Abstract
The occurrence of hybrid incompatibilities forms an important stage during the evolution of reproductive isolation. In early stages of speciation, males and females often respond differently to hybridization. Haldane's rule states that the heterogametic sex suffers more from hybridization than the homogametic sex. Although haplodiploid reproduction (haploid males, diploid females) does not involve sex chromosomes, sex-specific incompatibilities are predicted to be prevalent in haplodiploid species. Here, we evaluate the effect of sex/ploidy level on hybrid incompatibilities and locate genomic regions that cause increased mortality rates in hybrid males of the haplodiploid wasps Nasonia vitripennis and Nasonia longicornis. Our data show that diploid F1 hybrid females suffer less from hybridization than haploid F2 hybrid males. The latter not only suffer from an increased mortality rate, but also from behavioural and spermatogenic sterility. Genetic mapping in recombinant F2 male hybrids revealed that the observed hybrid mortality is most likely due to a disruption of cytonuclear interactions. As these sex-specific hybrid incompatibilities follow predictions based on Haldane's rule, our data accentuate the need to broaden the view of Haldane's rule to include species with haplodiploid sex determination, consistent with Haldane's original definition.
Keywords: hybrid incompatibilities, Haldane's rule, haplodiploidy, speciation, cytonuclear interaction, transmission ratio distorting loci
Introduction
The identification of hybrid incompatibilities, measured as increased sterility and mortality of hybrids, is a major goal in evolutionary biology as they cause strong selection pressure against the formation of hybrid offspring and thus are a potential driving force of speciation. Hybrid incompatibilities are caused by disrupted epistatic gene interactions in hybrids, also known as Dobzhansky–Muller (DM) interactions (Turelli and Orr, 1995), and in a few cases (mainly in Drosophila), hybrid incompatibility genes have been identified (reviewed by Presgraves, 2010). Although no general pattern in pathways and genes that are particularly prone to hybrid incompatibility has yet been discerned, it has long been known that ‘when in the offspring of two different animal races one sex is absent, rare or sterile, that sex is the heterozygous (heterogametic, that is, XY or ZW) sex' (Haldane, 1922). This pattern, referred to as Haldane's rule, is often observed in hybrids of recently diverged populations or species (Coyne and Orr, 1997). There are three main genetic theories that explain why especially the heterogametic sex should suffer from hybridization (Orr, 1997): The faster-X theory, the faster-male theory and the dominance theory. All three theories assume that, owing to their shared evolutionary history, epistatic genes are co-adapted within species but not between species. Thus, upon hybridization, these co-adapted genes are replaced by gene variants that have never been selected to interact properly with each other, and their interactions in hybrids are likely disrupted (DM interactions).
The faster-X theory assumes that beneficial recessive mutations accumulate more easily under haploidy. If true, genes on the sex chromosomes of diploid organisms should on average evolve faster than genes on the autosomes, because the sex chromosomes go through rounds of haploidy in the heterogametic sex (Charlesworth et al., 1987). Epistatic interactions of genes on a fast-evolving chromosome are more disruption-prone when brought into a foreign genetic background than genes on slowly evolving autosomes, because of their larger divergence. Therefore, the heterogametic sex is more likely to suffer from hybridization than the homogametic sex, because the latter can be saved by heterozygosity of the less compatible loci. Research has shown the complexity of sex chromosome evolution, as both supporting (for example, Musters et al., 2006; Baines et al., 2008) and opposing (for example, Betancourt et al., 2002; Thornton et al., 2006; Mank et al., 2010) evidence for faster evolution of the X-chromosome has been found (reviewed for Drosophila by Presgraves, 2008; Singh et al., 2009).
The faster-male theory states that male traits evolve faster than female traits owing to stronger sexual selection on males. This leads to Haldane's rule under male heterogamety, because more diverged male genomes have a higher chance to suffer from DM interactions than less diverged female genomes (Wu and Davis, 1993). Several studies have found support for the faster-male theory (Civetta and Singh, 1995; Meiklejohn et al., 2003; Zhang et al., 2004; Eads et al., 2007; Malone and Michalak, 2008). A second cause underlying the faster-male theory is that spermatogenesis is a sensitive process, easily disrupted by mutations that lead to male sterility, whereas mutations have less effect on oogenesis and hence female sterility (Wu and Davis, 1993). This leads to more cases of hybrid male sterility than female sterility (reviewed by Schilthuizen et al., 2011). One major problem of the faster-male theory is that it can only explain Haldane's rule under male heterogamety. Under female heterogamety other factors (such as dominance, see below) have to be invoked that overcome ‘faster-male' effects (Wu and Davis, 1993).
The dominance theory assumes that mutations are (partially) recessive and thus masked by heterozygosity (Turelli and Orr, 1995). In hybrids, gene interactions can be disrupted throughout the whole genome when diverged genes are forced to interact, but DM interactions are often rescued by dominance effects of the autosomes under diploidy. However, when the interactions involve sex-linked genes, the heterogametic sex is not saved by heterozygosity of the sex chromosome and thus has a higher chance to suffer from hybrid incompatibilities than the homogametic sex. This theory explains Haldane's rule for both male and female heterogamety, and is supported by several studies that have found a major effect of dominance in hybridizations that follow Haldane's rule (for example, True et al., 1996; Jiggins et al., 2001; Presgraves, 2003; Tao and Hartl, 2003; Bierne et al., 2006).
The general view is that Haldane's rule should be considered to be a composite phenomenon with multiple underlying mechanisms (Wu et al., 1996). What underlines this view is that some studies have found multiple mechanisms explaining their results on hybrid incompatibilities and Haldane's rule (for example, Hollocher and Wu, 1996). Although additional mechanisms have been proposed to explain Haldane's rule, the faster-male and the dominance theory are most supported (reviewed by Kulathinal and Singh, 2008). However, if hybrid incompatibilities are caused by recessive DM interactions, studying them can become a challenge because most incompatibilities are masked in diploid hybrid offspring. As a consequence, laborious introgression studies are necessary to investigate these recessive incompatibilities in diploid model organisms (Hollocher and Wu, 1996; True et al., 1996; Tao and Hartl, 2003; Tao et al., 2003; Masly and Presgraves, 2007).
Haplodiploid species, where fertilized and unfertilized eggs develop into females and males, respectively, are promising genetic models for studying hybrid incompatibilities when dominance effects are expected. Males express only one allelic variant, either dominant or recessive, and are thus very useful for the identification of negative epistatic gene interactions that cause hybrid incompatibilities on a genome-wide scale (Gadau et al., 1999; Ellison et al., 2008; Niehuis et al., 2008). Haldane himself referred to haplodiploids as a group in which the pattern that he had described, that is, Haldane's rule, should be prevalent. He stated: ‘groups in which the male sex is haploid are only extreme cases of the normal type, in that all the chromosomes here behave like sex chromosomes of other groups' (Haldane, 1922, page 101). Although recognized as a group that could be instrumental in unravelling the mechanisms that underlie Haldane's rule, haplodiploids have so far been deemed not to ‘fall under the Haldane's rule banner' because of their lack of heteromorphic sex chromosomes (Kulathinal and Singh, 2008). In line with the view of Haldane himself, Koevoets and Beukeboom (2009) argued that Haldane's observation of the differential effect of hybrid incompatibilities in males versus females should be expanded to include species with haplodiploid sex determination for two reasons: first, the inheritance of the complete haplodiploid genome is comparable to the inheritance of sex chromosomes in diploids; and second, all three mechanisms that explain Haldane's rule apply under haplodiploidy in that males are predicted to suffer more than females (that is, males would suffer under the dominance theory because of haploidy; under the faster-male theory because of faster evolution by sexual selection and under the faster-X theory because of faster evolution by natural selection). Moreover, having more genes (or chromosomes) that inherit in a haplodiploid manner (such as sex chromosomes) would lead to Haldane's rule faster. This is illustrated in Drosophila where sister species with larger X-chromosomes suffer from Haldane's rule faster than sister species with smaller X-chromosomes (Turelli and Begun, 1997).
Until now, only a single haplodiploid species pair (that is, Nasonia vitripennis and Nasonia giraulti) has been screened for sex-specific hybrid incompatibilities (Breeuwer and Werren, 1995). The authors found no incompatibilities in F1 hybrid females (although not systematically tested), but large mortality and slight sterility in F2 hybrid males. However, a problem when studying Haldane's rule in haplodiploids is that male and female hybrids are not formed in the same generation and thus cannot be tested under the same ploidy level. As in diploids, hybrid females in haplodiploids form in the F1 generation. Hybrid males, however, arise as offspring of hybrid females. Although we refer to these hybrids as F2 males, they are actually the first-generation male hybrids, and this is the generation that needs to be compared to F1 hybrid females when testing for the occurrence of hybrid incompatibilities (Koevoets and Beukeboom, 2009). Creating backcross F2 females is not a more appropriate comparison with F2 males, because it involves comparing hemizygous males to partially heterozygous females and confounds the comparison of males and females with regard to dominance effects.
The parasitoid wasp genus Nasonia has proven a valuable model system for studying complex genetic traits owing to its haplodiploid sex determination system and available genome sequences (Werren et al., 2010). The genus consists of four sister species, Nasonia vitripennis, Nasonia longicornis, Nasonia giraulti and Nasonia oneida (Darling and Werren, 1990; Raychoudhury et al., 2010a). All Nasonia species are reproductively isolated from each other owing to infections with different Wolbachia strains, which can be cured with antibiotics. Breeuwer and Werren (1995) found that F1 hybrid females from a cross between Wolbachia-cured N. vitripennis and N. giraulti are viable and fertile. Hybrid males had higher mortality rates than pure strain males, in particular hybrids with N. giraulti cytoplasm. The mortality occurred predominantly during larval development. The authors also found that surviving hybrids with N. vitripennis cytoplasm were mostly fertile, but those with N. giraulti cytoplasm were largely sterile. The asymmetry in the mortality and sterility rates of the hybrids was the first evidence for incompatibility between nuclear and cytoplasmic factors as a major cause of hybrid incompatibility in Nasonia.
Niehuis et al. (2008) investigated the increased mortality in hybrids of N. vitripennis and N. giraulti by analysing marker transmission ratio distortion in F2 hybrid males. Transmission ratio-distorting loci (TRDLs) deviate from the expected 1:1 transmission ratio of the parental alleles and can indicate differential mortality that relies on the genotype at the TRDL. The authors localized cytotype-dependent TRDLs on chromosomes 1, 2, 4 and 5 (Nasonia has five chromosomes). Overall, more TRDLs were incompatible with N. vitripennis cytoplasm than with N. giraulti cytoplasm. This is in contrast to the mortality found by Breeuwer and Werren (1995). Given this asymmetry in incompatibility, Niehuis et al. (2008) concluded that interactions between nuclear and cytoplasmic genes cause the mortality of F2 hybrid males. Clark et al. (2010) measured sterility in reciprocal F2 male hybrids and found disrupted mating behaviour and fewer sperm numbers than in pure species males, with N. giraulti cytoplasm leading to more sterility than N. vitripennis cytoplasm. By contrast, Bordenstein et al. (2001) found no increased mortality or sterility in hybrids between N. giraulti and N. longicornis. This can be explained by the short divergence time of these two sister species (0.2 mya; Campbell et al., 1993). The availability of four Nasonia species, with different divergence times and different levels of pre- and post-zygotic isolation, makes Nasonia particularly useful for speciation research, because it allows tracing the evolution of hybrid incompatibilities in this genus.
Here we use hybrids of N. vitripennis and N. longicornis to for the first time systematically compare female F1 and male F2 mortality and sterility levels, and to document sex-specific differences in hybrid incompatibilities between these haplodiploid species. Furthermore, we study and map hybrid incompatibilities in the F2 hybrid males and compare our data on mortality, sterility and the location of TRDLs with previous studies that used different interspecific crosses of Nasonia in order to infer the evolutionary history of the incompatibilities in this species complex.
Materials and methods
We used the N. vitripennis strain AsymC (origin: Leiden, The Netherlands) and the N. longicornis strain IV7R2 (origin: UT, USA) for the cross experiments. Both strains are cured from their Wolbachia infection and are highly inbred. Wasps were reared on Calliphora vicina hosts at 25 °C under constant light.
Mortality estimates
The experimental setup is summarized in Supplementary Figure S1. Virgin male (±24 h old) and female wasps (±72 h old and kept on hosts for feeding and to initiate egg-laying) were set up individually in four different crosses (♂ × ♀): N. vitripennis × N. vitripennis, N. longicornis × N. longicornis, N. longicornis × N. vitripennis and N. vitripennis × N. longicornis. These crosses are referred to as VV[V], LL[L], LV[V] and VL[L], respectively, with the first letter indicating the paternal genome complement, the second letter indicating the maternal genome complement, and the letters V and L with the square brackets indicating the cytotype. The mating pairs were left to mate for 24 h at 25 °C, after which the males were removed and the females provided with two hosts for 24 h at 25 °C (experimental day 1). The hosts were replaced every 24 h for 3 days, but only offspring from hosts of experimental days 2 and 3 were used for the experiment. Because there is high embryo mortality after egg counting owing to opening of the host puparium, the mating pairs were divided into two groups for each cross: one to count the number of oviposited eggs and one to count the number of eclosed adults. Hosts from the females in the egg-count group were submerged in Carnoy's fixative (ethanol:glacial acetic acid=1:3) and stored at −20 °C for at least 48 h. Hosts from the adult-count group were incubated at 25 °C and F1 adult offspring was counted in the black pupal stages (2–3 days prior to their eclosion after opening the host puparium) and separated into males and females to obtain virgins. It is assumed that wasp pupae collected in this stage would later eclose as adults, because the hybrid mortality occurs in earlier life stages (Breeuwer and Werren, 1995). Eclosion of counted wasps was near to 100%, but not systematically recorded. F1 virgin females were kept on fresh hosts upon eclosion for ±48 h for host feeding and to initiate egg-laying. After 48 h, three virgin females per mating pair were randomly assigned to one of three groups for (1) egg counting, (2) adult counting and (3) embryo collection (eggs) for DNA analysis. Hosts were replaced every 24 h for 3 days, from which only experimental days 2 and 3 were used for the experiment. After every 24 h, the hosts were removed and either submerged in Carnoy's fixative and stored at −20 °C for at least 48 h (group-1), replaced at 25 °C (group-2) or opened to remove the unhatched eggs (group-3). The F2 adult offspring were counted and collected 2–3 days prior to their eclosion. For the analysis of F1 and F2 mortality, the average offspring production per female was determined over all pupae parasitized on experimental days 2 and 3. Diapause larvae, larvae that were very retarded in their development and dead individuals were removed from the data set (<12%).
Sterility estimates
The level of F1 female sterility was determined by counting the proportion of F1 virgin females that produced F2 offspring. F2 male sterility was determined both behaviourally and spermatogenically. After being removed from the host as late instar pupae, F2 males were kept in plastic vials in groups of ∼15 individuals until the sterility experiment that took place 24 h after eclosion. The male's mating behaviour was observed in isolation with a virgin N. vitripennis female, except for N. longicornis males, which were offered a virgin N. longicornis female. Previous experiments have shown that N. vitripennis virgin females are more appropriate for testing the mating behaviour of VV[V], LV[V] and VL[L] F2 males because of the strong mating discrimination of N. longicornis virgin females, whereas LL[L] F2 males are better tested with N. longicornis virgins (Koevoets, unpublished data). Virgin females were put in test vials at least 1 h prior to the sterility experiment. The male was introduced to the female and observed for 10 min at 25 °C. Different behavioural categories were scored (Supplementary Table S2). Females that mated with a male within the 10-min observation period were isolated for 24 h after which they were given three hosts to screen their progeny. The absence of females in their progeny indicates male spermatogenic sterility, although other post-mating–pre-zygotic isolation factors (such as the inability to transfer sperm) could also explain a lack of female progeny. Therefore, the measure of spermatogenic sterility also includes these other post-mating–pre-zygotic isolation factors. All unmated females were discarded, and all tested males stored at −20 °C until DNA extraction.
Genotyping
F2 hybrid male embryos were collected ⩽24 h after oviposition by a virgin F1 female. The host puparium was removed and the embryos were transferred individually to 80-μl digestion buffer (Maniatis et al., 1982) (without proteinase-K) and ground with a sterile needle. DNA extraction was initiated by adding 20 μl of digestion buffer containing 40 μg of proteinase-K to the samples. Subsequent steps followed the standard high salt-chloroform protocol (Maniatis et al., 1982). The DNA was dissolved in 20 μl of MilliQ water. The DNA of the F2 hybrid adult males was extracted by following the regular high salt-chloroform protocol and the DNA was dissolved in 20 μl of MilliQ. Microsatellite markers were amplified by using the Qiagen multiplex PCR kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations (PCR profile: 15 min at 95 °C, followed by 30 cycles of 30 s at 94 °C, 1.5 min at TA and 1 min at 72 °C, followed by 45 min at 72 °C). We used eight multiplex sets of 6 or 7 microsatellite markers each (Supplementary Table S3). All stock DNA was diluted 10 × , from which 2 μl was used for embryonic DNA PCR and 1 μl for adult DNA PCR. All reactions were performed in 5-μl volumes using Applied Biosystems Veriti or Applied Biosystems 9700 thermocyclers. Fragments from embryonic PCR were diluted 20 times and from adult PCR 400 times, separated on the Applied Biosystems 3730 DNA Analyzer and analysed using GeneMapper v4.0 (Applied Biosystems, Carlsbad, CA, USA).
Statistical analyses
Linkage mapping
The linkage map was inferred by using the segregation data of 201 F2 hybrid embryos from both reciprocal hybrid crosses, using JoinMap (version 3.0; van Ooijen and Voorrips, 2001). Markers were grouped using a minimum logarithm of odds score of 7 and a maximum recombination fraction of 0.450. Markers with insufficient linkage were removed from the analysis. Following Niehuis et al. (2008), Haldane's mapping function was used to translate recombination fractions into map distances in centimorgans (cM).
Segregation bias
The segregation bias for all markers was tested by χ2 goodness of fit tests (df=1) against the expected segregation of 1:1 of parental alleles. Both Yates and Bonferroni corrections were applied. Furthermore, we used a Bayesian multipoint mapping approach as implemented in the software ANITA (Vogl and Xu, 2000). Interactions between nuclear markers were tested by χ2 goodness of fit tests (df=1), which detect deviations between the observed and the expected genotypes based on the allele frequencies in the two adult hybrid data sets. We refrained from testing makers that map to the same chromosome because of linkage. To assess significance, we simulated 10 000 random populations of 125 hybrid individuals each and with a marker distribution as in our experiments with the aid of a Perl script. For each population, we recorded the highest χ2-value and generated a frequency histogram. We then inferred the χ2 threshold value that was exceeded in less than 5% of the random populations (that is, 12.304).
Mortality
The mortality level was determined by an indirect egg-to-adult measurement by comparing the number of eggs produced in one group to the number of adults produced in another group, by using the Mann–Whitney U-test (MWU test) in SPSS 14.0. The survival probability (Z) was estimated from the ratio of the number of adults (Y) divided by the number of eggs (X). The variance in the survival probability was calculated from the variances of the number of eggs and adults according to Breeuwer and Werren (1995) using the formula
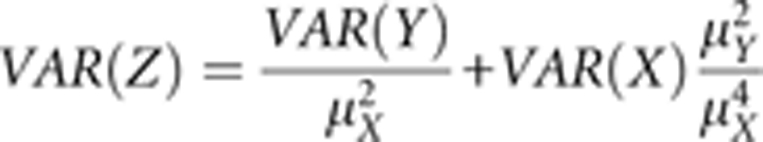 |
The properties of the normal distribution were used to determine confidence intervals for the different survival probabilities at α=0.05.
Sterility
The mating behaviour of males was scored, classified into seven successive categories and analysed following Clark et al. (2010). The transition probability was calculated as the frequency of males that perform a specific behaviour in category a that also perform the behaviour in category a+1. Deviations between crosses in the transition from one category to the next were determined by 2 × 4 χ2 tests with Bonferroni correction. When a significant difference was found, all the transitions were tested per cross in a pairwise manner, using 2 × 2 χ2 goodness of fit or Fisher's exact tests, depending on whether χ2 assumptions were violated (fewer than 50 samples, or expected values below 5). To compare the percentage sterility of the different crosses (for behavioural, spermatogenic and total sterility), we performed χ2-tests on proportions to see if the level of sterility depends on the cross type. When true, a Tukey-type multiple comparison was performed to test which crosses were significantly different (Zar, 1999). It was also determined whether there is a tendency for hybrids that carry more N. vitripennis alleles on each of the five chromosomes to be more or less often sterile. For this, the genotypes of fertile and (spermatogenically) sterile hybrids were compared. The data for all markers were merged per chromosome and the number of V alleles that was found per chromosome was tested for sterile versus fertile males of both hybrid types by using a 2 × 5 χ2 goodness of fit. All tests were performed by using Microsoft Excel 2003 and GraphPad online (GraphPad Software, La Jolla, CA, USA).
Results
Linkage map construction
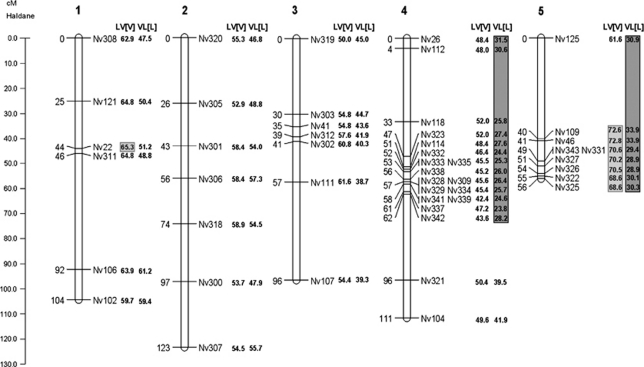
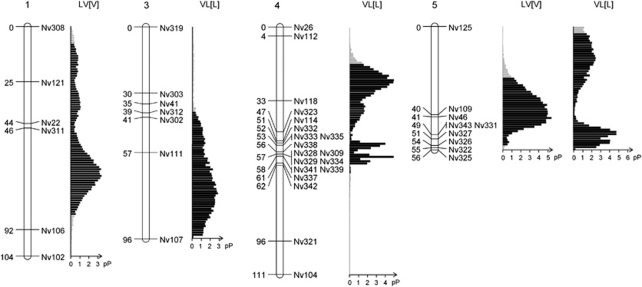
We inferred a linkage map based on the marker segregation data from 201 F2 hybrid male embryos after discarding individuals for which more than 20% of the markers failed to amplify. Data from the reciprocal hybrid crosses were pooled because we did not observe marker transmission ratio distortion at the embryonic stage in either mapping population (see below). We deliberately left two additional markers (that is, Nv324 and Nv344) in the final data set despite the fact that they did not map reliably, as we knew their position on the linkage map (chromosome-5 near marker Nv125). Five markers (out of the initial 55) were removed from the data set because they failed to amplify in more than 20% of the embryonic samples. The final linkage map consisted of 48 microsatellite markers (Supplementary Table S3), spread over five linkage groups (Figure 1), that were assigned to the five chromosomes of Nasonia using four anchored microsatellites (according to Rütten et al., 2004), The total map length was 490 cM and approximates the full Nasonia genome of 295 Mb (Werren et al., 2010).
Figure 1.
N. vitripennis and N. longicornis linkage map with allelic recovery rates. The linkage map is based on the microsatellite marker segregation in F2 hybrid embryos from pooled reciprocal hybrid crosses (n=201). Recombination distances are shown in Haldane centimorgans on the left and markers on the right of each chromosome. The recovery rates of the N. vitripennis and N. longicornis alleles in the F2 adult hybrids are shown next to the chromosomes for both crosses and are expressed as the percentage of V alleles. The markers with a significant segregation distortion are indicated in light grey (for LV[V]) and dark grey (for VL[L]) shaded boxes. Chromosome numbers are according to Rütten et al. (2004).
F1 mortality
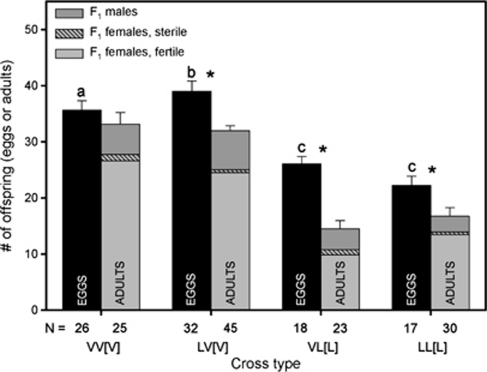
We found significant differences between the number of eggs laid and the number of emerging adults in all analysed crosses, except the N. vitripennis pure strain control (VV[V] Figure 2). As all strains that were used in our experiment are genetically homogeneous, all F1 female offspring from a given type of cross has the same genotype. This means that genetic variance contributes little to the phenotypic differences of the F1 hybrids and that the observed variance in their survival is largely environmental. This is also underlined by the significant mortality in the pure N. longicornis cross LL[L], which disappears in the next generation that was cultured on a different batch of hosts (see below). Because of haplodiploidy, F1 males from the VV[V] and LV[V] crosses are pure strain N. vitripennis, and F1 males produced in the LL[L] and VL[L] crosses are pure strain N. longicornis. The number of pure species males did not differ between crosses (MWU test VV[V] versus LV[V], P=0.269; LL[L] versus VL[L], P=0.971), which shows that F1 mortality is due to a reduction of F1 females (how this reconciles with the presence of sex-specific mortality will be discussed later). Table 1 shows the 95% confidence intervals for the F1 survival probabilities of the hybrid and pure species crosses. Based on non-overlapping confidence intervals, only F1 offspring from the VL[L] cross have lower survival than offspring from the other three crosses, the survival of the LV[V] cross did not differ significantly from either of the pure species.
Figure 2.
F1 mortality. F1 mortality in the hybrid and pure species crosses. Cross type on the x-axis represents the parental cross. Mortality is measured as indirect egg-to-adult survival probability. The black bars represent the number of eggs and the grey bars represent the number of adults (dark grey for male and light grey for female offspring). Female offspring is distinguished in sterile (patterned) and fertile (solid) females based on whether or not F1 females produced F2 males. Cross type represents ♂ × ♀[cytoplasm]. Sample sizes are indicated in the bars. * indicates significant difference between the number of eggs and adults per cross based on MWU test with α=0.05. The letters indicate different groups with regard to the number of eggs produced in the different crosses (MWU test, α=0.05).
Table 1. 95% confidence intervals of the survival probabilities for the hybrid and pure species crosses.
| Cross |
95% confidence interval |
|||
|---|---|---|---|---|
| F1 (female-biased) | F2 (male) | |||
| VV[V] | 85.8 | 99.6 | 85.5 | 99.9 |
| LV[V] | 76.9 | 87.2 | 72.2 | 84.2 |
| VL[L] | 49.8 | 61.3 | 68.8 | 81.9 |
| LL[L] | 66.9 | 83.9 | 106.3 | 124.2 |
Survival is calculated over the total offspring count: in the F1 these are male and female (sex ratio greatly biased towards female), and in the F2 these are only male.
F2 mortality
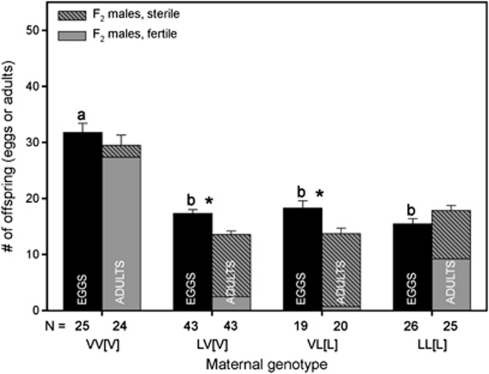
F2 hybrid males suffered from significant mortality (Figure 3). As the genome of F2 hybrid males is a product of recombination between the two chromosomal sets of the mother, each hybrid male has a unique nuclear genome. Figure 3 shows that F2 hybrid male mortality is slightly higher in the VL[L] hybrids than in the LV[V] hybrids, with the only genetic difference between them being their cytoplasm. Table 1 shows the 95% confidence intervals for the F2 survival probabilities of the hybrid and pure species crosses. The pure species F2 males have very high survival (in N. longicornis even over 100% owing to independent samples within and between groups, but the number of adults is not significantly higher than the number of eggs), whereas the hybrid F2 males have a lowered survival probability that is similar for both reciprocal hybrid crosses.
Figure 3.
F2 mortality. F2 mortality in the hybrid and pure species crosses. Cross type on the x-axis represents the grand parental cross. Mortality is measured as indirect egg-to-adult survival probability. The black bars represent the number of eggs and the grey bars represent the number of male offspring. Male offspring is distinguished in sterile (patterned) and fertile (solid) males based on the sterility experiment. Cross type represents ♂ × ♀[cytoplasm]. Sample sizes are indicated in the bars. * indicates significant difference between the number of eggs and adults per cross based on MWU test with α=0.05. The letters indicate different groups with regard to the number of eggs produced in the different crosses (MWU test, α=0.05).
F1 female sterility
Female sterility was estimated as the proportion of F1 females that did not produce F2 males. This proportion was used to infer the ratio of fertile and sterile F1 females in our data set (Figure 2). F1 female sterility did not differ significantly across all four crosses (P>0.05 χ2 on proportions (Zar, 1999)). The egg production of hybrid females was similar to pure N. longicornis females, but reduced compared with pure N. vitripennis females (Tukey test, P<0.05; see letters in Figure 3 for differences in F2 egg production by F1 females).
F2 male sterility
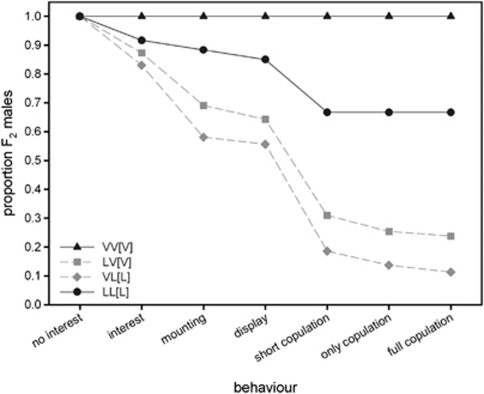
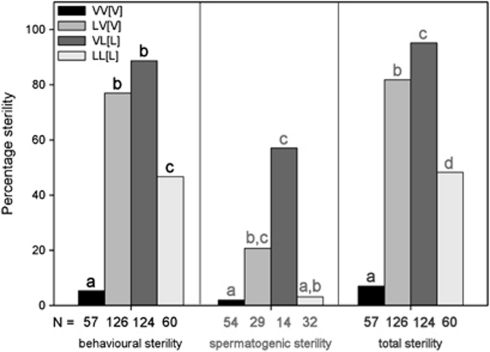
We tested a subset of F2 males for behavioural sterility by recording their courtship behaviour for seven different behavioural categories (Supplementary Table S2) in isolation with a single virgin female for 10 min. The results show that nearly all pure strain males showed normal courtship, whereas many hybrid males showed aberrant courtship behaviour (Figure 4). Table 2 shows the probabilities of males performing a typical sequence of courtship behaviours, ranging from showing interest in the female to performing a copulation with post-copulatory behaviour. When males show normal courtship, they transit from the first to subsequent categories with high probabilities. Hybrid males showed significantly lower transition rates for all categories, except from ‘mounting' to ‘display'. This means that hybrid males that were able to mount a female were also able to display courtship to that female. In almost all cases, the two pure crosses showed similar transition probabilities to each other, as did the two hybrid crosses. When combining all the aberrant courtship behaviours that did not lead to a successful copulation, we get a measure of behavioural sterility (Figure 5). The graph shows that hybrid males tend to be significantly more often sterile than males of the two pure species (Tukey-type multiple comparison, α=0.05)
Figure 4.
F2 male behavioural sterility. Behavioural sterility of F2 males from hybrid and pure species crosses measured as the ability of males to show different categories of the male mating behaviour. The cumulative proportion of males that showed a particular behaviour is plotted against the behavioural category.
Table 2. Transition probabilities between different categories of male courtship behaviour in the hybrids and pure species.
| F2 ♂♂ | n | Interest | Mounting | Display | Short cop. | Only cop. | Full copulation |
|---|---|---|---|---|---|---|---|
| VV | 57 | 1.00a | 1.00a | 1.00 | 1.00a | 1.00a | 1.00a |
| LV | 126 | 0.87b | 0.79b | 0.93 | 0.48b | 0.82b | 0.94a.b |
| VL | 124 | 0.83b | 0.70b | 0.96 | 0.33b | 0.74b | 0.82b |
| LL | 60 | 0.92a.b | 0.96a | 0.96 | 0.78c | 1.00a | 1.00a |
| P | 0.008 | <0.0001 | 0.241 | <0.0001 | <0.0001 | 0.002 |
The bold P-values indicate a significant difference (α=0.05; χ2 or Fisher's exact test) between the probabilities within a category. The letters indicate within a category which lines differ from each other in the transition from one category to the next; that is, the letters indicate per behavioural category which lines have the same courtship behaviour.
Figure 5.
F2 male sterility. F2 male sterility for three different types of sterility: behavioural sterility, spermatogenic sterility and total sterility (combining different sterilizing factors). The percentage of spermatogenic sterility is based on mated males only, because for the unmated males the spermatogenic sterility could not be determined. Sample sizes are indicated in the bars; the letters indicate a significant difference between the different types of males for a particular type of sterility based on a Tukey test for multiple comparisons (α=0.05) (different shadings of letters indicates which values have been tested against each other).
We estimated the level of spermatogenic sterility of F2 males by scoring the daughter production of those males that had successfully copulated (Figure 5). The estimated level of spermatogenic sterility in pure species males is less than 3%. The level of spermatogenic sterility in hybrid males is much higher, with the highest level for VL[L] males (57% compared with 21% for LV[V]). Using a Tukey-type multiple comparison to determine which crosses differed from each other, we found that spermatogenic sterility (Figure 5) of the two hybrid crosses is higher than that of the pure VV[V] cross, but the level of sterility in the LV[V] cross is equal to pure LL[L].
Combining behavioural and spermatogenic sterility gives a measure for total sterility of F2 males (Figure 5). A large percentage of hybrid F2 males are sterile, whereas the percentage of sterile F2 pure males is much smaller. All types of males differed significantly from each other in the percentage of total sterility (Tukey-type multiple comparison, α=0.05). Hybrids with N. longicornis cytoplasm had a higher level of sterility than hybrids with N. vitripennis cytoplasm. This is also reflected by the genotypic data (described in more detail below). When comparing the genotypes of fertile and sterile males from the two hybrid crosses for the occurrence of N. vitripennis alleles on the five different chromosomes, only the VL[L] data set shows a strong bias towards N. longicornis alleles in fertile males (P<0.0001). This effect is evident for loci on chromosomes 2–5, but strongest for chromosomes 2 and 5.
Transmission ratio-distorting loci
After the F2 hybrid males were tested for sterility, they were genotyped with the 50 microsatellite markers that were used to construct the linkage map (markers Nv324 and Nv344 were also used, although they could not be mapped). A total of 126 LV[V] and 124 VL[L] F2 males were analysed, from which only one LV[V] sample was removed because of bad amplification. Each marker was tested for a deviation from the expected ratio of 1:1 of parental alleles by using χ2-tests after Yates and Bonferroni corrections.
The marker segregation data of the LV[V] hybrids suggested a TRDL on chromosome-1 at marker Nv22 and at all markers on chromosome-5 in the region between and including Nv109 and Nv325. In all cases the distortion was in favour of the N. vitripennis (V) allele. No significant marker segregation bias was found on chromosomes 2, 3 and 4. Marker segregation data in hybrids from the reciprocal cross (VL[L]) suggested a TRDL on chromosome-4 from and including marker Nv26 to Nv342 and on chromosome-5 for all the markers. In all cases the distortion was in favour of the N. longicornis (L) allele. Chromosomes 1, 2 and 3 were not distorted in their marker segregation. The distortion is illustrated in Figure 1 as the percentage of V alleles in the F2 males of each hybrid cross. It is evident that in both hybrid crosses the microsatellite alleles are distorted towards the species that delivered the cytoplasm (including the mitochondria): N. vitripennis for LV[V] and N. longicornis for VL[L].
To estimate the number of TRDLs that could explain the observed distortion in the genotypic data, we applied a Bayesian mapping approach. For the F2 hybrids with N. vitripennis cytoplasm, it predicted a single TRDL on chromosome-1 with a posterior probability (pP) of 73% and a single TRDL on chromosome-5 with a pP of 75.2%. For the F2 hybrids with N. longicornis cytoplasm, the Bayesian analysis was much less distinct. The analysis suggested one or two TRDLs on chromosome-4 with a pP of 47.5% and 43.2% respectively, and an additional TRDL on chromosome-5 (pP of 59.4%). The analysis for chromosome-3 revealed the presence of one TRDL based on the Bayesian method (pP of 62.8%), whereas the χ2 failed to identify marker segregation distortion on that chromosome. The results of the Bayesian analysis with the number and location of the TRDLs are shown in Figure 6.
Figure 6.
Estimated position of predicted TRDLs in hybrid F2 males. Shown are density distributions of the pP for the position of the TRDLs in adult male F2 hybrids (chromosome-2 is not shown because it lacks TRDLs). Each distribution was calculated from 20 000 Markov chain Monte Carlo samples taken from the stationary phase and assuming a Poisson prior (λ=0.5) for the number of TRDL. The pP values for these distributions of single TRDLs were 73.00, 62.79, 47.46, 75.20 and 59.73%. The hybrid cross in which the TRDL distribution was found is indicated above the distribution. Chromosome numbers are according to Rütten et al. (2004).
We tested whether the genotype of one TRDL was dependent on the genotype of another TRDL by applying a χ2-test to the most distorted markers in our two F2 hybrid data sets. In F2 hybrids with N. vitripennis cytoplasm, we tested Nv22 (chromosome-1) and Nv46 (chromosome-4), and in F2 hybrids with N. longicornis cytoplasm, we tested Nv337 (chromosome-4) and Nv326 (chromosome-5). The comparison of the observed recombinant and non-recombinant genotypes to those expected when the two respective markers segregate independently, revealed in neither case any evidence for a two-way interaction (P=0.20 and 0.48 for N. vitripennis and N. longicornis cytoplasm, respectively). Given that the Bayesian mapping approach of TRDLs suggested slightly different positions of the TRDLs for hybrids with N. longicornis cytoplasm (on chromosomes 4 and 5, and an additional TRDL on chromosome-3) compared with the χ2 method, we conducted a 2 × 2 × 2 χ2-test on the genotypes of the markers Nv300, Nv46 and Nv302. Again, we found no evidence for an interdependence of the markers (P=0.16).
To assess the impact of nuclear–nuclear incompatibilities on hybrid mortality in general, we tested for significant deviations from the observed and expected ratio of recombinant and non-recombinant genotypes of all marker pairs for which the individual markers are on different chromosomes with the aid of a χ2-test. To account for linkage, and thus interdependence of markers on a given chromosome and multiple testing, we simulated 10 000 random mapping populations with exactly the same marker order and marker distances as in our experiments, but with no marker segregation bias. Analysing the simulated data sets, we found χ2-values larger than 12.304 for the conducted pairwise tests to occur in less than 5% of the cases. We therefore chose this value to assess significance in our F2 hybrid populations. None of the χ2-values from our F2 hybrid genotypes with N. longicornis cytoplasm exceeded the threshold value of 12.304. However, the χ2-tests of marker pairs in hybrids with N. vitripennis cytoplasm revealed a significant deviation from the expected values for marker pair Nv306 (chromosome-2) and Nv339 (chromosome-4) (χ2=12.778).
Discussion
We tested for the presence of sex-specific hybrid incompatibilities in the wasp genus Nasonia by investigating reciprocal crosses of N. vitripennis (V) and N. longicornis (L) to estimate mortality and sterility in males and females of pure strain and hybrid offspring. Compared with pure N. vitripennis, the F1 female offspring of the hybrid crosses and the pure N. longicornis cross suffered from significant mortality. This mortality is, however, due to environmental factors rather than a disruption of gene interactions in hybrid females. There are several arguments for this. First, mortality of the pure N. longicornis strain varied between generations and was not severe in the F2 control cross. This is most likely due to variable host quality. Second, as all F1 female offspring of a particular cross were genetically identical, any difference in mortality between individuals must have been environmentally induced. These observations are consistent with the biology of the species; N. vitripennis is a generalist parasitoid wasp and N. longicornis is a specialist of Protocalliphora blowfly pupae, a host species that we are unable to culture in the lab. N. longicornis is therefore more likely to be sensitive to fluctuations in host quality. In addition, the number of offspring produced by F1 hybrid females was lower than that of pure strain N. vitripennis, but it was equal to that of pure strain N. longicornis. A likely reason for this is that N. vitripennis females have eight ovarioles, whereas N. longicornis females and the reciprocal hybrids have only six each (E Geuverink, personal communication).
Our data revealed that first-generation hybrid male (F2) mortality was significantly larger than F2 pure male mortality, but much smaller than that reported for hybrids of N. vitripennis and N. giraulti (Breeuwer and Werren, 1995). This difference in hybrid mortality between species pairs could be due to the different Nasonia species that were used, but also due to differences in experimental conditions (such as host species), as environmental factors appear to also affect the level of F1 female mortality. This large effect of environmental conditions on the level of mortality obstructs the direct comparison of F1 and F2 mortality, because the subsequent generations experience different conditions. This is why in our experiment we always need to compare the hybrid mortality to the pure species mortality in the same generation in order to infer the level of mortality of the hybrids.
We found that F2 male sterility was higher in hybrids than in pure strain individuals, whereas F1 female sterility was not. The sterility test for pure LL[L] males differed from that of the other types of males in that N. longicornis rather than N. vitripennis virgin females were used. Nevertheless, the transition probabilities of the pure F2 males were in most cases similar, which shows that both pure strain transition probabilities are a good control for the two hybrids. The F2 hybrid male transition probabilities showed that hybrid males suffered greatly from behavioural sterility, similar to hybrids of N. vitripennis and N. giraulti (Clark et al., 2010). As the transition probabilities of both types of hybrids were equal, the cytoplasmic background does not affect the male's behaviour and the dysfunctions in courtship behaviour are most likely due to disruption of nuclear (courtship) genes. Hybrids with N. vitripennis cytoplasm suffered less from spermatogenic sterility than those with N. longicornis cytoplasm, which is in line with hybrids of N. vitripennis and N. giraulti (Clark et al., 2010). A role for cytoplasmic factors in inducing male (spermatogenic) sterility has been proposed previously (Ehrman, 1963; Mishra and Singh, 2005), but currently receives little scientific interest.
This is the first systematic study on sex-specific hybrid incompatibilities in a haplodiploid species pair. We found sex-specific hybrid incompatibilities, with heterozygous females suffering less than hemizygous males for hybrids of N. vitripennis and N. longicornis, which is largely consistent with limited data obtained from N. vitripennis and N. giraulti (Breeuwer and Werren, 1995). These results are in concordance with predictions by Haldane (1922) and Koevoets and Beukeboom (2009) about the presence of sex-specific hybrid incompatibilities in species with haplodiploid sex determination. We do, however, stress that the mechanisms that lead to Haldane's rule in haplodiploids can differ from those in diploids. As discussed below, cytonuclear incompatibilities seem to cause hybrid incompatibilities in Nasonia, and the disruption of the oxidative phosphorylation (OXPHOS) pathway is a likely candidate for causing hybrid mortality. If dominance has a large role in hybrid incompatibilities, then the cytonuclear interactions are unlikely to lead to hybrid incompatibilities in F1 hybrids of diploid species. This follows from the fact that the nuclear OXPHOS genes are predominantly located on the autosomes (more than 84% of the nuclear OXPHOS genes are located on the autosomes in Drosophila; based on MitoDrome: http://mitodrome.ba.itb.cnr.it). Thus recessive incompatibilities between the autosomal genes and the cytoplasm would always be masked by heterozygosity in diploid F1 hybrid offspring.
In this study we took the first step in studying Haldane's rule in Nasonia: We observed that diploid females suffer less from hybridization than haploid males. F1 female hybrids are mostly viable and fertile, whereas a significant percentage of hybrid males are inviable and the few surviving hybrid males are mostly sterile. F1 hybrid females and F2 hybrid males of haplodiploid species are, however, not fully comparable in their genetic make-up. F1 hybrid females inherit one intact genome complement from each parental species, that is, they are heterozygous for all diverged loci. F2 hybrid males inherit a single recombined chromosome set from their mother and each locus has either an N. vitripennis or an N. longicornis allele. This difference in genetic make-up between males and females can be uniquely exploited for finding the underlying mechanisms of Haldane's rule in Nasonia. If only dominance effects are responsible for the hybrid incompatibilities, then heterozygous hybrid females are not expected to suffer from incompatibilities. Our results are consistent with these dominance effects; however, if faster-male effects solely explain the incompatibilities in this cross, then hybrid females are not expected to suffer from incompatibilities either. As F1 diploid hybrid offspring possess one chromosome set from both parental species, the magnitude of hybrid incompatibilities in F1 hybrids is highly dependent on dominance effects, regardless of the sex of the hybrid. Therefore, the next step in studying Haldane's rule in Nasonia and to distinguish between the dominance theory and the faster-male theory is to generate partly homozygous hybrid females by backcrossing F1 hybrid females to both parental species. If dominance effects explain Haldane's rule in Nasonia, the obtained hybrid females should suffer from hybrid incompatibilities. However, if faster-male effects explain Haldane's rule instead, the obtained hybrid females should not suffer from hybrid incompatibilities. Note, however, that F2 females derived from backcrosses will still be heterozygous for, on average, 50% of their genome. Therefore, the resulting mortality/sterility will be obscured by heterozygosity if dominance has a large role. A specific TRDL in F2 haploid males will show a significant bias towards heterozygosity in F2 backcross hybrid females under the dominance theory. By contrast, the faster-male theory does not predict a bias at such TRDL in females. Our preliminary data suggest that the TRDLs that we identified in this study have different effects in F2 backcross females. This indicates that Haldane's rule is likely a composite phenomenon in Nasonia. A full analysis of these backcross F2 females will be published later.
We have identified the genomic regions that are involved in causing F2 hybrid male mortality by using microsatellite markers. Hybrid mortality in crosses between N. vitripennis and N. longicornis is highly genotype-specific. N. vitripennis nuclear genes on chromosomes 4 and 5 appear to be disrupted in their function if they are in N. longicornis cytoplasm and N. longicornis nuclear genes on chromosomes 1 and 5 are disrupted in their function if they are in N. vitripennis cytoplasm. This asymmetry of incompatibilities when studying reciprocal crosses is a common phenomenon for incompatibilities between few loci (Turelli and Moyle, 2007). Candidates for a stringent co-evolution of nuclear and cytoplasmic genes are those coding for the OXPHOS pathway, whose efficacy is most likely also dependent on environmental factors (Rawson and Burton, 2002). Nuclear and mitochondrial genes are interacting in four out of five OXPHOS complexes (only complex-II consists of nuclear products only). Ellison et al. (2008) found that OXPHOS complexes in Nasonia hybrids are less functional than those of pure species. Gibson et al. (2010) mapped the nuclear-encoded OXPHOS genes in Nasonia and found that chromosomes 1, 2, 4 and 5 encode for OXPHOS genes with non-synonymous substitutions among Nasonia species. Whether these substitutions are responsible for the observed hybrid mortalities needs to be validated by functional analysis of the genes.
A study on hybrids of N. vitripennis and N. giraulti suggested nuclear–nuclear hybrid incompatibilities being partially responsible for hybrid mortality (Breeuwer and Werren, 1995). Here, we screened for nuclear–nuclear incompatibilities by means of analysing linkage disequilibrium between markers from different chromosomes. Our data revealed one significant deviation from the expected ratio of recombinant and non-recombinant genotypes between two markers, one on chromosome-2 and one on chromosome-4 in hybrids with N. vitripennis cytoplasm. As we found no linkage disequilibrium between these two markers in the reciprocal cross, the interdependency might be part of a higher order incompatibility that includes a cytoplasmically inherited genetic factor. Niehuis et al. (2008) also found only evidence for cytonuclear incompatibilities between N. vitripennis and N. giraulti. Our data and those of Niehuis et al. (2008) indicate that cytonuclear incompatibilities have a primary role in hybrid incompatibilities in Nasonia. Whether or not this is a unique characteristic of the Nasonia genus remains to be investigated. However, given that the mitochondrial genome typically evolves faster than the nuclear genome and that cytonuclear incompatibilities in Nasonia are likely recessive, it appears likely that cytonuclear incompatibilities are more widespread and may have only remained undetected in diploid organisms because of dominance effects (but see Burton et al., 2006).
We have located TRDLs responsible for the hybrid incompatibilities between N. vitripennis and N. longicornis, which are in congruence with previous results from N. vitripennis and N. giraulti. Our data suggest nuclear loci on chromosomes 4 and 5 whose N. vitripennis allele is incompatible with an allospecific cytoplasm (that is, that of N. longicornis). Niehuis et al. (2008) postulated a similar locus on chromosome-4 only. Furthermore, we identified nuclear loci on chromosomes 1 and 5, which are incompatible with N. vitripennis cytoplasm when having an allospecific nuclear allele. Niehuis et al. (2008) again found similar results when studying N. vitripennis and N. giraulti. Although the currently available mapping data are not accurate enough to evaluate the evolution of hybrid incompatibilities in the Nasonia genus in detail, it appears that most of the mapped hybrid incompatibilities evolved prior to the split of N. longicornis and N. giraulti. This is supported by the lack of hybrid incompatibilities between these species (Bordenstein et al., 2001). The true nature of the gene interactions can ultimately only be fully assessed by identifying the genes that underlie the hybrid incompatibilities.
We have found a large effect of the cytotype on F2 hybrid male sterility and mortality, indicative of nuclear mitochondrial crosstalk (illustrated by our genetic analysis). Cytotype did not affect F1 hybrid female sterility and mortality. The disruption of the nuclear–mitochondrial crosstalk could explain our finding of sex-specific hybrid incompatibilities in Nasonia. Different Nasonia species are infected by different strains of Wolbachia and these Wolbachia species seem to have caused mitochondrial sweeps during their spread (Raychoudhury et al., 2010b). This could have strengthened the co-evolution of nuclear and mitochondrial genes in Nasonia. Furthermore, cytonuclear incompatibilities can have different effects on male and female mortality (for example, different energy requirements) and sterility (no role of mitochondria reported during oogenesis).
We have studied hybrid incompatibilities in F1 and F2 female and male hybrids of N. vitripennis and N. longicornis, and we have found that this genus of haplodiploid wasps follows a sex-specific pattern described by Haldane's rule. Hybrid incompatibilities in Nasonia cause both mortality and sterility, and the specific level is largely controlled by the cytoplasm, suggesting cytonuclear incompatibilities. Although haplodiploids have been ignored so far when studying Haldane's rule, our data suggest that haplodiploids at least follow the predictions of the rule in that the heterozygous sex suffers less than the hemizygous sex. We encourage other researchers to test these sex-specific hybrid incompatibilities in more haplodiploid systems in order to identify the generality of Haldane's rule and to facilitate the identification of the genetic mechanisms underlying Haldane's rule.
Acknowledgments
We are grateful to S Ferber for help with hosting the wasps; V de Haan and B Verheijen for the pilot of the sterility experiment; T Schwander for help with statistical analyses; J Gadau for discussions on the experimental setup and three anonymous reviewers for valuable comments on the manuscript. This work has been made possible by Grant ALW 816.01.004 and Pioneer Grant ALW 833.02.003 of the Netherlands Organisation for Scientific Research to LWB, and a Feodor Lynen Research Fellowship for Postdoctoral Researchers of the Alexander von Humboldt Foundation to ON.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Baines JF, Sawyer SA, Hartl DL, Parsch J. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol Biol Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Presgraves DC, Swanson WJ. A test for faster X evolution in Drosophila. Mol Biol Evol. 2002;19:1816–1819. doi: 10.1093/oxfordjournals.molbev.a004006. [DOI] [PubMed] [Google Scholar]
- Bierne N, Bonhomme F, Boudry P, Szulkin M, David P. Fitness landscapes support the dominance theory of post-zygotic isolation in the mussels Mytilus edulis and M. galloprovincialis. Proc Royal Soc B Biol Sci. 2006;273:1253–1260. doi: 10.1098/rspb.2005.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, O'Hara FP, Werren JH. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 2001;409:707–710. doi: 10.1038/35055543. [DOI] [PubMed] [Google Scholar]
- Breeuwer JAJ, Werren JH. Hybrid breakdown between 2 haplodiploid species—the role of nuclear and cytoplasmic genes. Evolution. 1995;49:705–717. doi: 10.1111/j.1558-5646.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- Burton RS, Ellison CK, Harrison JS. The sorry state of F-2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am Nat. 2006;168:S14–S24. doi: 10.1086/509046. [DOI] [PubMed] [Google Scholar]
- Campbell BC, Steffen-Campbell JD, Werren JH. Phylogeny of the Nasonia species complex (Hyemoptera: Pteromalidae) inferred from an internal transcribed spacer (ITS2) and 28S rDNA sequences. Insect Mol Biol. 1993;2:225–237. doi: 10.1111/j.1365-2583.1994.tb00142.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex-chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Civetta A, Singh RS. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J Mol Evol. 1995;41:1085–1095. doi: 10.1007/BF00173190. [DOI] [PubMed] [Google Scholar]
- Clark ME, O'Hara FP, Chawla A, Werren JH. Behavioural and spermatogenic hybrid male breakdown in Nasonia. Heredity. 2010;104:289–301. doi: 10.1038/hdy.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. ‘Patterns of speciation in Drosophila' revisited. Evolution. 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- Darling DC, Werren JH. Biosystematics of Nasonia (Hymenoptera, Pteromalidae)—2 new species reared from birds nests in North-America. Ann Entomol Soc Am. 1990;83:352–370. [Google Scholar]
- Eads BD, Colbourne JK, Bohuski E, Andrews J.2007Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex BMC Genomics 8464doi: 10.1186/1471-2164-8-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrman L. Apparent cytoplasmic sterility in Drosophila paulistorum. Proc Natl Acad Sci USA. 1963;49:155–157. doi: 10.1073/pnas.49.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Niehuis O, Gadau J. Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J Evol Biol. 2008;21:1844–1851. doi: 10.1111/j.1420-9101.2008.01608.x. [DOI] [PubMed] [Google Scholar]
- Gadau J, Page RE, Werren JH. Mapping of hybrid incompatibility loci in Nasonia. Genetics. 1999;153:1731–1741. doi: 10.1093/genetics/153.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JD, Niehuis O, Verrelli BC, Gadau J. Contrasting patterns of selective constraints in nuclear-encoded genes of the oxidative phosphorylation pathway in holometabolous insects and their possible role in hybrid breakdown in Nasonia. Heredity. 2010;104:310–317. doi: 10.1038/hdy.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Hollocher H, Wu CI. The genetics of reproductive isolation in the Drosophila simulans clade: X vs autosomal effects and male vs female effects. Genetics. 1996;143:1243–1255. doi: 10.1093/genetics/143.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Linares M, Naisbit RE, Salazar C, Yang ZH, Mallet J. Sex-linked hybrid sterility in a butterfly. Evolution. 2001;55:1631–1638. doi: 10.1111/j.0014-3820.2001.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Koevoets T, Beukeboom LW. Genetics of postzygotic isolation and Haldane's rule in haplodiploids. Heredity. 2009;102:16–23. doi: 10.1038/hdy.2008.44. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Singh RS. The molecular basis of speciation: from patterns to processes, rules to mechanisms. J Genet. 2008;87:327–338. doi: 10.1007/s12041-008-0055-x. [DOI] [PubMed] [Google Scholar]
- Malone JH, Michalak P. Physiological sex predicts hybrid sterility regardless of genotype. Science. 2008;319:59. doi: 10.1126/science.1148231. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J.1982Molecular Cloning: a Laboratory Manual11th edn.Cold Spring Harbor Laboratory Press: New York [Google Scholar]
- Mank JE, Nam K, Ellegren H. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 2010;27:661–670. doi: 10.1093/molbev/msp282. [DOI] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci USA. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Singh BN. Why hybrid males are sterile in Drosophila. Curr Sci India. 2005;89:1813–1819. [Google Scholar]
- Musters H, Huntley MA, Singh RS. A genomic comparison of faster-sex, faster-X, and faster-male evolution between Drosophila melanogaster and Drosophila pseudoobscura. J Mol Evol. 2006;62:693–700. doi: 10.1007/s00239-005-0165-5. [DOI] [PubMed] [Google Scholar]
- Niehuis O, Judson AK, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 2008;178:413–426. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Haldane's rule. Annu Rev Ecol Syst. 1997;28:195–218. [Google Scholar]
- Presgraves DC. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics. 2003;163:955–972. doi: 10.1093/genetics/163.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Rawson PD, Burton RS. Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc Natl Acad Sci USA. 2002;99:12955–12958. doi: 10.1073/pnas.202335899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychoudhury R, Desjardins CA, Buellesbach J, Loehlin DW, Grillenberger BK, Beukeboom LW, et al. Behavioural and genetic characteristics of a new species of Nasonia. Heredity. 2010a;104:278–288. doi: 10.1038/hdy.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychoudhury R, Grillenberger BK, Gadau J, Bijlsma R, van de Zande L, Werren JH, et al. Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial–Wolbachia sweep in North America. Heredity. 2010b;104:318–326. doi: 10.1038/hdy.2009.160. [DOI] [PubMed] [Google Scholar]
- Rütten KB, Pietsch C, Olek K, Neusser M, Beukeboom LW, Gadau J. Chromosomal anchoring of linkage groups and identification of wing size QTL using markers and FISH probes derived from microdissected chromosomes in Nasonia (Pteromalidae: Hymenoptera) Cytogenet Genome Res. 2004;105:126–133. doi: 10.1159/000078019. [DOI] [PubMed] [Google Scholar]
- Schilthuizen M, Giesbers MCWG, Beukeboom LW. Haldane's rule in the 21st century. Heredity. 2011;107:95–102. doi: 10.1038/hdy.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Larracuente AM, Sackton TB, Clark AG. Comparative genomics on the Drosophila phylogenetic tree. Annu Rev Ecol Evol Syst. 2009;40:459–480. [Google Scholar]
- Tao Y, Hartl DL. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane's rule. Evolution. 2003;57:2580–2598. doi: 10.1111/j.0014-3820.2003.tb01501.x. [DOI] [PubMed] [Google Scholar]
- Tao Y, Xhen SN, Hartl DL, Laurie CC. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003;164:1383–1397. doi: 10.1093/genetics/164.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: no evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Begun DJ. Haldane's rule and X-chromosome size in Drosophila. Genetics. 1997;147:1799–1815. doi: 10.1093/genetics/147.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. The dominance theory of Haldane's rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen JW, Voorrips RE. JoinMap® Version 3.0: Software for the Calculation of Genetic Linkage Maps. Plant Research International: Wageningen; 2001. [Google Scholar]
- Vogl C, Xu SZ. Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics. 2000;155:1439–1447. doi: 10.1093/genetics/155.3.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Davis AW. Evolution of postmating reproductive isolation—the composite nature of Haldane's rule and its genetic bases. Am Nat. 1993;142:187–212. doi: 10.1086/285534. [DOI] [PubMed] [Google Scholar]
- Wu CI, Johnson NA, Palopoli MF. Haldane's rule and its legacy: why are there so many sterile males. Trends Ecol Evol. 1996;11:281–284. doi: 10.1016/0169-5347(96)10033-1. [DOI] [PubMed] [Google Scholar]
- Zar JH.1999Biostatistical Analysis4th edn.Prentice-Hall: Englewood, Upper Saddle river, NJ [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.