, 18 and 23, and one for body mass on chromosome 26) using genome-wide significance thresholds (Logarithm of odds (LOD) >3.31 and >1.88, respectively). We also confirmed the presence of a horn dimension QTL in bighorn sheep at the only position known to contain a similar QTL in domestic sheep (on chromosome 10 near the horns locus; nominal P<0.01) and highlighted a number of regions potentially containing weight-related QTLs in both species. As expected for sexually dimorphic traits involved in male–male combat, loci with sex-specific effects were detected. This study lays the foundation for future work on adaptive genetic variation and the evolutionary dynamics of sexually dimorphic traits in bighorn sheep.
Keywords: adaptive variation, animal model, domestic sheep, Ovis aries, sexual dimorphism, sexual selection
Introduction
Dissecting the genetic architecture of ecologically important traits is key to understanding evolution as well as the mechanisms allowing the maintenance of adaptive genetic variation (Ellegren and Sheldon, 2008; Nadeau and Jiggins, 2010; Slate et al., 2010). While a variety of approaches can be used to identify relevant loci (Stinchcombe and Hoekstra, 2007; Ellegren and Sheldon, 2008), there is growing interest in performing genomic studies using free-living pedigreed populations (Slate et al., 2010). Apart from enabling work on species that may not be amenable to controlled experiments, the study of wild populations is motivated by unparalleled opportunities to address topics requiring fitness estimates that are minimally influenced by experimental conditions (Ellegren and Sheldon, 2008; Clutton-Brock and Sheldon, 2010; Slate et al., 2010). These include the genetic architecture of fitness in natural environments (Ellegren and Sheldon, 2008), the evolutionary dynamics of sexually selected traits (Chenoweth and McGuigan, 2010), evolutionary stasis (for example, Gratten et al., 2008) and sexually antagonistic genetic variation (Bonduriansky and Chenoweth, 2009; Slate et al., 2010). However, apart from work in humans, studies on the genetic architecture of ecologically important traits in free-living populations remain rare because of difficulties in maintaining multigenerational pedigrees and assembling adequate genotype–phenotype data sets (Slate, 2005; Slate et al., 2010).
The bighorn sheep (Ovis canadensis), a mountain ungulate endemic to Western North America, has been the focus of numerous ecological and evolutionary quantitative genetic investigations (for example, Coltman et al., 2003, 2005; Poissant et al., 2008; Réale et al., 2009) and is emerging as an excellent ecological model for studies of evolution in the wild. Two traits of interest, because of their links with fitness, are horn size and body mass. Both traits are sexually selected in males (Coltman et al., 2002), and body mass is associated with offspring survival (Feder et al., 2008) and female lifetime reproductive fitness (Poissant et al., 2008). In the Ram Mountain study population, male horn size and body mass also experience negative directional selection through trophy hunting (Coltman et al., 2003). The identification of chromosomal regions containing genes influencing these traits (quantitative trait loci, QTLs) would therefore open a unique window of opportunity to study the evolutionary dynamics of adaptive molecular variation in wild bighorn sheep.
The study of horn size and body mass in bighorn sheep is also motivated by the presence of notable sexual dimorphism (Poissant et al., 2008). In theory, the evolution of sexual dimorphism depends on the presence of sex-specific genetic variance (Lande, 1980). Although such variance has been documented in a large number of organisms (Poissant and Coltman 2009; Poissant et al., 2010a), including bighorn sheep (Poissant et al., 2008), little is known about its molecular underpinning and micro-evolutionary dynamics. Dissecting the genetic architecture of sexually dimorphic traits in bighorn sheep would thus also provide insights into the molecular mechanisms facilitating the independent evolution of males and females.
Differentiating real QTLs from false positives is a major challenge in any QTL mapping study (Lander and Kruglyak, 1995), especially for studies of wild populations in which sample sizes are typically limited (Slate et al., 2010). In bighorn sheep, data interpretation could be facilitated by previous research in domestic sheep (Ovis aries, ∼3 million years divergence, Bunch et al., 2006). Indeed, one QTL has already been mapped for horn size in domestic sheep (that is, on chromosome 10 near the horns locus, Johnston et al. 2010). A large number of QTLs have also been identified for weight-related traits (reviewed in Cavanagh et al., 2010). QTLs often appear to be conserved across species (for example, Reid et al., 2005; Moghadam et al., 2007) but expectations for fitness-related traits are unclear, in particular because selection is expected to reduce genetic variation through the fixation of advantageous alleles (Falconer, 1989).
We performed a genome-wide scan for horn dimension (volume, length, base circumference) and body mass QTLs in wild bighorn sheep from Ram Mountain, Alberta, Canada, using a recently developed microsatellite genetic linkage map (Poissant et al., 2010b). As sexually antagonistic selection resulting from sexual selection in one sex is expected to promote the accumulation of sex-specific genetic variance (Poissant et al., 2010a), we searched for QTLs influencing both sexes similarly as well as QTLs having sex-specific effects. We also tested for QTL colocalization between bighorn sheep and domestic sheep to assist with data interpretation and assess whether the same loci could be involved in similar micro-evolutionary processes across species.
Materials and methods
Study population
The Ram Mountain bighorn sheep population is native to a small isolated mountain range located about 50 km east of the Canadian Rockies in Alberta, Canada (52°N, 115°W, elevation 1080–2170 m). This study is based on data collected from 1970 to 2009. Techniques used to capture, mark, measure and monitor animals were described in detail by Jorgenson et al. (1993). Briefly, animals were captured in a corral trap baited with salt from late May to September or early October each year. Almost all animals were marked early in life, so their exact age was known. Individuals captured for the first time as adults were aged by counting horn growth rings. Marked sheep were subsequently monitored throughout their lifetime.
Phenotypic data
Most females and young males were captured multiple (>3) times each year, while males 3 years and older were typically caught one to three times per season, usually in June or July. At each capture, sheep were weighed and the size of their horns was measured. Horn measurements included length along the outside curvature and horn base circumference. As in Poissant et al. (2008), horn volume was subsequently calculated assuming a conical shape using the average horn base circumference of both horns and the length of the longest horn to reduce the influence of horn breakage. Horn length measurements of females with two severely broken horns were excluded. We focused on phenotypes measured in adults aged 2 to 10 to reduce the potentially confounding influence of maternal effects (Wilson et al., 2005; Kruuk and Hadfield, 2007) and age × QTL interactions (Poissant and Coltman 2009).
Pedigree information
Over the entire study period, maternity was inferred in the field using suckling behavior. Genetic analyses (described below) showed that this technique is accurate in >99% of cases. Since 1988, the collection of DNA samples permitted formal genetic parentage analyses. These were based on ∼30 microsatellite loci (for details, see Coltman et al., 2005) and the 95% confidence threshold in Cervus (Marshall et al., 1998). In addition, the software Colony (Wang, 2004) was used to infer sibships resulting from sires that were not DNA sampled (for details, see Coltman et al., 2005). The accuracy of parts of the pedigree was also recently assessed using >200 microsatellite loci used for linkage map construction (details below). The current pedigree contains 803 maternal links resulting from 236 dams (mean number of offspring±1 s.d.=3.40±2.52) and 454 paternal links resulting from 70 sampled and 36 unsampled sires (mean number of offspring per sire=4.28±4.40).
Only parts of the full pedigree are informative for QTL mapping purposes because genome-wide genotypes have only been obtained for a subset of individuals. We therefore based our QTL mapping analyses on a restricted pedigree composed of 310 fully typed animals (172 females and 138 males). We also included animals that were either untyped (n=18) or only typed at markers used for initial parentage analyses (n=41) if they helped to connect fully typed animals in the pedigree (that is, parents). The QTL mapping pedigree included 201 females and 159 males connected by 301 maternal links (mean number of offspring per dam±1 s.d.=2.59±1.49) and 259 paternal links (mean number of offspring per sire=4.05±3.28).
Bighorn sheep linkage map
The bighorn sheep linkage map is based on information from two wild pedigreed populations (Ram Mountain, Alberta, Canada, and National Bison Range, MT, USA) and contains 247 microsatellites ordered along all 26 autosomes and the X chromosome (Poissant et al., 2010b). A total of 241 markers have been genotyped in the Ram Mountain population, and all but three (OarFCB11, BMS1247 and BMS1948, which are located near telomeres of chromosomes 2, 5 and 21 in domestic sheep, respectively) are positioned in the species map. In this study, we used recombination fractions from the integrated species map instead of the Ram Mountain population-specific map because they are likely more accurate (Poissant et al., 2010b). Map distances used in this study differ slightly from those presented in Poissant et al. (2010b) because recombination fractions were converted to centimorgans using Haldane's rather than Kosambi's mapping function to accommodate downstream QTL mapping analyses (that is, identity-by-descent (IBD) estimation, details below). These new map distances are presented in Supplementary Appendix S1. Additional details about markers, laboratory techniques, map construction and map characteristics are available in Poissant et al. (2009, 2010b).
Quantitative genetic analyses
Phenotypic variance was partitioned into additive genetic and other components using the animal model and restricted maximum likelihood implemented in the program ASReml 3.1. (Gilmour et al., 2009). The animal model is a form of mixed model incorporating pedigree information where the phenotype of each individual is modelled as the sum of its additive genetic value and other random and fixed effects. The method has a long history in animal breeding and is now commonly used for studies of free-living populations because of its ability to optimize the use of information in complex and incomplete pedigrees (Wilson et al., 2010).
In a typical animal model:
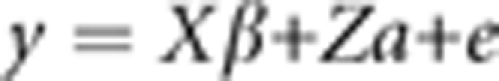 |
y is the vector of individual phenotypes, X and Z are incidence matrices relating fixed and random effects to each individual, β is a vector of fixed effects, a is a vector of polygenic (additive genetic) effects and e is the vector of residual errors.
We initially analyzed male and female traits separately because the genetic architecture of sexually dimorphic traits is expected to be partly independent between the sexes (Poissant et al., 2010a). However, doing so considerably reduces the amount of phenotypic information included in any given analysis and diminishes the probability of detecting QTLs influencing both sexes similarly. All analyses were therefore repeated treating male and female traits as a single trait. We standardized each trait in each age/sex class to an s.d. of one (that is, trait value divided by age- and sex-specific s.d.) before analysis because phenotypic variance differed between the sexes and increased with age, especially in males.
In sex-specific analyses, fixed effects included age (factor), date of capture (continuous, second-order polynomial, with 24 May as day 0), and the age × date interaction. In analyses where male and female homologous traits were combined, fixed effects also included sex and all possible interactions.
We extended the basic animal model described above in all analyses with the addition of permanent environmental (identity), year of capture and year of birth random effects. The permanent environmental effect was included to account for inter-individual variation resulting from non-genetic causes (for example, horn breakage) as well as dominance and epistasis. The year of capture and year of birth effects were fitted to account for common environmental conditions (Kruuk and Hadfield, 2007). Phenotypic variance (Vp) was therefore partitioned into five components after having taken fixed effects into account (described below): additive genetic (Va), permanent environmental (Vpe), year of capture (Vy), year of birth (Vyob) and residual (Vr). All components were retained in final models even when not significant to prevent biasing Va upwardly (Wilson et al., 2010).
Heritability (h2) and other ratios were obtained by dividing individual variance components by Vp where Vp=Va+Vpe+Vy+Vyob+Vr. Covariances and correlations were obtained using bivariate models. Significance of (co)variance components and ratios was tested using likelihood ratio tests contrasting models including and excluding individual random effects. To test if correlations were significantly smaller than one, we used a similar approach where unconstrained models were contrasted to models in which correlations were constrained to one. All analyses were performed using the full Ram Mountain pedigree as well as the more restricted QTL mapping pedigree for comparative purposes.
QTL mapping
Variance component analysis
We mapped QTLs using a variance component approach (George et al., 2000, Slate 2005). This was done by extending the animal model described above with the addition of a QTL variance component (that is, random effect) estimated using pairwise estimates of IBD for specific genomic locations. IBD matrices were estimated every 2cM (Haldane's mapping function) as well as for unassigned markers using pedigree information, genotypes and map distances with the software Loki (Heath, 1997). Loki does not estimate proper IBD matrices for the sex chromosomes (Lange and Sobel, 2006), but we are unaware of software that will do so in large complex pedigrees. We therefore adopted the approach of Beraldi et al. (2007a, 2007b) and estimated IBD matrices for the X chromosome with Loki by treating the Y chromosome as a non-variable X chromosome. After a burn-in period of 50 cycles, 1 million iterations were performed with statistics being stored every two iterations. Significance of QTL effects was determined using logarithm of odds (LOD) scores calculated as
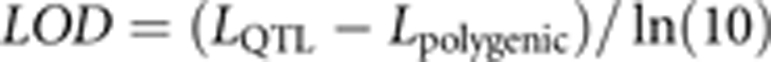 |
where L was the log likelihood of models with and without a QTL component. As linkage maps of bighorn sheep and domestic sheep are very similar (Poissant et al., 2010b), we adopted significance thresholds previously calculated for domestic sheep by Johnston et al. (2010) based on the formula from Lander and Kruglyak (1995). QTL were therefore considered suggestive and significant when LOD scores were >1.88 and 3.31, respectively. Following Lander and Botstein (1989), 95% confidence interval for QTL positions were approximated using the one-LOD drop-off method.
Cross-species QTL colocalization
Following Lander and Kruglyak (1995), we tested for QTL colocalization between bighorn sheep and the closely related domestic sheep using a nominal P<0.01 threshold (equivalent to LOD >1.175). More specifically, a QTL was considered to be colocalized between species when a position with LOD >1.175 in bighorn sheep was located within the 95% confidence interval of a significant domestic sheep QTL. This approach is valid for horn size because only one QTL has been mapped for this trait in domestic sheep to date (that is, on chromosome 10 near the horns locus, Johnston et al., 2010). On the other hand, it is anticonservative for body mass because of the large number of QTLs that have been mapped in domestic sheep for this trait. Results for body mass should therefore be interpreted with caution. Domestic sheep weight-related QTL information was obtained from Cavanagh et al. (2010) and references therein, Beraldi et al. (2007b), Margawati et al. (2006, 2009) and Hadjipavlou and Bishop (2008).
Sex × QTL interactions
As differences between results from univariate sex-specific models can be artifacts of small sample sizes (Curtsinger, 2002), we explicitly tested for sex × QTL interactions using bivariate animal models. More specifically, we compared the likelihood of models where QTL (co)variance components were left unconstrained with models where QTL variances were constrained to be equal between the sexes and the cross-sex QTL correlation was constrained to one. We tested for significance of sex × QTL interactions using likelihood ratio tests assuming a χ2 distribution with two degrees of freedom. These tests were restricted to regions identified as potentially containing a QTL using univariate analyses.
Results
All traits showed significant additive genetic, year, year of birth and permanent environmental variance after accounting for fixed effects when analyzing the entire data set for both sex-specific and sexes-combined analyses (Table 1). Similar results were observed when analyzing the smaller QTL mapping data set, except that year of birth and permanent environmental effects were not all significant (Supplementary Appendix S2). The proportion of phenotypic variance explained by each component was similar between data sets, except that heritability tended to be higher in the QTL mapping data set (0.18–0.38 versus 0.21–0.50). In sex-specific analyses, year of capture and year of birth together explained ∼20–40% of the phenotypic variance while permanent environmental effects explained ∼20–25%. In the sexes-combined analyses, year of capture and year of birth explained ∼20–25% of the phenotypic variance while permanent environmental effects explained ∼30–50%.
Table 1. Proportion of phenotypic variance after having accounted for fixed effects (Vp) explained by additive genetic (h2), year, year of birth and permanent environmental effects in adult bighorn sheep for horn volume (cm3), horn length (cm), horn base circumference (cm) and body mass (kg).
| Trait | Ind. | Obs. | Mean (s.d.) | Transformed data mean (s.d.) | Vp | h2 | Year | Year of birth | Perm. env |
|---|---|---|---|---|---|---|---|---|---|
| Male traits | |||||||||
| Horn volume | 261 | 1711 | 1546 (1057) | 3.36 (3.20) | 0.78 (0.08) | 0.27 (0.12)** | 0.08 (0.02)*** | 0.31 (0.07)*** | 0.21 (0.11)** |
| Horn length | 262 | 1718 | 52.19 (18.41) | 7.20 (7.09) | 0.79 (0.08) | 0.26 (0.13)* | 0.14 (0.03)*** | 0.26 (0.07)*** | 0.24 (0.12)** |
| Horn base circ. | 261 | 1715 | 30.60 (6.91) | 10.12 (15.87) | 0.76 (0.08) | 0.30 (0.11)*** | 0.06 (0.02)*** | 0.30 (0.07)*** | 0.19 (0.11)* |
| Body mass | 262 | 1708 | 72.29 (16.84) | 6.69 (2.01) | 0.59 (0.05) | 0.34 (0.12)** | 0.14 (0.04)*** | 0.13 (0.05)*** | 0.22 (0.11)** |
| Female traits | |||||||||
| Horn volume | 311 | 4028 | 111.6 (32.0) | 4.57 (1.81) | 0.96 (0.08) | 0.28 (0.10)*** | 0.06 (0.02)*** | 0.17 (0.05)*** | 0.33 (0.09)*** |
| Horn length | 313 | 4089 | 22.54 (4.21) | 7.99 (3.01) | 1.04 (0.09) | 0.22 (0.11)* | 0.05 (0.01)*** | 0.15 (0.06)*** | 0.49 (0.11)*** |
| Horn base circ. | 313 | 4292 | 13.55 (1.08) | 14.76 (5.49) | 0.90 (0.07) | 0.38 (0.08)*** | 0.08 (0.02)*** | 0.12 (0.04)*** | 0.16 (0.06)*** |
| Body mass | 318 | 4791 | 60.10 (9.37) | 8.33 (2.33) | 0.57 (0.04) | 0.20 (0.07)*** | 0.16 (0.04)*** | 0.10 (0.04)*** | 0.22 (0.06)*** |
| Sexes combined | |||||||||
| Horn volume | 572 | 5739 | — | — | 0.88 (0.06) | 0.19 (0.06)*** | 0.05 (0.01)*** | 0.18 (0.05)*** | 0.41 (0.06)*** |
| Horn length | 575 | 5807 | — | — | 0.91 (0.06) | 0.18 (0.06)*** | 0.06 (0.02)*** | 0.17 (0.05)*** | 0.47 (0.07)*** |
| Horn base circ. | 574 | 6007 | — | — | 0.88 (0.05) | 0.23 (0.06)*** | 0.07 (0.02)*** | 0.15 (0.04)*** | 0.31 (0.05)*** |
| Body mass | 580 | 6499 | — | — | 0.59 (0.03) | 0.24 (0.06)*** | 0.13 (0.03)*** | 0.07 (0.03)*** | 0.28 (0.05)*** |
Standard errors generated by ASReml are presented in parentheses. Number of individuals and observations included in each analysis as well as trait means (s.d. in parentheses) prior and following data transformation (see Materials and methods section) are also presented. Estimates are based on the entire Ram Mountain data set; equivalent estimates for the smaller QTL mapping data set are presented in Supplementary Appendix S2. Significance of ratios was assessed using likelihood ratio tests (*P<0.05, **P<0.01, ***P<0.001).
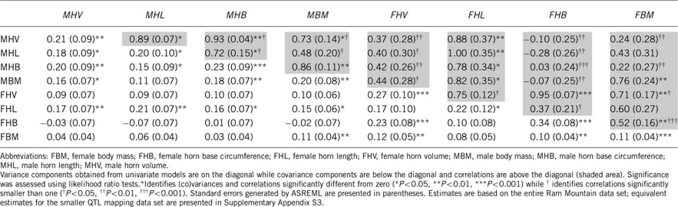
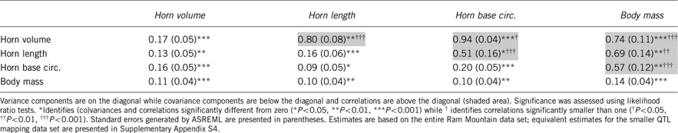
Genetic correlation estimates were generally positive (31 of 34). The only (nonsignificant) negative estimates were between female horn base circumference and male traits. Most genetic correlations were significantly smaller than one (24 of 34, Tables 2 and 3). Of the four cross-sex genetic correlations involving homologous male and female traits, two were significantly smaller than one and close to zero (horn volume and horn base circumference) while two were large and not significantly different from one (horn length and body mass). Estimates obtained using the full and the smaller QTL mapping data set were similar (Tables 2 and 3, Supplementary Appendix S3, Supplementary Appendix S4).
Table 2. Additive genetic (co)variances and correlations for sex-specific fitness-related traits in adult bighorn sheep.
Table 3. Additive genetic (co)variances and correlations for fitness-related traits in adult bighorn sheep.
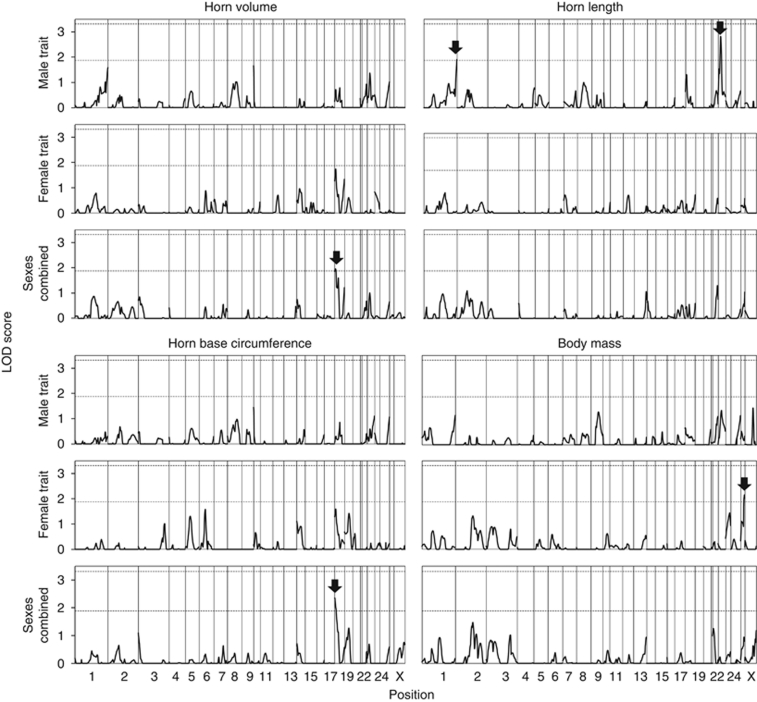
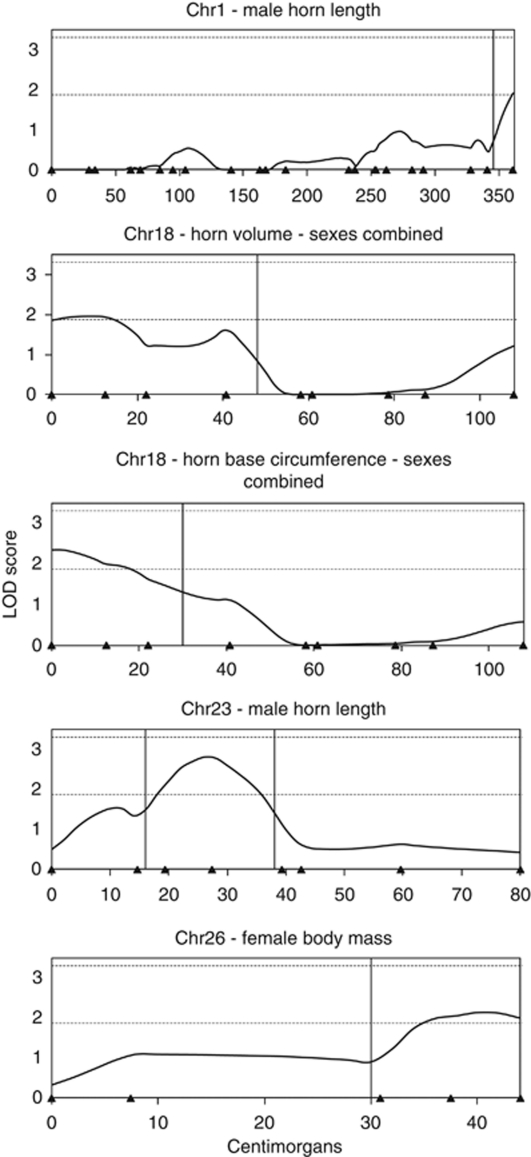
The QTL analysis did not result in the identification of significant QTLs when using the genome-wide significance threshold (LOD >3.31). However, five suggestive QTLs deserving further attention were detected (LOD >1.88, Table 4, Figures 1 and 2). Two of these were identified using male-specific analyses (horn length on chromosomes 1 and 23), one was identified using female-specific analyses (body mass on chromosome 26) and two were identified using sexes-combined analyses (horn volume and base circumference colocalized on chromosome 18). Estimates of individual QTL effects were generally large and comprised most or all of the additive genetic variance (Table 4).
Table 4. Genomic position of putative QTL for fitness-related traits in the Ram Mountain bighorn sheep population and their estimated parameters (VQTL, phenotypic variance explained by the QTL after having accounted for fixed effects; q2, proportion of phenotypic variance explained by the QTL after having accounted for fixed effects; h2, residual heritability after having fitted the QTL effect).
| Trait | LOD | Chr. | Pos. (cM)a | Closest marker | 1-LOD drop (cM) | 1.5-LOD drop (cM) | VQTL | q2 | h2 | QTL × sex (P-value) | Domestic sheep QTL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male trait | |||||||||||
| Horn volume | 1.66 | 10 | 0 | OarSEJ10, 11 | 0–6 | 0–12 | 0.36 (0.11) | 0.39 (0.10) | 0.00 (0.00) | 0.03 | (1) |
| Horn length | 1.91* | 1 | 361 | BMS2263 | 346–361 | 248–361 | 0.50 (0.09) | 0.60 (0.08) | 0.00 (0.00) | 0.35 | — |
| 2.82* | 23 | 26 | AGLA269 | 16–38 | 6–40 | 0.65 (0.12) | 0.73 (0.06) | 0.00 (0.00) | 0.22 | — | |
| Horn base circ. | 1.45 | 10 | 0 | OarSEJ10, 11 | 0–6 | — | 0.32 (0.12) | 0.37 (0.12) | 0.06 (0.16) | 0.02 | (1) |
| Body mass | 1.35 | 23 | 37 | RT9 | 16–80 | — | 0.25 (0.09) | 0.45 (0.13) | 0.00 (0.00) | 0.03 | (2–4) |
| Female trait | |||||||||||
| Body mass | 1.32 | 2 | 190 | BM81124 | 166–296 | — | 0.11 (0.04) | 0.22 (0.07) | 0.00 (0.00) | 0.81 | (5–7) |
| 1.44 | 24 | 44 | BP28 | 0–54 | — | 0.12 (0.04) | 0.24 (0.07) | 0.00 (0.00) | 0.16 | (8–9) | |
| 2.15* | 26 | 40 | JMP58 | 30–44 | 2–44 | 0.13 (0.04) | 0.26 (0.07) | 0.00 (0.00) | 0.06 | (10) | |
| Sexes combined | |||||||||||
| Horn volume | 1.95* | 18 | 9 | ILSTS52 | 0–48 | 0–52 | 0.30 (0.09) | 0.33 (0.08) | 0.00 (0.00) | 0.27 | — |
| Horn base circ. | 2.35* | 18 | 1 | SRCRSP5 | 0–30 | 0–46 | 0.30 (0.08) | 0.33 (0.07) | 0.00 (0.00) | 0.37 | — |
| Body mass | 1.47 | 2 | 190 | BM81124 | 156–242 | — | 0.12 (0.04) | 0.21 (0.06) | 0.00 (0.00) | 0.81 | (5–7) |
Abbreviation: QTL, quantitative trait locus.
Map distances are based on Haldane's mapping function (see Supplementary Appendix S1) and therefore not directly comparable to distances presented in Poissant et al. (2010b) where Kosambi's mapping function was used. (1) Horn morphology, Johnston et al. (2010), (2) body weight, Margawati et al. (2006), (3) carcass weight, no 95% CI, Margawati et al. (2009), (4) growth rate, Raadsma et al. (2009), (5) muscle development, Laville et al. (2004), (6) carcass weight, no 95% CI, Margawati et al. (2009), (7) weight, suggestive, Walling et al. (2004), (8) muscle mass, Campbell et al. (2003), (9) body weight and growth rate, Raadsma et al. (2009) and (10) body weight and growth rate, Raadsma et al. (2009). *Denotes suggestive QTLs (LOD >1.88). Regions with nominal P-values >0.01 (LOD >1.175) that did not exceed genome-wide significance thresholds are also presented when co-located with putatively homologous domestic sheep QTL. All regions with LOD >1.175 are presented in Supplementary Appendix S5.
Figure 1.
LOD scores along the 26 autosomes and the X chromosome for the presence of horn volume, length, base circumference and body mass QTL in the Ram Mountain bighorn sheep population. Dashed horizontal lines depict genome-wide thresholds used to identify suggestive (LOD>1.88) and significant (LOD >3.31) QTLs. Arrows highlight suggestive QTLs.
Figure 2.
LOD scores along chromosomes for the five suggestive QTLs (LOD >1.88) in the Ram Mountain bighorn sheep population. Triangles on the X axis depict marker positions. Dashed horizontal lines depict genome-wide thresholds used to identify suggestive (LOD >1.88) and significant (LOD >3.31) QTLs. Vertical lines depict 1-LOD 95% confidence intervals.
Our test for the cross-species QTL colocalization confirmed the presence of a horn size QTL on chromosome 10 near the horns locus across sheep species (nominal P=0.003 and 0.005 for male horn volume and base circumference, respectively, Table 4). Similar cross-species comparisons for body mass identified four putative cases of cross-species QTL colocalization. These included one of the suggestive QTLs identified on chromosome 26 and three regions with LOD <1.88 located on chromosomes 2, 23 and 24 (Table 4). As noted earlier, the use of a nominal P<0.01 test for the colocalization of body mass QTLs is anticonservative and results should be interpreted with caution.
We tested for the presence of sex × QTL interactions using bivariate models (Table 4). Significant sex-specific QTL effects were observed for the two horn dimension QTLs co-located on chromosome 10 (horn volume and base circumference) as well as a putative body mass QTL on chromosome 23 (all P<0.05). A near-significant sex × QTL interaction was also observed for the suggestive body mass QTL on chromosome 26 (P=0.06).
Discussion
We studied the genetic architecture of fitness-related traits in free-living bighorn sheep from Ram Mountain, Alberta, Canada. In addition to estimating sex-specific and cross-sex quantitative genetic parameters for horn volume, length, base circumference and body mass, we tested for the presence of QTLs influencing these traits, colocalization of QTLs between bighorn sheep and domestic sheep, and sex × QTL interactions.
Horn size
Significant additive genetic variance was detected for all sex-specific horn dimension traits, indicating that QTL detection was possible. The cross-sex genetic correlation for horn base circumference was one of the lowest ever estimated for a pair of homologous male and female traits (Poissant et al., 2010a) while the cross-sex genetic correlation for horn length was large and not significantly different from one. This suggests that the genetic decoupling of male and female horn volume reported in Poissant et al. (2008) and this study may in most part be attributable to the evolution of sex-specific genetic variance for horn base circumference. The reason for this is unclear but horn base circumference may have experienced greater sexually antagonistic selection than horn length. While most studies of sexual selection in sheep have focused on male horn length (for example, Coltman et al., 2002; Preston et al., 2003), there is no obvious reason to expect sexual selection to act on horn length more than horn base circumference. Horn base circumference is likely more important than horn length for fighting because males clash their horns near the base. The observed pattern would also be consistent with the presence of sexually antagonistic selection on horn volume or horn mass rather than base circumference or length since a change in horn base circumference will have a greater influence on horn volume than a proportional change in horn length.
We identified six putative horn dimension QTLs including four that were suggestive at the genome-wide level (LOD >1.88). The other two did not surpass genome-wide significance thresholds but were significantly colocalized with the only horn size QTL mapped in domestic sheep to date (LOD >1.175, Johnston et al., 2010). The six putative QTLs likely only represented four loci because horn volume and base circumference QTLs were colocalized on chromosomes 10 and 18. The detection of overlapping QTLs for different horn dimension traits was to be expected given strong positive phenotypic correlations among these traits. The other two QTLs were for horn length and located on chromosomes 1 and 23. The chromosome 10 QTL overlapped with the horns locus, a locus controlling discrete horn phenotypes in domestic sheep (that is, presence versus absence of horns, Montgomery et al., 1996). On the other hand, no putative QTL overlapped with loci known to influence discrete horn polymorphisms in other bovid genera (Georges et al., 1993; Vaiman et al., 1996; Asai et al., 2004). This suggests that different genes may be responsible for quantitative and discrete variation in horn morphology among bovids.
Body mass
The presence of additive genetic variance for both sex-specific traits indicated that QTL detection was possible in both sexes. The cross-sex genetic correlation for body mass was large and not significantly different from one, indicating that the detection of QTLs influencing variation similarly in both sexes was likely.
The genome scan for body mass QTLs yielded a single suggestive QTL on chromosome 26. This locus was colocalized with domestic sheep body weight and growth rate QTLs identified by Raadsma et al. (2009). Three additional regions (out of 5) with LOD scores smaller than genome-wide significance thresholds appeared to be colocalized with domestic sheep weight-related QTLs. In domestic sheep, these regions contain QTLs for body weight and muscularity (chromosome 2, Laville et al., 2004; Walling et al., 2004; Margawati et al., 2009), body weight and growth rate (chromosome 23, Margawati et al., 2006, 2009; Raadsma et al., 2009) and body weight, growth rate and muscle mass (chromosome 24, Campbell et al., 2003; Raadsma et al., 2009). Possible candidate genes in these regions (genes known to influence weight-related traits in sheep or other species) identified by Raadsma et al. (2009) include myostatin, beta-3-adrenergic receptor, melanocortin 4 receptor, erythropoietin, elastin and fibrosin genes. Myostatin (also known as growth differentiation factor 8, GDF8), located in the center of the region on chromosome 2, is perhaps the most promising of these genes because it has been linked to muscle development in domestic sheep (Clop et al., 2006; Kijas et al., 2007) and cattle (Casas et al., 1999). Although anti-conservative, our test for the colocalization of body mass QTL nonetheless highlighted chromosomal areas deserving further investigation and suggested that the genetic architecture of body mass may be partially conserved across species.
Sex × QTL interactions
Most QTLs were only identified in one of the two sex-specific analyses. In addition, significant sex × QTL interactions were detected for the horn size QTLs located on chromosome 10. A significant interaction was also detected for the putative body mass QTL on chromosome 23 but this result remains speculative because of uncertainties regarding the presence of a QTL at that position. QTLs with sex-specific effects have been documented in a variety of organisms (for example, Nuzhdin et al., 1997; Farber and Medrano, 2007; Moghadam et al., 2007) including domestic sheep (Raadsma et al., 2009), but to our knowledge had never been documented in a free-living wildlife population. The presence of sex-specific QTL effects in bighorn sheep adds to the accumulating evidence suggesting that sexual selection alters the genetic architecture of quantitative traits by promoting the accumulation of sex-specific genetic variance (Moller 1993; Wilkinson 1993; Bonduriansky and Rowe, 2005; Wright et al., 2008; Robinson et al., 2009).
QTL number and effect sizes
Our genome-wide analysis yielded no significant and a modest number of suggestive QTLs. Such results are similar to the ones obtained in the three other QTL mapping experiments performed using free-living wildlife populations to date. In Soay sheep, Ovis aries, analyses of over 10 traits yielded only one significant (jaw length) and 7 suggestive QTLs (Beraldi et al., 2007a, 2007b, Johnston et al., 2010). In red deer, Cervus elaphus, a test for birth weight QTLs yielded a single suggestive QTL (Slate et al., 2002). Finally, in great reed warblers, Acrocephalus arundinaceus, analyses of wing length and tarsus length resulted in the identification of a single significant QTL (Tarka et al., 2010). This study as well as the ones just mentioned demonstrated that QTL mapping in free-living wildlife populations was feasible. However, it is becoming clear that larger sample sizes and marker densities will be needed to improve QTL detection.
All QTLs appeared to explain all or most of the additive genetic variation. Such results are typical of QTL studies in free-living wildlife populations (Slate et al., 2010) and likely a consequence of small sample sizes (Beavis, 1998) combined with the upward bias occurring when QTL effects are estimated in the population in which they were discovered (Goring et al., 2001). Further research based on larger sample sizes will be necessary to obtain more reliable estimates (Slate et al., 2010).
Future directions
Upcoming research will focus on refining genome-wide QTL searches and test if QTLs identified in this study are valid. This will be accomplished by increasing the number of animals analyzed as well as marker coverage using single-nucleotide polymorphisms identified using the ovine OvineSNP50 BeadChip (Miller et al., 2011) and/or novel approaches based on next-generation sequencing technologies such as restriction-site-associated DNA sequencing (Baird et al., 2008). Linkage disequilibrium methods will also be used to test the presence of QTL in additional non-pedigreed populations.
Acknowledgments
JP's doctoral research was supported by graduate scholarships and research grants from Alberta Ingenuity (AI), the Natural Sciences and Engineering Council of Canada (NSERC), the Alberta Conservation Association (ACA) and the University of Alberta. We thank Marco Festa-Bianchet for his comments and suggestions as well as Jon Jorgenson and the numerous graduate students and field assistants who worked at Ram Mountain over the years.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Asai M, Berryere TG, Schmutz SM. The scurs locus in cattle maps to bovine chromosome 19. Anim Genet. 2004;35:34–39. doi: 10.1111/j.1365-2052.2003.01079.x. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis WD.1998QTL analyses: power, precision, and accuracyIn: Paterson AH (ed).Molecular Dissection of Complex Traits CRC Press: New York; 145–162. [Google Scholar]
- Beraldi D, McRae AF, Gratten J, Pilkington JG, Slate J, Visscher PM, et al. Quantitative trait loci (QTL) mapping of resistance to strongyles and coccidia in the free-living Soay sheep (Ovis aries) Int J Parasitol. 2007a;37:121–129. doi: 10.1016/j.ijpara.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Beraldi D, McRae AF, Gratten J, Slate J, Visscher PM, Pemberton JM. Mapping quantitative trait loci underlying fitness-related traits in a free-living sheep population. Evolution. 2007b;61:1403–1416. doi: 10.1111/j.1558-5646.2007.00106.x. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflicts. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R, Rowe L. Intralocus sexual conflict and the genetic architecture of sexually dimorphic traits in Prochyliza xanthostoma (Diptera: piophilidae) Evolution. 2005;59:1965–1975. [PubMed] [Google Scholar]
- Bunch TD, Wu C, Zhang YP, Wang S. Phylogenetic analysis of snow sheep (Ovis nivicola) and closely related taxa. J Hered. 2006;97:21–30. doi: 10.1093/jhered/esi127. [DOI] [PubMed] [Google Scholar]
- Campbell AW, Bain WE, McRae AF, Broad TE, Johnstone PD, Dodds KG, et al. Bone density in sheep: genetic variation and quantitative trait loci localization. Bone. 2003;33:540–548. doi: 10.1016/s8756-3282(03)00228-x. [DOI] [PubMed] [Google Scholar]
- Casas E, Keele JW, Fahrenkrug SC, Smith TP, Cundiff LV, Stone RT. Quantitative analysis of birth, weaning, and yearling weights and calving difficulty in Piedmontese crossbreds segregating an inactive myostatin allele. J Anim Sci. 1999;77:1686–1692. doi: 10.2527/1999.7771686x. [DOI] [PubMed] [Google Scholar]
- Cavanagh C, Jonas E, Hobbs M, Thomson P, Tammen I, Raadsma H. Mapping quantitative trait loci (QTL) in sheep. III. QTL for carcass composition traits derived from CT scans and aligned with a meta-assembly for sheep and cattle carcass QTL. Genet Sel Evol. 2010;42:36. doi: 10.1186/1297-9686-42-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth SF, McGuigan K. The genetic basis of sexually selected variation. Annu Rev Ecol Evol Syst. 2010;41:81–101. [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T, Sheldon BC. Individuals and populations: the role of long-term, individual-based studies of animals in ecology and evolutionary biology. Trends Ecol Evol. 2010;25:562–573. doi: 10.1016/j.tree.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Coltman DW, Festa-Bianchet M, Jorgenson JT, Strobeck C. Age-dependent sexual selection in bighorn rams. Proc R Soc B. 2002;269:165–172. doi: 10.1098/rspb.2001.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman DW, O'Donoghue P, Jorgenson JT, Hogg JT, Strobeck C, Festa-Bianchet M. Undesirable consequences of trophy hunting. Nature. 2003;426:655–658. doi: 10.1038/nature02177. [DOI] [PubMed] [Google Scholar]
- Coltman DW, O'Donoghue P, Hogg JT, Festa-Bianchet M. Selection and genetic (co)variance in bighorn sheep. Evolution. 2005;59:1372–1382. [PubMed] [Google Scholar]
- Curtsinger JW. Sex specificity, life-span QTLs, and statistical power. J Gerontol A Biol Sci Med Sci. 2002;57:B409–B414. doi: 10.1093/gerona/57.12.b409. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Sheldon B. Genetic basis of fitness differences in natural populations. Nature. 2008;452:169–175. doi: 10.1038/nature06737. [DOI] [PubMed] [Google Scholar]
- Falconer DS.1989Introduction to Quantitative Genetics3rd edn.Wiley: New York [Google Scholar]
- Farber CR, Medrano JF. Fine mapping reveals sex bias in quantitative trait loci affecting growth, skeletal size and obesity-related traits on mouse chromosomes 2 and 11. Genetics. 2007;175:349–360. doi: 10.1534/genetics.106.063693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder C, Martin JGA, Festa-Bianchet M, Bérubé C, Jorgenson J. Never too late? Consequences of late birthdate for mass and survival of bighorn lambs. Oecologia. 2008;156:773–781. doi: 10.1007/s00442-008-1035-9. [DOI] [PubMed] [Google Scholar]
- George AW, Visscher PM, Haley CS. Mapping quantitative trait loci in complex pedigrees: a two-step variance component approach. Genetics. 2000;156:2081–2092. doi: 10.1093/genetics/156.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges M, Drinkwater R, King T, Mishra A, Moore SS, Nielsen D, et al. Microsatellite mapping of a gene affecting horn development in Bos taurus. Nat Genet. 1993;4:206–210. doi: 10.1038/ng0693-206. [DOI] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. ASReml User Guide. Release 3.0. VSN International Ltd: Hemel Hempstead, UK; 2009. [Google Scholar]
- Goring HHH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genome-wide scans. Am J Hum Genet. 2001;69:1357–1369. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Wilson AJ, McRae AF, Beraldi D, Visscher PM, Pemberton JM, et al. A localized negative genetic correlation constrains microevolution of coat color in wild sheep. Science. 2008;319:318–320. doi: 10.1126/science.1151182. [DOI] [PubMed] [Google Scholar]
- Hadjipavlou G, Bishop SC. Age-dependant quantitative trait loci affecting growth traits in Scottish Blackface sheep. Anim Genet. 2008;40:165–175. doi: 10.1111/j.1365-2052.2008.01814.x. [DOI] [PubMed] [Google Scholar]
- Heath SC. Markov chain monte carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Beraldi D, McRae AF, Pemberton JM, Slate J. Horn type and horn length genes map to the same chromosomal region in Soay sheep. Heredity. 2010;104:196–205. doi: 10.1038/hdy.2009.109. [DOI] [PubMed] [Google Scholar]
- Jorgenson JT, Festa-Bianchet M, Wishart WD. Harvesting bighorn ewes: consequences for population size and trophy ram production. J Wildl Manage. 1993;57:429–435. [Google Scholar]
- Kijas J, McCulloch R, Edwards J, Oddy VH, Lee S, van der Werf J. Evidence for multiple alleles effecting muscling and fatness at the Ovine GDF8 locus. BMC Genet. 2007;8:80. doi: 10.1186/1471-2156-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk LEB, Hadfield JD. How to separate genetic and environmental causes of similarity between relatives. J Evol Biol. 2007;20:1890–1903. doi: 10.1111/j.1420-9101.2007.01377.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution. 1980;34:292–305. doi: 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Sobel E. Variance component models for X-linked QTLs. Genet Epidemiol. 2006;30:380–383. doi: 10.1002/gepi.20158. [DOI] [PubMed] [Google Scholar]
- Laville E, Bouix J, Sayd T, Bibe B, Elsen JM, Larzul C, et al. Effects of a quantitative trait locus for muscle hypertrophy from Belgian Texel sheep on carcass conformation and muscularity. J Anim Sci. 2004;82:3128–3137. doi: 10.2527/2004.82113128x. [DOI] [PubMed] [Google Scholar]
- Margawati ET, Indriawati, Subandriyo, Fullard K, Raadsma H. Detection of quantitative trait loci (QTL) affecting carcass traits in backcross family of Indonesian thin tail sheep. J Biotechnol Res Tropical Region. 2009;2:1–4. [Google Scholar]
- Margawati ET, Raadsma HW, Martojo H, Subandriyo, Muladno Quantitative trait loci (QTL) analysis for production traits of birth weight and weight 360 days in backcross sheep. Hayati. 2006;13:31–35. [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Miller JM, Poissant J, Kijas JW, Coltman DW, the International Sheep Genomics Consortium A genome-wide set of SNPs detects population substructure and long range linkage disequilibrium in wild sheep. Mol Ecol Res. 2011;11:314–322. doi: 10.1111/j.1755-0998.2010.02918.x. [DOI] [PubMed] [Google Scholar]
- Moghadam HK, Poissant J, Fotherby H, Haidle L, Ferguson MM, Danzmann RG. Quantitative trait loci for body weight, condition factor and age at sexual maturation in Arctic charr (Salvelinus alpinus): comparative analysis with rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar) Mol Genet Genomics. 2007;277:647–661. doi: 10.1007/s00438-007-0215-3. [DOI] [PubMed] [Google Scholar]
- Moller AP. Sexual selection in the barn swallow Hirundo rustica. III. Female tail ornaments. Evolution. 1993;47:417–431. doi: 10.1111/j.1558-5646.1993.tb02103.x. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Henry HM, Dodds KG, Beattie AE, Wuliji T, Crawford AM. Mapping the horns (Ho) locus in sheep: a further locus controlling horn development in domestic animals. J Hered. 1996;87:358–363. doi: 10.1093/oxfordjournals.jhered.a023014. [DOI] [PubMed] [Google Scholar]
- Nadeau NJ, Jiggins CD. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 2010;26:484–492. doi: 10.1016/j.tig.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Pasyukova EG, Dilda CL, Zeng ZB, Mackay TFC. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc Natl Acad Sci USA. 1997;94:9734–9739. doi: 10.1073/pnas.94.18.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J, Coltman DW. The ontogeny of cross-sex genetic correlations: an analysis of patterns. J Evol Biol. 2009;22:2558–2562. doi: 10.1111/j.1420-9101.2009.01862.x. [DOI] [PubMed] [Google Scholar]
- Poissant J, Hogg JT, Davis CS, Miller JM, Maddox JF, Coltman DW. Genetic linkage map of a wild genome: genomic structure, recombination and sexual dimorphism in bighorn sheep. BMC Genomics. 2010b;11:524. doi: 10.1186/1471-2164-11-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J, Shafer ABA, Davis CS, Mainguy J, Hogg JT, Coté SD, et al. Genome-wide cross-amplification of domestic sheep microsatellites in bighorn sheep and mountain goats. Mol Ecol Res. 2009;9:1121–1126. doi: 10.1111/j.1755-0998.2009.02575.x. [DOI] [PubMed] [Google Scholar]
- Poissant J, Wilson AJ, Coltman DW. Sex-specific genetic variance and the evolution of sexual-dimorphism: a systematic review of cross-sex genetic correlations. Evolution. 2010a;64:97–107. doi: 10.1111/j.1558-5646.2009.00793.x. [DOI] [PubMed] [Google Scholar]
- Poissant J, Wilson AJ, Festa-Bianchet M, Hogg JT, Coltman DW. Quantitative genetics and sex-specific selection on sexually dimorphic traits in bighorn sheep. Proc R Soc B. 2008;275:623–628. doi: 10.1098/rspb.2007.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BT, Stevenson IR, Pemberton JM, Coltman DW, Wilson K. Overt and covert competition in a promiscuous mammal: the importance of weaponry and testes size to male reproductive success. Proc R Soc B. 2003;270:633–640. doi: 10.1098/rspb.2002.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raadsma H, Thomson P, Zenger K, Cavanagh C, Lam M, Jonas E, et al. Mapping quantitative trait loci (QTL) in sheep. I. A new male framework linkage map and QTL for growth rate and body weight. Genet Sel Evol. 2009;41:34. doi: 10.1186/1297-9686-41-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réale D, Martin J, Coltman DW, Poissant J, Festa-Bianchet M. Male personality, life-history strategies and reproductive success in a promiscuous mammal. J Evol Biol. 2009;22:1599–1607. doi: 10.1111/j.1420-9101.2009.01781.x. [DOI] [PubMed] [Google Scholar]
- Reid DP, Szanto A, Glebe B, Danzmann RG, Ferguson MM. QTL for body weight and condition factor in Atlantic salmon (Salmo salar): comparative analysis with rainbow trout (Oncorhynchus mykiss) and Arctic charr (Salvelinus alpinus) Heredity. 2005;94:166–172. doi: 10.1038/sj.hdy.6800590. [DOI] [PubMed] [Google Scholar]
- Robinson MR, Wilson AJ, Pilkington JG, Clutton-Brock TH, Pemberton JM, Kruuk LEB. The impact of environmental heterogeneity on genetic architecture in a wild population of soay sheep. Genetics. 2009;181:1639–1648. doi: 10.1534/genetics.108.086801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol Ecol. 2005;14:363–380. doi: 10.1111/j.1365-294X.2004.02378.x. [DOI] [PubMed] [Google Scholar]
- Slate J, Santure AW, Feulner PGD, Brown EA, Ball AD, Johnston SE, et al. Genome mapping in intensively studied wild vertebrate populations. Trends Genet. 2010;26:275–284. doi: 10.1016/j.tig.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Slate J, Visscher PM, MacGregor S, Stevens D, Tate ML, Pemberton JM. A genome scan for quantitative trait loci in a wild population of Red Deer (Cervus elaphus) Genetics. 2002;162:1863–1873. doi: 10.1093/genetics/162.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2007;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Tarka M, Akesson M, Beraldi D, Hernandez-Sanchez J, Hasselquist D, Bensch S, et al. A strong quantitative trait locus for wing length on chromosome 2 in a wild population of great reed warblers. Proc R Soc B. 2010;277:2361–2369. doi: 10.1098/rspb.2010.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiman D, Koutika O, Oustry A, Elsen JM, Manfredi E, Fellous M, et al. Genetic mapping of the autosomal region involved in XX sex-reversal and horn development in goats. Mamm Genome. 1996;7:133–137. doi: 10.1007/s003359900033. [DOI] [PubMed] [Google Scholar]
- Walling GA, Visscher PM, Wilson AD, McTeir BL, Simm G, Bishop SC. Mapping of quantitative trait loci for growth and carcass traits in commercial sheep populations. J Anim Sci. 2004;82:2234–2245. doi: 10.2527/2004.8282234x. [DOI] [PubMed] [Google Scholar]
- Wang J. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GS. Artificial sexual selection alters allometry in the stalk- eyed fly Cyrtodiopsis dalmanni (Diptera: Diopsidae) Genet Res. 1993;62:213–222. [Google Scholar]
- Wilson AJ, Kruuk LEB, Coltman DW. Ontogenetic patterns in heritable variation for body size: using random regression models in a wild ungulate population. Am Nat. 2005;166:E177–E192. doi: 10.1086/497441. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Réale D, Clements MN, Morrissey MM, Postma E, Walling CA, et al. An ecologist's guide to the animal model. J Anim Ecol. 2010;79:13–26. doi: 10.1111/j.1365-2656.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- Wright D, Kerje S, Brandstrom H, Schutz K, Kindmark A, Andersson L, et al. The genetic architecture of a female sexual ornament. Evolution. 2008;62:86–98. doi: 10.1111/j.1558-5646.2007.00281.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.