Abstract
Wolbachia are endosymbiotic bacteria known to manipulate the reproduction of their hosts. Some populations of the parasitoid wasp Asobara japonica are infected with Wolbachia and reproduce parthenogenetically, while other populations are not infected and reproduce sexually. Wolbachia-infected A. japonica females regularly produce small numbers of male offspring. Because all females in the field are infected and infected females are not capable of sexual reproduction, male production seems to be maladaptive. We investigated why these females nevertheless produce males. We tested three hypotheses: high rearing temperatures could result in higher offspring sex ratios (more males), low Wolbachia titer of the mother could lead to higher offspring sex ratios and/or the Wolbachia infection is of relatively recent origin and not enough time has passed to allow complete coadaptation between Wolbachia and host. In all, 33% of the Wolbachia-infected females produced males and 56% of these males were also infected with Wolbachia. Neither offspring sex ratio nor male infection frequency was significantly affected by rearing temperature or Wolbachia concentration of the mother. The mitochondrial DNA sequence of one of the uninfected populations was identical to that of two of the infected populations. Therefore, the initial Wolbachia infection of A. japonica must have occurred recently. Mitochondrial sequence variation among the infected populations suggests that the spread of Wolbachia through the host populations involved horizontal transmission. We conclude that the occasional male production by Wolbachia-infected females is most likely a maladaptive side effect of incomplete coevolution between symbiont and host in this relatively young infection.
Keywords: Asobara japonica, Wolbachia, parthenogenesis, sex ratio, quantitative PCR, mitochondrial DNA
Introduction
Wolbachia are maternally inherited, intracellular, symbiotic bacteria belonging to the order Rickettsiales within the α-Proteobacteria. It has been estimated that Wolbachia infect about 66% of all insect species, and either some or all individuals per species (Hilgenboecker et al., 2008). To enhance its own transmission, Wolbachia can induce various alterations of the reproduction of its host, such as cytoplasmic incompatibility, feminization, male killing and parthenogenesis induction (PI). Wolbachia-induced parthenogenesis is most commonly found in haplodiploid organisms, such as Hymenoptera (Werren, 1997; Stouthamer et al., 1999; Werren et al., 2008). In uninfected haplodiploid organisms, fertilized eggs develop into diploid daughters and unfertilized eggs develop into haploid sons (arrhenotoky). In haplodiploids, PI Wolbachia cause diploidization of the haploid eggs by alteration of meiotic and/or mitotic processes (Huigens and Stouthamer, 2003; Pannebakker et al., 2004) resulting in the production of daughters from unfertilized eggs (thelytoky). In most cases of Wolbachia-induced parthenogenesis, the infection is fixed and the whole host population consists of females (Huigens and Stouthamer, 2003). Males tend to be absent or rare in such populations. Here, we identify an exception to this pattern and examine its cause.
Asobara japonica (Hymenoptera: Braconidae) is a larval–pupal parasitoid of drosophilid flies and naturally occurs in Japan. Populations of A. japonica on the main islands of Japan exhibit highly female-biased sex ratios (92.7–99.2% females), whereas population sex ratios on the smaller southern islands are not biased (Mitsui et al., 2007). The populations on the main islands are infected with parthenogenesis-inducing Wolbachia, while the populations on the smaller southern islands are not (Kremer et al., 2009). Also, during routine culturing in our lab, Wolbachia-infected A. japonica females regularly produce small numbers of male offspring and in rare cases even male-biased sex ratios. The production of males in parthenogenetic populations of A. japonica seems to be maladaptive, because parthenogenetic females are not capable of sexual reproduction (Kremer et al., 2009).
The occasional male production suggests that transmission of Wolbachia from mother to daughter is not always 100% efficient. In several hosts, infected females loose their Wolbachia when exposed to high temperatures (for example Pijls et al., 1996; Clancy and Hoffmann, 1998; Pintureau et al., 1999; Hurst et al., 2000). Eggs laid at high temperatures would then contain low Wolbachia concentrations, which may cause the effect of Wolbachia to be reduced or lost (Clancy and Hoffmann, 1998; Hurst et al., 2000). In PI Wolbachia-infected females this would lead to male offspring production. A similar effect would be predicted if Wolbachia concentrations are reduced for reasons other than temperature. In contrast, Mouton et al. (2006) found increased Wolbachia densities in the parasitoid wasp Leptopilina heterotoma when reared at high temperatures, but this did not influence the effect of Wolbachia on its host.
An alternative explanation for inefficient transmission of Wolbachia might be that the Wolbachia infection is relatively young. While vertical transmission is the main transmission mode of Wolbachia within established hosts, horizontal transmission plays a major role in the spread of Wolbachia in(to) novel hosts (Hurst et al., 1992; O'Neill et al., 1992; Rousset et al., 1992; Werren et al., 1995; Vavre et al., 1999; Huigens et al., 2000, 2004; Kraaijeveld et al., 2011a). This predicts that upon invasion of a new host population, Wolbachia would initially be selected for efficient horizontal transmission, as well as efficient vertical transmission. This would be followed by adaptation to vertical transmission only once most of the host population is infected. For example, the parasitoid wasp Leptopilina boulardi is infected with a symbiont that manipulates the superparasitism behavior of its host in order to enhance its own horizontal transmission (Varaldi et al., 2003). Successful experimental horizontal transmission of Wolbachia often leads to unstable infections in the new host and reduced or altered expression of the reproductive manipulation (Grenier et al., 1998; Heath et al., 1999; Huigens et al., 2004; Jaenike, 2007). An explanation for such poor vertical transmission might be residual incompatibilities or asynchronies between Wolbachia and the new host (Heath et al., 1999).
In this paper, we investigate why PI Wolbachia-infected A. japonica females produce males. First, we quantified how often and in what numbers male offspring are produced by infected females and whether male production is affected by rearing temperature. We predicted that higher rearing temperatures would result in higher offspring sex ratios (more males). Second, we examined whether male offspring production is influenced by Wolbachia titer of the mother. Quantitative PCR (qPCR) was used to measure Wolbachia concentrations of A. japonica females. We predicted that lower Wolbachia concentrations would lead to higher offspring sex ratios. Last, we used mitochondrial DNA (mtDNA) sequences to date the initial infection of A. japonica with Wolbachia. We hypothesized that the Wolbachia infection in A. japonica is relatively young.
As far as we know, this is the first example from the field of a possible relation between incomplete Wolbachia–host coadaptation and a relatively recent Wolbachia infection.
Materials and methods
A. japonica strains
Five Wolbachia-infected thelytokous strains of the parasitoid wasp A. japonica (Hymenoptera: Braconidae) were used in all experiments: Sapporo, Hirosaki, Sendai, Tokyo and Kagoshima. In addition, two uninfected arrhenotokous strains were used for the mtDNA analysis: Amami and Iriomote. The strains were kindly provided by M.T. Kimura from cultures derived from field samples, collected along the entire length of Japan, described in Mitsui et al. (2007) and Murata et al. (2009). Maps of the sampled locations can be found in both papers. These strains were subsequently maintained in our lab under a partial inbreeding regime. Each generation, three females per strain were allowed to parasitize about 100–200 2-day-old (first or second instar) larvae of Drosophila melanogaster in glass jars with a medium of agar covered by a layer of 2 ml baker's yeast suspension and kept at 25 °C, light:dark 16:8, 65% relative humidity.
Temperature experiment
To test whether temperature affected male production, we examined the offspring sex ratio of Wolbachia-infected thelytokous strains of A. japonica at two temperatures. One-week-old females from five thelytokous strains (Sapporo, Hirosaki, Sendai, Tokyo and Kagoshima) were placed individually in a glass jar with a medium of agar covered by a layer of 2 ml baker's yeast suspension, in which they were allowed to parasitize about 100–200 2-day-old (first or second instar) larvae of D. melanogaster. As thelytokous A. japonica females are not capable of sexual reproduction (Kremer et al., 2009), we assumed that all females used in the sex ratio experiment were virgins. For each strain, half of the females were kept at 25 °C, light:dark 16:8, 65% relative humidity, while the other half were kept at 20 °C, light:dark 16:8, 65% relative humidity. We chose the normal rearing temperature as the highest temperature, because we previously observed that males are produced at that temperature. After 10–13 days, mothers were removed from the jars. The number of male and female offspring was counted several times a week for the next 7 weeks. To count the offspring, wasps were anaesthetized with CO2. Females could be distinguished from males by their ovipositor, which permanently and prominently protrudes from the posterior abdomen. Mothers and offspring were stored separately in 70% ethanol in 1.5 ml eppendorf tubes until DNA extraction.
DNA extraction
DNA extractions were performed using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol, using mini spin columns. Before starting the DNA extraction, each wasp was transferred to a new 1.5 ml eppendorf tube. After evaporation of remaining ethanol, tissue lysis buffer (ATL) was added to the tube and the wasp was crushed using a plastic pestle. The tissue was incubated overnight in proteinase K at 56 °C. The DNA was dissolved in 100 μl elution buffer (AE).
Wolbachia detection
All male offspring were tested for Wolbachia infection by amplifying the Wolbachia-specific wsp and ftsZ genes, using the primers wsp-81F/wsp-691R (Braig et al., 1998; Zhou et al., 1998) and ftsZ-F/ftsZ-R (Holden et al., 1993; Sinkins et al., 1995), respectively. PCRs were performed in a total volume of 20 μl, containing 1 × PCR-buffer (Qiagen), 62.5 μ dNTPs, 1 unit Taq polymerase, 250 n forward primer, 250 n reverse primer and 2 μl DNA template. A PTC-200 DNA Engine Thermal Cycler PCR machine (MJ Research, Waltham, MA, USA) was used for all PCRs. PCR conditions for the wsp gene were as follows: 3 min at 94 °C, then 35 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C, and finally 5 min at 72 °C. PCR conditions for the ftsZ gene were as follows: 3 min at 94 °C, then 35 cycles of 45 s at 94 °C, 1 min at 55 °C and 1 min at 72 °C, and finally 5 min at 72 °C. All PCR products were run on a 2% agarose gel and visualized using ethidium bromide staining.
qPCR analysis
To test whether offspring sex ratios produced by Wolbachia-infected females were affected by Wolbachia titer, we measured Wolbachia concentration of the mothers from the temperature experiment using qPCR. We also tested whether the Wolbachia concentration of the mother affected the percentage of Wolbachia-infected males among her offspring.
qPCRs were performed for the Wolbachia-specific wsp and gatB genes, with the nuclear ITS2 gene as a control. To optimize the qPCRs, we designed new primers for all three genes. Primer sequences for wsp and gatB were based on the sequences for wAjap described in Kraaijeveld et al. (2011b), while the primer sequences for ITS2 were based on the sequence for A. japonica described in Kremer et al. (2009). The following primers were used: wsp-wAjap-F: 5′-GAG GCA AAA TTT ACG CCA GA-3′ and wsp-wAjap-R: 5′-AAC TAG CCC TGA AAT TGC TGT TA-3′, producing a 60-bp amplicon; gatB-wAjap-F: 5′-GAA GCA AAG AGG ATG CAA GC-3′ and gatB-wAjap-R: 5′-TCC TGG CTT ACC TCA ACA GG-3′, producing a 73-bp amplicon; ITS2-Ajap-F: 5′-GGC AAG CAC AAT CAA GGT CT-3′ and ITS2-Ajap-R: 5′-ACA AAA ACA AAT TTT GCG GC-3′, producing a 93-bp amplicon. Each qPCR was performed in a total volume of 10 μl, containing 1 × SybrGreen Mastermix (Roche, Penzberg, Germany), 300 n forward primer, 300 n reverse primer and 2 μl DNA template. A LightCycler 480 Real-Time PCR System (Roche) was used for all qPCRs. qPCR conditions for all genes were as follows: 10 min at 95 °C, then 45 cycles of 10 s at 95 °C, 30 s at 60 °C and 20 s at 72 °C, and finally 5 min at 72 °C. Each qPCR was performed in triplicate.
The resulting Ct values were used to calculate the relative quantity of the focal gene (relative gene quantity=mean PCR efficiency (mean overall Ct value − mean sample Ct value)). To correct for the total amount of DNA, we calculated the ratio between the Wolbachia gene and the control gene (ratio=relative quantity Wolbachia gene/relative quantity control gene).
mtDNA analysis
To estimate the age of the Wolbachia infection, we sequenced a part of the mtDNA of A. japonica from both infected and uninfected strains. We sequenced part of the NADH 1 dehydrogenase (ND1) gene, using the primers ND1-F (Smith and Kambhampati, 1999) and ND1-R (Smith et al., 1999), producing a 447-bp amplicon, and the cytochrome oxidase 1 (CO1) gene, using the primers CO1-1775F/CO1-2413R and CO1-2222F/CO1-2773R (Scheffer and Grissell, 2003), together producing a 997-bp amplicon. PCRs were performed in a total volume of 25 μl, containing 1 × PCR-buffer (Qiagen), 750 μ (extra) MgCl2, 200 μ dNTPs, 1.25 units Taq polymerase, 320 n forward primer, 320 n reverse primer and 2.5 μl DNA template. A PTC-200 DNA Engine Thermal Cycler PCR machine (MJ Research) was used for all PCRs. PCR conditions for both genes were as follows: 5 min at 94 °C, then 40 cycles of 1 min at 94 °C, 1 min at 50 °C and 1 min at 72 °C, and finally 10 min at 72 °C. All PCR products were run on a 2% agarose gel and visualized using ethidium bromide staining.
Sequencing was performed by Macrogen Inc. (Seoul, Korea). Sequences were checked with Sequencher software (version 4.2; Gene Codes, Ann Arbor, MI, USA) and aligned with BioEdit software (version 7.0.9; Hall, 1999).
Haplotype diversity and nucleotide diversity were calculated using DnaSP software (version 5.10; Librado and Rozas, 2009). The haplotype diversity (Hd), the nucleotide diversity (π), the average number of synonymous substitutions per synonymous site between populations (ks) and the average number of nucleotide substitutions per site between populations (Dxy) were determined. Median joining haplotype networks were drawn using Network software (version 4.6.0.0; Bandelt et al., 1999).
To calculate the divergence time between strains, we used the estimates for the CO1 gene mutation rate in the parasitoid wasp Nasonia (Hymenoptera: Pteromalidae), described in Raychoudhury et al. (2010). However, mutation rates can vary considerably between species. Therefore, the calculated divergence times are only very rough estimates and must be interpreted with caution. The mitochondrial mutation rate in Nasonia was estimated to be 3.5–13 times higher than in D. melanogaster (6.2 × 10−8 mutations per site per generation; Haag-Liautard et al., 2008), that is 2.2 × 10−7–8.1 × 10−7 mutations per site per generation (Raychoudhury et al., 2010). The divergence time (in generations) between two populations can be calculated by dividing ks (the average number of synonymous substitutions per synonymous site between populations) by this mutation rate (the number of mutations per site per generation).
Statistical analysis
Statistical analyses were performed in R software (version 2.12. 1; R Developmental Core Team, 2010). Generalized linear models with a binomial error distribution and an empirically estimated scale parameter were used to test for differences in sex ratio and male infection frequency. In the sex ratio model, the number of males was used as the response variable and clutch size as the binomial denominator. In the male infection frequency model, the number of infected males was used as the response variable and the total number of males as the binomial denominator. Significance of explanatory variables (Wolbachia concentration of the mother, rearing temperature and A. japonica strain) was tested by dropping (interactions between) explanatory variables from the model and comparing the resulting change in deviance using an F-test. As the Wolbachia concentration of the mother was measured twice, using two different genes for Wolbachia, the correlation between the two estimates was calculated using the Pearson's product moment correlation method.
Results
Temperature experiment
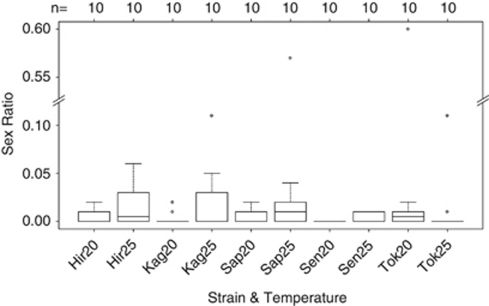
In total, 100 individual females were used: 20 females from each of the five thelytokous strains, of which 10 were kept at 25 °C and the remaining 10 at 20 °C. All females produced offspring (ranging from 5 to 133 individuals). In all, 67 of these females produced only daughters, while 33 females produced both daughters and sons (ranging from 1 to 69 sons per clutch). The mean sex ratio was 2%, ranging from 0 to 60% per clutch. Most of the male producing females produced small numbers of males, with sex ratios ranging from 1 to 11%. Only two females produced higher sex ratios (Sapporo at 25 °C: clutch size=121, 57% males and Tokyo at 20 °C: clutch size=45, 60% males). There were no significant differences in sex ratio between rearing temperatures, strains, or their interaction (Figure 1, Supplementary Table 1; overall: F9,90=1.77, P=0.09; interaction: F4,90=1.18, P=0.33; temperature: F1,98=1.08, P=0.30; strain: F4,95=1.46, P=0.22).
Figure 1.
Sex ratio (proportion males) per strain and rearing temperature (20 °C and 25 °C). Number of clutches (n=sample size) are indicated above the graph. The horizontal dark lines represent the median sex ratios, the bottom and top of the boxes indicate the 25th and 75th percentiles, the whiskers show up to 1.5 times the interquartile range and the dots represent outliers. Hir, Hirosaki; Kag, Kagoshima; Sap, Sapporo; Sen, Sendai; Tok, Tokyo.
Wolbachia detection
The two Wolbachia-specific genes used in the qPCRs were amplified in all samples. Therefore, all mothers were infected with Wolbachia.
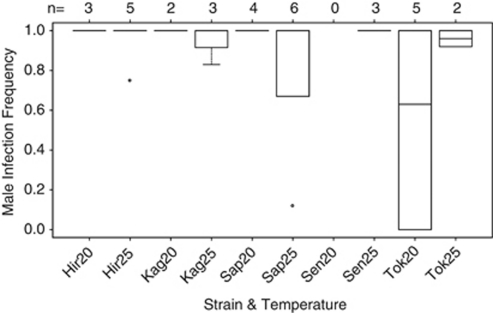
In total, 177 sons were produced by 33 thelytokous females. Of these, 100 (56%, produced by 31 females) were infected with Wolbachia. The male infection frequency per clutch ranged from 0 to 100% (mean 88%). There was a significant interaction effect between rearing temperatures and strains on male infection frequency (Figure 2; overall: F8,24=5.99, P<0.001; interaction: F3,24=4.05, P=0.02). However, this effect appeared to be spurious. When we removed two outlying data points with extreme sex ratios, there were no significant differences in male infection frequency between rearing temperatures, strains or their interaction (Figure 2, Supplementary Table 1; overall: F8,22=1.41, P=0.25; interaction: F3,22=2.16, P=0.12; temperature: F1,29=0.58, P=0.45; strain: F4,26=0.71, P=0.59).
Figure 2.
Male infection frequency (proportion infected males) per strain and rearing temperature (20 °C and 25 °C). Number of clutches (n=sample size) are indicated above the graph. The horizontal dark lines represent the median male infection frequencies, the bottom and top of the boxes indicate the 25th and 75th percentiles, the whiskers show up to 1.5 times the interquartile range and the dots represent outliers. Hir, Hirosaki; Kag, Kagoshima; Sap, Sapporo; Sen, Sendai; Tok, Tokyo.
qPCR analysis
The Wolbachia concentration of 64 females from the temperature experiment was measured. Half of these females produced both daughters and sons, while the other half produced only daughters (gatB: nwith sons=32, nwithout sons=32; wsp: nwith sons=30, nwithout sons=31). The two estimates for Wolbachia concentration (gatB/ITS2 and wsp/ITS2) correlated significantly with each other (r=0.92, t=17.90, df=59, P<0.0001).
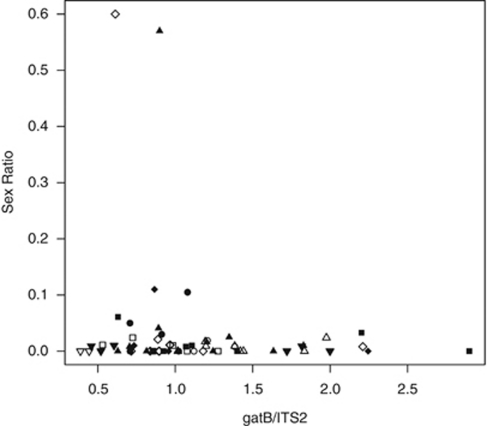
There were no significant relations between sex ratio and Wolbachia concentration of the mother, rearing temperatures, strains or their interactions (gatB: Figure 3; overall: F7,56=0.08, P=0.99; Wolbachia: F1,62=1.20, P=0.28; temperature: F1,62=0.75, P=0.39; strain: F1,62=0.15, P=0.70; none of the interactions between explanatory variables were significant; wsp: overall: F7,53=0.30, P=0.95; Wolbachia: F1,59=0.32, P=0.58; temperature: F1,59=0.47, P=0.50; strain: F1,59=0.04, P=0.85; none of the interactions between explanatory variables were significant).
Figure 3.
Relation between sex ratio (proportion males) and Wolbachia concentration (measured as the ratio between the Wolbachia-specific gene gatB and the control gene ITS2) per rearing temperature (white symbols: 20 °C, black symbols: 25 °C) and strain (squares: Hirosaki, circles: Kagoshima, upward triangles: Sapporo, downward triangles: Sendai, diamonds: Tokyo).
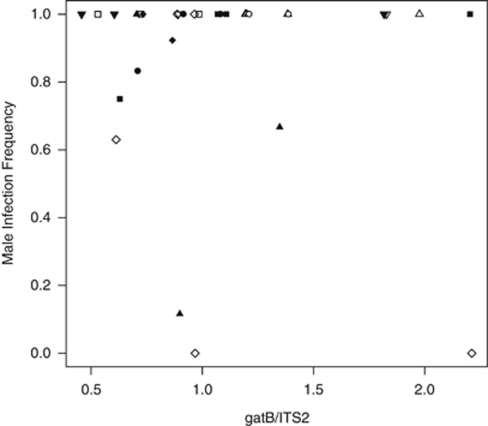
Also, there were no significant relations between male infection frequency and Wolbachia concentration of the mother, rearing temperatures, strains or their interactions (gatB: Figure 4; overall: F7,24=1.05, P=0.42; Wolbachia: F1,30=1.17, P=0.29; temperature: F1,30=2.20, P=0.15; strain: F1,30=0.03, P=0.86; none of the interactions between explanatory variables were significant; wsp: overall: F7,22=1.24, P=0.33; Wolbachia: F1,28=0.01, P=0.91; temperature: F1,28=3.73, P=0.06; strain: F1,28=0.53, P=0.47; none of the interactions between explanatory variables were significant).
Figure 4.
Relation between male infection frequency (proportion infected males) and Wolbachia concentration (measured as the ratio between the Wolbachia-specific gene gatB and the control gene ITS2) per rearing temperature (white symbols: 20 °C, black symbols: 25 °C) and strain (squares: Hirosaki, circles: Kagoshima, upward triangles: Sapporo, downward triangles: Sendai, diamonds: Tokyo).
mtDNA analysis
We sequenced part of the mtDNA of seven females, representing five thelytokous and two arrhenotokous strains of A. japonica. The sequences have been submitted to the GenBank database (CO1 accession numbers: JF430425–JF430431; ND1 accession numbers: JF430432–JF430438).
We found five unique mitochondrial haplotypes (Hd=0.8571). Two thelytokous strains (Hirosaki and Sendai) and the arrhenotokous strain Amami exhibited the same DNA sequence and thus shared the same haplotype, while the other four strains (Sapporo, Tokyo, Kagoshima and Iriomote) all exhibited unique haplotypes (Figure 5). These five haplotypes contained 55 polymorphic sites (π=0.0111), of which 50 were synonymous and five were non-synonymous mutations. The arrhenotokous strain Iriomote differed with 51–53 mutations from all the other haplotypes (ks=0.1741; Dxy=0.0360). Within these other four haplotypes there were zero to three mutations between strains and four polymorphic sites (π=0.0011; three synonymous and one non-synonymous mutations).
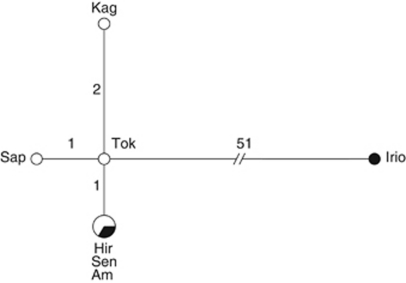
Figure 5.
Haplotype network based on 997 bp of the mitochondrial CO1 gene and 447 bp of the mitochondrial ND1 gene of A. japonica. Each circle represents one haplotype. The size of the circle represents the haplotype frequency. White circles indicate haplotypes found in thelytokous strains, black circles indicate haplotypes found in arrhenotokous strains, mixed black and white circles indicate haplotypes shared by thelytokous and arrhenotokous strains. Strain names are given for the haplotype in which they were found. Lines show mutational routes between haplotypes. The length of the line and the numbers on the line indicate the number of mutations between haplotypes. Am, Amami; Hir, Hirosaki; Irio, Iriomote; Kag, Kagoshima; Sap, Sapporo; Sen, Sendai; Tok, Tokyo.
The five haplotypes could be distinguished within the CO1 gene sequence (Hd=0.8571). Within the CO1 gene, the five haplotypes contained 40 polymorphic sites (π=0.0118; 37 synonymous and 3 non-synonymous mutations). The arrhenotokous strain Iriomote differed with 36–38 mutations from all the other haplotypes (ks=0.1788; Dxy=0.0371). Within these other four haplotypes there were zero to three mutations between strains and four polymorphic sites (π=0.0016; three synonymous and one non-synonymous mutations). Only two haplotypes were found for the ND1 gene sequence (Hd=0.2857). All thelytokous strains (Sapporo, Hirosaki, Sendai, Tokyo and Kagoshima) and the arrhenotokous strain Amami exhibited the same ND1 haplotype, while the second ND1 haplotype was only represented by the arrhenotokous strain Iriomote. The two ND1 haplotypes differed by 15 polymorphic sites (π=0.0096; 13 synonymous and 2 non-synonymous mutations; ks=0.1625; Dxy=0.0336).
The mean divergence time, based only on the CO1 gene, between the arrhenotokous strain Iriomote and the other six strains (ks=0.1788) was estimated to be between 8.2 × 105 and 2.2 × 105 generations ago. Because the arrhenotokous strain from Amami exhibited the same mitochondrial haplotype as two thelytokous strains, we concluded that these strains have not (yet) diverged in their mtDNA.
Discussion
PI Wolbachia-infected A. japonica females regularly produced small numbers of male offspring and rarely male-biased offspring sex ratios. Slightly more than half of these males were infected with Wolbachia. Neither offspring sex ratio nor male infection frequency was affected by strain, rearing temperature or Wolbachia concentration of the mother.
Within seven strains of A. japonica, we found five unique mitochondrial haplotypes. Four of these haplotypes, represented by six strains, were closely related, while the uninfected arrhenotokous strain from Iriomote was very different from the rest. The uninfected arrhenotokous strain from Amami exhibited the same haplotype as two thelytokous strains from Hirosaki and Sendai. More or less the same pattern, based on 645 bp of the mitochondrial CO1 gene, was found by Murata et al. (2009).
Fertile crosses between males and females from Iriomote and Amami, and between (natural and cured) males from Kagoshima and females from Amami indicate that individuals from these strains belong to the same species (Murata et al., 2009; Kraaijeveld et al., 2011b). However, Murata et al. (2009) also found indications for weak asymmetrical sexual isolation, suggesting that Iriomote and Amami have been geographically isolated for a long time. Based on the mDNA sequences, we estimated that the divergence time between Iriomote and the other strains was between 8.2 × 105 and 2.2 × 105 generations ago. However, given that these estimates are extrapolated from mutation rates in a different species (Nasonia), they should be interpreted with caution.
The uninfected arrhenotokous strain from Amami exhibited the same mitochondrial haplotype as two thelytokous strains. There was also very little mitochondrial variation between Amami and the other three thelytokous strains. Moreover, there is no variation between the Wolbachia strains in the five thelytokous A. japonica strains (Kraaijeveld et al., 2011b). This suggests that the Wolbachia infection in A. japonica is relatively young. An alternative possibility might be that the strain from Amami was infected with Wolbachia before, but has lost its infection. However, this seems unlikely because thelytokous females that have been cured from their Wolbachia infection with antibiotics are not capable of sexual reproduction (Kremer et al., 2009).
New bacterial symbiont infections can spread rapidly in host populations. Invasions of Wolbachia in Drosophila simulans in California and Rickettsia in Bemisia tabaci in Arizona have been reported in which the infection frequency increased from 0% to near fixation in <100 generations (Turelli and Hoffmann, 1991; Himler et al., 2011). The mitochondrial variation among the five thelytokous strains suggests that multiple infection events have occurred and that the Wolbachia infection has spread (partly) via horizontal transmission. The five infected thelytokous strains were collected from the two Japanese main islands, with 1580 km distance between the northernmost location Sapporo and the southernmost location Kagoshima (for maps of the locations see Mitsui et al., 2007 and Murata et al., 2009). The distance between the two main Japanese islands is very small (20 km) and the Wolbachia infection probably spread easily between the two islands. However, the distance between the island of Amami and the main islands of Japan may be too large (290 km) for the Wolbachia infection to invade Amami. No geographical gradient could be distinguished from the mitochondrial data. Although Kagoshima and Amami are geographically closest to each other (370 km), based on their mtDNA they are the least closely related within the ‘thelytokous' population. Also, Hirosaki and Sendai are geographically distant from Amami (1680 km and 1520 km, respectively), but based on their mtDNA they are closely related.
The occasional male production by PI Wolbachia-infected A. japonica females is most likely due to a relatively young Wolbachia infection. There may have been too little time to allow complete coevolution between Wolbachia and A. japonica. Incomplete coadaptation may be caused by remaining incompatibilities or asynchronies between Wolbachia and its host (Heath et al., 1999). The incomplete adaptation of Wolbachia to its host A. japonica may lead to incomplete diploidization of the haploid eggs, so that part of the eggs remains haploid and develop into sons. As thelytokous A. japonica females are not capable of sexual reproduction (Kremer et al., 2009), these males will have zero fitness.
The inability of thelytokous A. japonica females to reproduce sexually could be due to selection against the maintenance of costly sexual traits, or due to accumulation of neutral mutations (Pijls et al., 1996). However, this interpretation seems at odds with the recent origin of the Wolbachia infection in this species. The spread of parthenogenesis-inducing Wolbachia may be facilitated by the concomitant spread of ‘functional virginity mutations' (Stouthamer et al., 2010). Mutations that prevent females from fertilizing their eggs will have a selective advantage in the presence of PI Wolbachia-infected females because they induce the bearer to produce more sons which will have many mating opportunities. Virginity mutations may thus explain both the rapid spread of PI Wolbachia through the population of A. japonica and the inability of A. japonica females to reproduce sexually.
We conclude that the occasional male production by PI Wolbachia-infected A. japonica females is not due to high rearing temperatures or low Wolbachia concentrations of the mother, but most likely is a maladaptive side effect of the relatively young age of the Wolbachia infection. Wolbachia possibly is not (yet) fully adapted to its host A. japonica.
Data Archiving
Data of the temperature experiment, Wolbachia detection and qPCR experiment have been deposited at Dryad: doi: 10. 5061/dryad.5qm51. Sequence data have been deposited at GenBank: CO1 accession numbers: JF430425–JF430431; ND1 accession numbers: JF430432–JF430438.
Acknowledgments
We would like to thank Rolf Vossen from the department of Human Genetics at the Leiden University Medical Center for his help with the qPCRs.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Braig HR, Zhou W, Dobson SL, O'Neill SL. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol. 1998;180:2373–2378. doi: 10.1128/jb.180.9.2373-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Hoffmann AA. Environmental effects on cytoplasmic incompatibility and bacterial load in Wolbachia-infected Drosophila simulans. Entomol Exp Appl. 1998;86:13–24. [Google Scholar]
- Grenier S, Pintureau B, Heddi A, Lassablière F, Jager C, Louis C, et al. Successful horizontal transfer of Wolbachia symbionts between Trichogramma species. Proc R Soc Lond B. 1998;265:1441–1445. [Google Scholar]
- Haag-Liautard C, Coffey N, Houle D, Lynch M, Charlesworth B, Keightley PD. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6:1706–1714. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Heath BD, Butcher RDJ, Whitfield WGF, Hubbard SF. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol. 1999;9:313–316. doi: 10.1016/s0960-9822(99)80139-0. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?—a statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himler AG, Adachi-Hagimori T, Bergen JE, Kozuch A, Kelly SE, Tabashnik BE, et al. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science. 2011;332:254–256. doi: 10.1126/science.1199410. [DOI] [PubMed] [Google Scholar]
- Holden PR, Brookfield JFY, Jones P. Cloning and characterization of an ftsZ homologue from a bacterial symbiont of Drosophila melanogaster. Mol Gen Genet. 1993;240:213–220. doi: 10.1007/BF00277059. [DOI] [PubMed] [Google Scholar]
- Huigens ME, de Almeida RP, Boons PAH, Luck RF, Stouthamer R. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc R Soc Lond B. 2004;271:509–515. doi: 10.1098/rspb.2003.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huigens ME, Luck RF, Klaassen RHG, Maas MFPM, Timmermans MJTN, Stouthamer R. Infectious parthenogenesis. Nature. 2000;405:178–179. doi: 10.1038/35012066. [DOI] [PubMed] [Google Scholar]
- Huigens ME, Stouthamer R.2003Parthenogenesis associated with WolbachiaIn: Bourtzis K, Miller TA (eds).Insect Symbiosis CRC Press: Boca Raton, Florida; 247–266. [Google Scholar]
- Hurst GGD, Hurst LD, Majerus MEN. Selfish genes move sideways. Nature. 1992;356:659–660. doi: 10.1038/356659a0. [DOI] [PubMed] [Google Scholar]
- Hurst GDD, Johnson AP, v.d. Schulenburg JHG, Fuyama Y. Male-killing Wolbachia in Drosophila: a temperature-sensitive trait with a threshold bacterial density. Genetics. 2000;156:699–709. doi: 10.1093/genetics/156.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J. Spontaneous emergence of a new Wolbachia phenotype. Evolution. 2007;61:2244–2252. doi: 10.1111/j.1558-5646.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld K, Franco P, de Knijff P, Stouthamer R, van Alphen JJM. Clonal genetic variation in a Wolbachia-infected asexual wasp: horizontal transmission or historical sex. Mol Ecol. 2011a;20:3644–3652. doi: 10.1111/j.1365-294X.2011.05150.x. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld K, Reumer BM, Mouton L, Kremer N, Vavre F, van Alphen JJM. Does a parthenogenesis-inducing Wolbachia induce vestigial cytoplasmic incompatibility. Naturwissenschaften. 2011b;98:175–180. doi: 10.1007/s00114-010-0756-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Charif D, Henri H, Bataille M, Prévost G, Kraaijeveld K, et al. A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity. 2009;103:248–256. doi: 10.1038/hdy.2009.63. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Mitsui H, van Achterberg K, Nordlander G, Kimura MT. Geographical distributions and host associations of larval parasitoids of frugivorous Drosophilidae in Japan. J Nat Hist. 2007;41:1731–1738. [Google Scholar]
- Mouton L, Henri H, Bouletreau M, Vavre F. Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology. 2006;132:1–8. doi: 10.1017/S0031182005008723. [DOI] [PubMed] [Google Scholar]
- Murata Y, Ideo S, Watada M, Mitsui H, Kimura MT. Genetic and physiological variation among sexual and parthenogenetic populations of Asobara japonica (Hymenoptera: Braconidae), a larval parasitoid of drosophilid flies. Eur J Entomol. 2009;106:171–178. [Google Scholar]
- O'Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannebakker BA, Pijnacker LP, Zwaan BJ, Beukeboom LW. Cytology of Wolbachia-induced parthenogenesis in Leptopilina clavipes (Hymenoptera: Figitidae) Genome. 2004;47:299–303. doi: 10.1139/g03-137. [DOI] [PubMed] [Google Scholar]
- Pijls JWAM, van Steenbergen HJ, van Alphen JJM. Asexuality cured: the relations and differences between sexual and asexual Apoanagyrus diversicornis. Heredity. 1996;76:506–513. [Google Scholar]
- Pintureau B, Chapelle L, Delobel B. Effects of repeated thermic and antibiotic treatments on a Trichogramma (Hym., Trichogrammatidae) symbiont. J Appl Entomol. 1999;123:473–483. [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2010. [Google Scholar]
- Raychoudhury R, Grillenberger BK, Gadau J, Bijlsma R, van de Zande L, Werren JH, et al. Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial-Wolbachia sweep in North America. Heredity. 2010;104:318–326. doi: 10.1038/hdy.2009.160. [DOI] [PubMed] [Google Scholar]
- Rousset F, Bouchon D, Pintureau B, Juchault P, Solignac M. Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc R Soc Lond B. 1992;250:91–98. doi: 10.1098/rspb.1992.0135. [DOI] [PubMed] [Google Scholar]
- Scheffer SJ, Grissell EE. Tracing the geographical origin of Megastigmus transvaalensis (Hymenoptera: Torymidae): an African wasp feeding on a South American plant in North America. Mol Ecol. 2003;12:415–421. doi: 10.1046/j.1365-294x.2003.01725.x. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, Braig HR, O'Neill SL. Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proc R Soc Lond B. 1995;261:325–330. doi: 10.1098/rspb.1995.0154. [DOI] [PubMed] [Google Scholar]
- Smith PT, Kambhampati S. Status of the Cotesia flavipes species complex (Braconidae: Microgastrinae) based on mitochondrial 16S rRNA and NADH 1 dehydrogenase gene sequence. J Kansas Entomol Soc. 1999;72:306–314. [Google Scholar]
- Smith PT, Kambhampati S, Völkl W, Mackauer M. A phylogeny of aphid parasitoids (Hymenoptera: Braconidae: Aphidiinae) inferred from mitochondrial NADH 1 dehydrogenase gene sequence. Mol Phylogenet Evol. 1999;11:236–245. doi: 10.1006/mpev.1998.0575. [DOI] [PubMed] [Google Scholar]
- Stouthamer R, Breeuwer JAJ, Hurst GDD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- Stouthamer R, Russell JE, Vavre F, Nunney L. Intragenomic conflict in populations infected by parthenogenesis inducing Wolbachia ends with irreversible loss of sexual reproduction. BMC Evol Biol. 2010;10:229. doi: 10.1186/1471-2148-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- Varaldi J, Fouillet P, Ravallec M, López-Ferber M, Boulétreau M, Fleury F. Infectious behavior in a parasitoid. Science. 2003;302:1930. doi: 10.1126/science.1088798. [DOI] [PubMed] [Google Scholar]
- Vavre F, Fleury F, Lepetit D, Fouillet P, Boulétreau M. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol Bio Evol. 1999;16:1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084. [DOI] [PubMed] [Google Scholar]
- Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev: Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia—reproductive parasites of arthropods. Proc R Soc Lond B. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Zhou W, Rousset F, O'Neill S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc Lond B. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.