Abstract
The development of most autoimmune diseases includes a strong heritable component. This genetic contribution to disease ranges from simple Mendelian inheritance of causative alleles to the complex interactions of multiple weak loci influencing risk. The genetic variants responsible for disease are being discovered through a range of strategies from linkage studies to genome-wide association studies. Despite the rapid advances in genetic analysis, substantial components of the heritable risk remain unexplained, either owing to the contribution of an as-yet unidentified, “hidden,” component of risk, or through the underappreciated effects of known risk loci. Surprisingly, despite the variation in genetic control, a great deal of conservation appears in the biological processes influenced by risk alleles, with several key immunological pathways being modified in autoimmune diseases covering a broad spectrum of clinical manifestations. The primary translational potential of this knowledge is in the rational design of new therapeutics to exploit the role of these key pathways in influencing disease. With significant further advances in understanding the genetic risk factors and their biological mechanisms, the possibility of genetically tailored (or “personalized”) therapy may be realized.
Genetic risk variants associated with various autoimmune diseases, from rheumatoid arthritis to type 1 diabetes, are often similar. For example, most autoimmune diseases, if not all, are associated with the human leukocyte antigen (HLA) region.
Autoimmune diseases affect a significant proportion of the population, with >4% of the European population suffering from one or more of these disorders (Vyse and Todd 1996; Cooper et al. 2009; Eaton et al. 2010). Although all autoimmune diseases share similarities in the basic immunological mechanisms, in other aspects, such as clinical manifestation and age of onset, individual diseases vary widely. A few rare autoimmune diseases with Mendelian inheritance patterns within families occur including APS-1 (autoimmune polyendocrine syndrome type 1), IPEX (immunodysregulation, polyendocrinopathy, and enteropathy X-linked) syndrome, and ALPS (autoimmune lymphoproliferative syndrome). Most autoimmune diseases are, however, multifactorial in nature, with susceptibility controlled by multiple genetic and environmental factors.
The genetic component of more common autoimmune diseases can be calculated in several different manners, including the sibling recurrence risk (λs) and the twin concordance rate. The sibling recurrence risk is the ratio of the lifetime risk in siblings of patients to the lifetime population risk, whereas the twin concordance rate measures the proportion of the siblings of affected twins that are also affected. Most common autoimmune diseases, such as multiple sclerosis (MS), type 1 diabetes (T1D), rheumatoid arthritis (RA), and inflammatory bowel disease (IBD) are characterized by a sibling recurrence risk between 6 and 20 (Vyse and Todd 1996), and concordance rates of 25%–50% in monozygotic twins and 2%–12% in dizygotic twins (Cooper et al. 1999). A substantial proportion of relatives may also have subclinical evidence of autoimmunity without developing clinically overt disease. For example, 19% of healthy siblings of MS patients show antibody production in the cerebrospinal fluid, compared to 4% of unrelated healthy controls (Haghighi et al. 2000), whereas 4% of healthy first-degree relatives display lesions that are indistinguishable from those seen in patients and are not seen in unrelated healthy controls (De Stefano et al. 2006). Furthermore, comorbidity with the development of several autoimmune diseases in the same patient and clustering of several autoimmune diseases within families above what is expected by chance appear common (Cooper et al. 2009; Zhernakova et al. 2009). Together these data show a strong genetic component to autoimmune disease development.
STRATEGIES TO IDENTIFY GENETIC FACTORS UNDERLYING AUTOIMMUNE DISEASE
The human genome consists of 3 billion base pairs, and any two genomes typically differ by 0.1% (Kruglyak and Nickerson 2001). Common variants or polymorphisms, where both alleles have a frequency of >1% in the population, account for most (90%) of these differences (Kruglyak and Nickerson 2001; The International HapMap Consortium 2003). Rare variants, each found in <1% of the population, make up the remaining 10% of genetic polymorphism, but are far more numerous than common variants in total counts, given the large size of the human population. In all, 10–15 million common variants and billions of rare sequence variants constitute the human genetic diversity, making the identification of those variants relevant to particular autoimmune diseases challenging (Sawcer 2008).
Different strategies are tailored to each category of variants. Linkage studies search for segregation or coinheritance of a genomic region with disease in families and have been very successful in Mendelian diseases. This strategy has identified mutations in the autoimmune regulator gene (AIRE), the forkhead box P3 gene (FOXP3), and the Fas gene (TNFRSF6) causing APS-1 (The Finnish-German APECED Consortium 1997; Nagamine et al. 1997), IPEX (Bennett et al. 2001; Wildin et al. 2001), and ALPS (Fisher et al. 1995) syndromes, respectively. In most common autoimmune diseases, in contrast, few extended families with Mendelian inheritance patterns exist. Linkage studies have summed signals over multiple smaller families, including affected sibling pairs, instead. These have shown an overall increased allele sharing between affected siblings, confirming the shared genetic risk observed in epidemiological studies, but have only been able to pinpoint precisely those regions exerting large effects on disease risk, such as mutations in NOD2 in Crohn’s disease (CD) (Hugot et al. 2001; Ogura et al. 2001). Indeed, unrealistically high numbers of affected sibling pairs are required to detect the modest effect sizes that we now know are typical for multifactorial disorders by the linkage approach (The International Multiple Sclerosis Genetics Consortium 2005).
Association testing is more suited to detect the contribution of genetic loci to common autoimmune diseases. An association study compares a group of patients and healthy controls and searches for genetic variants that differ in frequency between both groups. Variations in the human leukocyte antigen (HLA) region were the first polymorphisms investigated in association studies (in the 1970s) and turned out to play a major role in most autoimmune diseases, even though precise understanding of the effects is still under investigation owing to the highly polymorphic nature, exceptional linkage disequilibrium, and high gene density of this region (Fernando et al. 2008). Through the investigation of candidate genes, a handful of non-HLA genes influencing risk of autoimmune disease were identified, e.g., the CTLA4 and PTPN22 genes in T1D (Ueda et al. 2003; Bottini et al. 2004). A major breakthrough came, however, around 2007 with the advent of high-throughput genotyping technology (microarrays), the information from the mapping of the human genome and interindividual genetic variation (International HapMap project), and the realization that the collection of large study populations and the establishment of collaborations is necessary. With these tools, it became possible to undertake genome-wide association studies (GWAS). GWAS are based on the “common disease–common variant” hypothesis, and screen the vast majority (>80%) of common genetic variation for its contribution to disease susceptibility (Hirschhorn and Gajdos 2011). Because of a low prior probability of association amongst the multiple tests performed (typically a few hundred thousand to a million variants screened capture >80% of common variation in the genome), a conservative significance threshold needs to be applied to avoid false positives. It has been shown that results reaching P values <5 × 10−8 (genome-wide significance level) in a well-powered study of at least 2000 cases and 2000 controls have a high likelihood of being true positive and of replicating in later studies (The Wellcome Trust Case Control Consortium 2007; Sawcer 2008). To date >1200 genomic regions influencing risk for 210 multifactorial diseases have been identified (http://www.genome.gov/gwastudies/ up to 12/2010) and autoimmune diseases have been exceptionally successful, with >30 risk regions each identified in T1D (Barrett et al. 2009), CD (Franke et al. 2010), ulcerative colitis (UC) (Anderson et al. 2011), RA (Stahl et al. 2010), and MS (The International Multiple Sclerosis Genetics Consortium 2011). This suggests a substantial role of common genetic variation in susceptibility to autoimmune diseases, and contrasts with other multifactorial diseases, such as neurodegenerative disorders. The important role of common variation in autoimmune diseases is thought to reflect a history of adaption and selection for variability in regions controlling the immune system, as a defense against intrapopulation pathogen spread (Corona et al. 2010; Casto and Feldman 2011).
THE EMERGING GENETIC ARCHITECTURE OF COMMON AUTOIMMUNE DISEASES
GWAS have revealed important conclusions on the genetic architecture of autoimmune diseases. The genetic architecture refers to the number of risk variants, their frequencies in the population (risk allele frequency), and the risks on disease they confer (odds ratio). The odds ratio estimates the fold change in risk for carriers versus noncarriers of a risk allele. Most variants identified follow a multiplicative model, i.e., carrying two copies of the risk allele amounts to the square of the risk of carrying one copy. The genetic architecture of multifactorial autoimmune diseases as emerging from GWAS appears to follow the L shape predicted on the basis of theoretical modeling and animal data (Barton and Keightley 2002). Few if any regions exert a large effect on the risk of disease (odds ratio >3), and a handful have modest effect sizes (odds ratios >2) (Hindorff et al. 2009). In most autoimmune diseases these important loci include the HLA region (Fernando et al. 2008); other examples are NOD2 and IL23R in CD (Hugot et al. 2001; Ogura et al. 2001; Duerr et al. 2006), and PTPN22 in T1D and RA (Begovich et al. 2004; Bottini et al. 2004). Subsequently, there is a long tail of loci with odds ratios of around 1.2 or smaller (Hindorff et al. 2009). Based on currently identified loci, theoretical models predict that many more risk variants are hidden below the genome-wide significance threshold in current GWAS and that a large number of susceptibility loci, possibly thousands, with very small effect sizes may contribute to multifactorial disease (Park et al. 2010). Indeed, the presence of such a polygenic or “en masse” effect has already been shown in MS (The International Multiple Sclerosis Genetics Consortium et al. 2010).
Most of the already identified risk variants are common in the general population (Hindorff et al. 2009), although examples of both common and less common variants in the same gene influencing risk have been reported, e.g., for IL23R in CD (Momozawa et al. 2011) and IFIH1 in T1D (Nejentsev et al. 2009). For many variants, the risk allele is even more common than the other, protective, allele. This appears counterintuitive at first instance but underlines once again the multifactorial nature of most autoimmune diseases. Immune and infectious agents have been recognized as amongst the strongest selection pressures in human evolution and several autoimmune-associated genes show signs of positive selection, favoring either the protective or risk allele, depending on the case (Hindorff et al. 2009; Corona et al. 2010; Casto and Feldman 2011). Whereas Mendelian disease mutations are controlled by negative selection, risk variants in complex diseases may at least in part be tied to evolutionary adaptations (Blekhman et al. 2008). Pleiotropic effects of these variants may contribute to adaptation and selection, or environmental changes in modern societies may expose the disease risk associated with the variants (Manolio et al. 2009). As an example, whereas the majority of the individuals carry the T1D risk alleles at IFIH1, the rare protective alleles are functionally deleterious and may reduce the ability to mediate an immune response against enterovirus infection (Nejentsev et al. 2009).
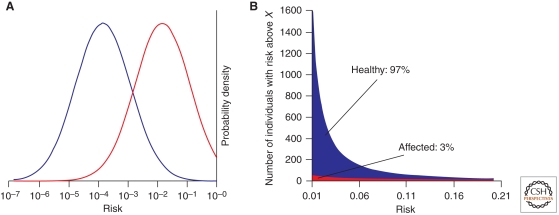
Under a polygenic model with many variants independently contributing to disease, the logarithm of the risk is normally distributed in the population (Fig. 1) (Pharoah et al. 2002; Clayton 2009; Sawcer et al. 2010). The genetic risk or burden can be thought of as the number of risk alleles weighted by their effect on disease risk. Most individuals carry a mix of protective and risk alleles and will have an average genetic burden. However, a small subset of individuals differs substantially in genetic risk, e.g., individuals in the 99th percentile versus the first percentile. The distributions of the logarithm of the risk in the general population and in individuals who will effectively go on to develop the disease have the same shape with the average genetic burden being higher in patients than in the general population. However, the distributions overlap to an extent that is dependent on the heritability of the disease (Sawcer et al. 2010). For a typical autoimmune disease with a sibling recurrence risk (λs) of 10, approximately two-thirds of the patients have a genetic risk that is higher than the 95th percentile of the genetic risk in healthy controls (Sawcer et al. 2010). Families with multiple affected relatives appear to share common risk alleles with sporadic patients, but may have a higher genetic load (D’Netto et al. 2009; Gourraud et al. 2011).
Figure 1.
Genetic risk distribution in cases and controls. (A) The effectiveness of a hypothetical “diagnostic chip” measuring all genetic variants (including currently unknown variants) can be assessed by considering the distribution of genetic risk at birth among the general population (blue) and affected individuals (red) under a multiplicative model with many risk variants involved (Pharoah et al. 2002; Clayton 2009; Sawcer et al. 2010). The probability distribution of risk is calculated for a population with a prevalence (=mean risk in the general population) of 1/1000 and a λs of 10. The variance in risk observed (=2* ln λs) is the same for the general population and for individuals who go on to develop the disease. The distribution in cases is, however, shifted toward a higher risk, with a magnitude equal to the variance. (B) The number of false-negative and false-positive diagnostic calls is dictated by the absolute number of healthy (blue) and affected (red) individuals at any given level of risk. Under this scenario a diagnostic chip with a false-negative rate of 50% (i.e., capable of detecting 50% of affected patients) would have only a 3% true-positive rate, as it would produce a positive result for all individuals in the highest 1.6% risk bracket, of which 97% would still come from the healthy population (false positives).
A consequence of the polygenic model for complex diseases is that patients are inevitably highly heterogeneous in terms of the particular set of risk alleles they carry (Janssens and van Duijn 2008; Sawcer et al. 2010). It has been suggested that this may translate in different genetically determined disease mechanisms in subgroups of patients (heterogeneity) or a common disease mechanism that is complemented by additional pathways that are more or less predominant in different subgroups (complexity). This concept of heterogeneity or complexity is an important emerging question. A first example of such complexity is found in RA, where the strong association with the HLA-DRB1 risk alleles sharing a specific epitope at positions 70–74 (shared epitope alleles) is specific to the subset of patients producing antibodies to citrullinated peptide antigens (ACPAs) (van der Helm-van Mil et al. 2007; Padyukov et al. 2011). The two forms of the disease are clinically indistinguishable at onset but ACPA positivity is the factor most predictive of future prognosis (van der Helm-van Mil et al. 2007).
In summary, in contrast to rare Mendelian mutations causing childhood-onset autoimmune diseases, the genetic architecture of multifactorial autoimmune diseases is highly complex. Susceptibility rather than causality results from the combined effects of many variants, most of them common in the general population, that each exert a small effect on risk. Many different combinations of risk alleles are able to independently generate a high level of disease risk, without individual loci being necessary or sufficient for the development of disease.
FROM GENETICS TO IMMUNOLOGY
Despite the enormous diversity of genetic variants contributing to susceptibility from individual to individual with the same autoimmune disease, a high degree of similarity is observed between autoimmune diseases in the pool of genetic variants associated with disease. These genetic associations can be dissected into three biologically distinct classes. First, for most autoimmune diseases, the strongest genetic associations are with the HLA locus. Although some HLA haplotype linkages are shared between autoimmune diseases, most HLA associations seem to be specific for a disease (Trowsdale 2011). Second, many of the strong loci that have been associated with one autoimmune disease are also found to be involved in multiple other autoimmune diseases (Makaroff et al. 2008). Typically, the allele of these shared variants that causes risk in one autoimmune disease also causes risk in other autoimmune diseases, indicating that the alleles participate in a shared immunological process common to the development of multiple, clinically distinct, autoimmune diseases (Makaroff et al. 2008). There is a degree of complexity, however, in which precise mechanisms may differ between subgroups of autoimmune diseases. For example, variants in the PTPN22 gene have been associated, with various degrees of risk, with T1D (Smyth et al. 2004), autoimmune thyroid disease (AITD) (Smyth et al. 2004), and RA (The Wellcome Trust Case Control Consortium 2007), among other diseases but are protective against CD (Barrett et al. 2009). Third, a smaller number of associations are observed that are specific to a single autoimmune disease, with no measured impact on other autoimmune diseases, suggesting that they drive a target organ-specific pathway toward disease. These three classes will be discussed in turn.
HLA Associations in Autoimmune Disease
Most, if not all, autoimmune conditions appear associated with the HLA region, if sufficiently large study populations are investigated, and this region is the strongest genetic component in most of these diseases. The HLA region spans approximately 4 Mb, and is the most gene-dense region in the human genome, containing around 250 genes of which 40% are predicted to exert a function in the immune system (The MHC Sequencing Consortium 1999). The region is characterized by an exceptionally high degree of polymorphism, with >1000 alleles known for HLA-A and -B, for example, and the most extensive degree of linkage disequilibrium within the genome. These features have hampered fine mapping of the functional variants in this region, even decades after the first reports of association. The classic HLA class I (HLA-A, -B, and -C) and class II (HLA-DP, -DQ, and -DR) gene clusters involved in antigen processing and presentation are best characterized and show the strongest association.
Unlike many other autoimmune risk variants (see below), most HLA variants have associations with specific autoimmune diseases, with different variants providing susceptibility to different diseases (Trowsdale 2011). For example, the HLA-DR2 haplotype is protective against T1D, yet predisposes toward MS and SLE (Brand et al. 2005). The unique amino acid sequences of HLA class I and class II alleles determine to a large extent the ability to respond to an antigen, whether foreign or self, and the nature of that response. It is thought that this binding of specific peptides or peptide modifications provides an explanation for their contribution to autoimmune disease. Alterations in binding efficiency may change autoimmune risk in several different manners. By increasing the binding efficiency of otherwise sequestered peptide, HLA risk variants may allow the presentation of potentially immunogenic peptides. Alternatively, by reducing the binding efficiency of particular peptides, HLA risk variants may allow autoreactive T-cell clones to escape tolerance mechanisms. A likely example of the first is the T1D risk allele of HLA-DQ8, which has enhanced binding of insulin peptides, resulting in peripheral activation of autoreactive T cells (Faas and Trucco 1994; Wen et al. 2002). An example of the latter is the MS risk allele of HLA-DR2, which shows poor binding of myelin basic protein, likely causing a failure in thymic tolerance mechanisms and activation of weakly binding autoreactive T cells in the periphery (Li et al. 2005; Maynard et al. 2005). Although HLA has the strongest linkage to particular autoimmune diseases, it is likely that most of the general autoimmune genetic susceptibility component is mediated by non-HLA risk alleles (see below), with HLA risk variants altering the target of autoimmunity. This phenomenon is observed in the nonobese diabetic mouse model, where the replacement of the T1D MHC risk variant, H2g7, with H2h4 reroutes the autoimmune response from the pancreas to the thyroid (Braley-Mullen et al. 1999).
Common Autoimmune Risk Variants
Many of the gene variants associated with risk in one autoimmune disease have been shown to also have an association with the development of additional autoimmune diseases. This shared association, and the observation that multiple autoimmune diseases track in the same patients and same families, has led to the model of a cluster of gene variants that decrease immunological tolerance. A large number of genes fall into the category of general autoimmune risk variants, however, it is likely that only a few key immunological pathways are affected.
One immunological phenomenon that is commonly associated with autoimmunity is that of partial T-cell immunodeficiency (Liston et al. 2008). Many of the rare Mendelian autoimmune diseases have a dual association of partial T-cell immunodeficiency and autoimmunity or immune dysregulation, of which Omenn syndrome can be considered a classical example. Partial loss-of-function mutations in RAG1, RAG2, DCLRE1C, IL7RA, and RMRP all produce lymphopenia and result in excessive T-cell activation (Liston et al. 2008). Although these are all examples of single gene variants being sufficient to produce Mendelian autoimmunity, milder gene variants are likely to produce a partial T-cell immunodeficiency, which, in combination with additional genetic insults, may predispose to autoimmunity. The best example of this phenomenon is the association of IL7RA polymorphisms with several common autoimmune diseases, including MS and T1D (Gregory et al. 2007; Todd et al. 2007). The variants that result in Omenn syndrome show severe loss of function, whereas the variant associated with common autoimmune disease alters splicing efficiency, creating a soluble competitive antagonist form of the receptor (Gregory et al. 2007). This reduction in available IL-7 in turn reduces thymic output of T cells (Broux et al. 2010), replicating on a less severe form the immunological defect present in Omenn syndrome. Similarly, the gain-of-function PTPN22 allele associated with multiple autoimmune diseases results in a reduction in T-cell receptor (TCR) signaling (Smerdel et al. 2004; Vang et al. 2005), and may therefore also contribute to partial immunodeficiency.
Another Mendelian autoimmune disease, IPEX, shows a separate mechanism by which risk variants may increase susceptibility to autoimmunity. IPEX is caused by mutants in FOXP3 (Bennett et al. 2001; Wildin et al. 2001), which in turn eliminate the production of regulatory T cells (Fontenot et al. 2003; Hori et al. 2003; Khattri et al. 2003). The absence of regulatory T cells results in autoreactive T cells undergoing uncontrolled activation, driving multiorgan autoimmunity (Kim et al. 2007). Again, this Mendelian disease finds parallels in common autoimmunity. IL-2 is important for the survival of regulatory T cells (Sadlack et al. 1993; Kramer et al. 1995; Suzuki et al. 1995, 1999; Willerford et al. 1995; Almeida et al. 2002), and variants in IL2RA are associated with T1D and MS. It has been shown that the T1D risk variant of IL2RA decreases receptor expression, which may result in defective regulatory T-cell biology (Dendrou et al. 2009). At the functional level, CTLA4 is a key mechanism by which regulatory T cells suppress disease (Wing et al. 2008), so it is notable that a splice variant of CTLA4 associated with autoimmune diseases has reduced production of a tolerogenic soluble isoform (Ueda et al. 2003).
A third cluster of risk variants is observed in cytokine and cytokine signaling genes, including STAT4, TNFA, IL12B, IL23R, and IL10 (Makaroff et al. 2008). Because many autoimmune diseases are strongly associated with a particular effector T-cell lineage (e.g., Th1, Th2, or Th17), it is likely that these variants alter the ease with which naïve T cells enter a pathogenic lineage. For example, IBD and psoriasis are both associated with Th17 infiltrate and both show associations with a disease risk allele in IL23R. As IL-23 is critical for the differentiation of Th17 cells (McGeachy et al. 2007), it is likely that this risk variant increases the generation of Th17 cells, and hence the development of IBD and psoriasis. In addition to the three pathways discussed above, there are multiple other cases where the known biology of associated risk variants suggests a common function in impeding tolerance. However, the examples of immunodeficiency, regulatory T cells, and effector T-cell differentiation are sufficient to show the general rule.
Disease-Specific Risk Variants
In addition to HLA and the shared autoimmune variants, additional risk variants are associated with single autoimmune diseases. Unlike the shared autoimmune variants, which are largely in genes expressed within leukocytes, the disease-specific risk variants are often expressed by the target organ. Thus, for example, polymorphisms in INS are associated with T1D, ATG16L1 variants with IBD, and KAZALD1 variants with RA. There are two main biological mechanisms by which these variants may drive disease, and often it is not clear for a given variant which mechanism is active (Makaroff et al. 2008).
The first mechanism is that of a target antigen gene variant which results in poorer tolerance induction. Studies of the rare Mendelian autoimmune disease APS-1 have shown that thymic expression of target organ antigens is essential to prevent autoimmunity (Liston et al. 2003; Liston et al. 2004a; DeVoss et al. 2006). In the case of APS-1, mutations in the gene AIRE destroy entire networks of thymic antigen expression, driving disease against multiple targets (The Finnish-German APECED Consortium 1997; Nagamine et al. 1997). More pertinent to the common autoimmune diseases is when expression of single targets is reduced in the thymus. The best example of this phenomenon is the T1D INS risk variant, which has a two- to threefold decrease in thymic expression of insulin (Vafiadis et al. 1997; Taubert et al. 2007).
The second mechanism by which disease-specific variants can drive autoimmunity is through the modification of the target organ. For example, ATG16L1 is involved in the barrier function of the gut, and so the association of variants with IBD risk may represent a failure to prevent inflammatory bacterial infections (Rioux et al. 2007). In a similar vein, KAZALD1 is involved in joint regeneration, so the observed association of variants with RA risk (The Wellcome Trust Case Control Consortium 2007) may represent reduced ability of the joints to heal after autoimmune attack. Until further biological analysis of the mechanism of disease-specific variants is completed, explanations of the pathway to risk will largely remain “Just So” stories, with plausible models that invoke differentiation tolerance induction or differences in target organ function.
FROM IMMUNOLOGY TO GENETICS
Just as genetic data have been used to further understand the biology of autoimmunity, immunological data can be used to further understand the genetics of autoimmunity. Although most GWAS have used clinical presentation as the phenotype, it is becoming increasingly recognized that GWAS can equally use intermediate biological phenotypes as the mapping subject, such as immunological parameters. These “endophenotypes” are quantitative measures that have a biologically plausible mechanism leading to disease. Endophenotypes are thought to be more closely related to gene function than clinical end point phenotypes. As subtraits, endophenotypes are expected to have a simpler genetic architecture than the final clinical manifestation, with fewer influencing variants each of larger effect, making them easier to dissect (Weidinger et al. 2010). Pilot studies in mice suggest that this is indeed the case (Liston et al. 2004b). Furthermore, endophenotypes can be measured within the healthy population, providing more scope for understanding the genetics of the less common autoimmune diseases and eliminating the confounding secondary effects of autoimmune disease on biological processes. GWAS of immunological traits underlying asthma, i.e., IgE production and eosinophil numbers, have been successful in identifying association with single-nucleotide polymorphisms (SNPs) involved in asthma risk, providing critical immunological information of the mechanism of these SNPs (Weidinger et al. 2008; Gudbjartsson et al. 2009). Similar studies of immunological parameters associated with autoimmune diseases are under way, and not only have the potential to provide mechanistic information on already known SNPs, but also the capacity to identify new SNPs with greater power.
MISSING OR HIDDEN HERITABILITY?
Despite enormous progress over recent years, only a fraction of the variance in risk between individuals that has been estimated from epidemiological studies can be explained by known genetic risk factors. Typically, several tens of variants explain at most 25% of the variance (Park et al. 2010). The remainder is described as the “missing heritability” and represents a current challenge in complex genetics (Manolio et al. 2009). Several explanations can be offered. First, additional genetic risk factors may remain to be discovered, having been hidden from previous studies by technical constraints. Alternatively, the known genetic component may account for far more of the variance in susceptibility than currently recognized.
Several different sources can be postulated to make up the hidden genetic risk:
The identification of weak effect common variants is limited by study design. Although the variants with the strongest effect on disease susceptibility have been identified, larger studies may identify more variants with a more modest effect (Park et al. 2010; Yang et al. 2010). The effect of these variants can be predicted even without identification, but is unlikely to increase the proportion of variance explained above 50% (Park et al. 2010; The International Multiple Sclerosis Genetics Consortium et al. 2010; Yang et al. 2010).
The effect of less common (1%–5%) and rare (<1%) variants is yet to be fully calculated. If “common” diseases actually constitute a heterogeneous set of clinically similar diseases, multiple distinct rare variants may be having a large effect on susceptibility, while simultaneously evading detection by linkage and GWAS approaches (Smith and Lusis 2002). For example, rare mutations in the exonuclease gene TREX1 are found in 0.5% of SLE patients (Namjou et al. 2011), and mutations leading to defective expression of the sialic acid acetylesterase (SIAE) are found in 2.4% of patients with autoimmune disease (Surolia et al. 2010). However, the linkage studies in affected sibling pairs provide upper estimates for the effect size that can be expected from a yet unknown rare variant (Sawcer 2008; Forabosco et al. 2009). A rare variant with a frequency of 0.2% and an odds ratio of 20 would result in >60% allele sharing in siblings and should have been detected in linkage studies using a few hundred affected sib pairs (Risch and Merikangas 1996). Simulation studies suggest that there are fewer rare variants with large effect sizes (odds ratios >3) in autoimmune diseases than in other multifactorial disorders (Nejentsev et al. 2009; Momozawa et al. 2011). However, rare variants with weak to moderate effects may have a substantial cumulative effect and are unlikely to be detected using any association approach.
The contribution of other types of human genetic variation than sequence variation is yet to be thoroughly investigated. Variation such as copy number variation (CNV), inversions, and complex rearrangements could have an impact on disease susceptibility that has not been picked up in association studies that focus on SNPs. Early data suggest that the net contribution of CNV will be less than that observed with SNPs (The Wellcome Trust Case Control Consortium 2010). The effect of other non-SNP genetic variation will be harder to assess, especially inherited epigenetic variation, which could conceivably have a large impact on disease susceptibility (Meda et al. 2011).
Alternatively, there are multiple explanations by which the effect of known genetic variation on susceptibility may be systematically underestimated:
Measures of familial clustering such as the sibling recurrence risk (λs) used for estimation of the total variance tend to be biased upward by an overestimated lifetime risk in siblings in the numerator and an underestimated lifetime risk in the general population in the denominator (Guo 1998; Sawcer et al. 2010).
Gene–gene or gene–environment interactions may contribute to part of the missing heritability. The observation of monozygotic concordance rates of less than 100% (typically 25%–50% [Cooper et al. 1999]) shows the influence of either environmental or stochastic events (such as TCR rearrangement) in determining disease development. As environmental variation between dizygotic twins is lower than environmental variation of the population at large, the genetic contribution of risk loci influenced by environmental effects may be reduced in GWAS studies, compared to the total genetic risk calculated from twin concordance rates. Known examples of gene–gene interaction include the risk alleles at HLA-C and ERAP1 in psoriasis (Strange et al. 2010), whereas gene–environment interactions have been observed in RA (Mahdi et al. 2009). Many additional gene–environment interactions are predicted, with variants in key infection-sensing genes, such as NOD2 (Franke et al. 2010), IFIH1 (Smyth et al. 2006; Nejentsev et al. 2009), and FUT2 (McGovern et al. 2010), being associated with autoimmunity.
Limitations inherent in current GWAS may underreport the association effect. For example, the SNPs found to be associated with disease may not be the functional SNPs, and thus the effect will be diluted by imperfect correlation (Yang et al. 2010). Similarly, the most associated SNP may not capture the entire contribution from a locus if there are several risk alleles clustered, as observed frequently in both mice (Fraser et al. 2010) and humans (Ioannidis et al. 2009; Zhernakova et al. 2009). In particular, risk variants at the same locus that are in negative linkage disequilibrium with each other may mask each other’s effect, again as observed in mice (McDuffie 2000). Extensive fine-mapping studies investigating regions of association, such as those using a custom genotyping array covering autoimmune-associated genes developed by the Immunochip consortium of autoimmune disease research groups (Cortes and Brown 2011), are ongoing and will elucidate more precisely the contribution of associated loci.
CONCLUDING REMARKS
Enormous advances have been made in dissecting the genetics of autoimmunity over the past decade, both in identifying genetic risk factors and in understanding the cellular basis for their effect. The ultimate aim of these studies was, of course, to mitigate disease in the clinic. There are three main ways in which our increased understanding of the genetics of autoimmunity has potential for translation—through the identification of drug targets, through genetic identification of at-risk individuals, and through personalized medicine.
With a shown impact on disease development, genes identified to contain disease-associated variation constitute high-quality candidates for therapeutic drug design. Examples of drug design following genetic association are highly limited, however, the reverse shows that the principle of associated genes being high-quality candidates is valid. Several examples of the latter exist, such as the identification of two genes that are targets for current treatments in recent genome-wide association screens for MS (The International Multiple Sclerosis Genetics Consortium 2007; The International Multiple Sclerosis Genetics Consortium and The Wellcome Trust Case Control Consortium 2 2011). With genetic data maturing, the forward drug-design strategy is anticipated to be productive in the near future.
The prospect of translating genetic knowledge into diagnostic identification of at-risk individuals is less rosy. The large component of “missing” genetic risk obviously limits diagnostic accuracy (Kraft et al. 2009). However, even if all genetic risk variants were known the potential for diagnostics is limited. A hypothetical “diagnostic chip” containing all genetic risk factors may give a reasonable sensitivity (proportion of people who are correctly classified as high risk among those individuals who actually develop the disease) and specificity (proportion of people who are correctly classified as low risk amongst people who do not develop the disease) (Clayton 2009). However, positive and negative predictive values (proportion of people classified as high risk who develop the disease and the proportion of people classified as low risk who do not develop the disease) are limited by the low prevalence of most autoimmune diseases, such that even the hypothetical perfect chip would have low diagnostic value (see Fig. 1) (Kraft and Hunter 2009; Sawcer et al. 2010). These predictive values are not expected to be much better than those based on traditional risk factors, such as a family history of the disease (De Jager et al. 2009; Sawcer et al. 2010), except within specific cohorts (Ziegler and Nepom 2010). The value of a diagnostic chip may be increased if gene–environment interactions are understood and both genetic and environmental data was available for individual risk assessment, which is one possible avenue to broad diagnostic utility.
Last, our understanding of the genetics of autoimmunity may translate to personalized medicine, i.e., providing each patient with the right treatment at the right time. Studies correlating genetic factors with treatment response or side effects are in their childhood as yet, and the collection of large study populations and detailed clinical follow-up is a challenge. One of the first examples suggests that genetically determined increased expression levels of interleukin-21 contribute to the development of secondary autoimmune diseases, an important side effect on treatment of MS patients with alemtuzumab (Jones et al. 2009). If further correlations like this are elucidated, the genetic profile of individual autoimmune patients may be used to recommend or contraindicate particular treatment regimes.
ACKNOWLEDGMENTS
A.G. is supported by the Belgian Neurological Society, the Belgian Charcot Foundation, Wetenschappelijk Onderzoek Multiple Sclerose (WOMS)—Vlaanderen, and Onderzoeksfonds K.U. Leuven/Research Fund K.U. Leuven (OT/11/087). A.L. is supported by the Juvenile Diabetes Research Foundation, European Research Council, FWO, and VIB.
Footnotes
Editors: Diane Mathis and Alexander Y. Rudensky
Additional Perspectives on Immune Tolerance available at www.cshperspectives.org
REFERENCES
- Almeida AR, Legrand N, Papiernik M, Freitas AA 2002. Homeostasis of peripheral CD4+ T cells: IL-2R α and IL-2 shape a population of regulatory cells that controls CD4+ T cell numbers. J Immunol 169: 4850–4860 [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, et al. 2011. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43: 246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. 2009. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41: 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Keightley PD 2002. Understanding quantitative genetic variation. Nat Rev Genet 3: 11–21 [DOI] [PubMed] [Google Scholar]
- Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, et al. 2004. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 27: 20–21 [DOI] [PubMed] [Google Scholar]
- Blekhman R, Man O, Herrmann L, Boyko AR, Indap A, Kosiol C, Bustamante CD, Teshima KM, Przeworski M 2008. Natural selection on genes that underlie human disease susceptibility. Curr Biol 18: 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, et al. 2004. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36: 337–338 [DOI] [PubMed] [Google Scholar]
- Braley-Mullen H, Sharp GC, Medling B, Tang H 1999. Spontaneous autoimmune thyroiditis in NOD.H-2h4 mice. J Autoimmun 12: 157–165 [DOI] [PubMed] [Google Scholar]
- Brand O, Gough S, Heward J 2005. HLA, CTLA-4 and PTPN22: The shared genetic master-key to autoimmunity? Expert Rev Mol Med 7: 1–15 [DOI] [PubMed] [Google Scholar]
- Broux B, Hellings N, Venken K, Rummens JL, Hensen K, Van Wijmeersch B, Stinissen P 2010. Haplotype 4 of the multiple sclerosis-associated interleukin-7 receptor α gene influences the frequency of recent thymic emigrants. Genes Immun 11: 326–333 [DOI] [PubMed] [Google Scholar]
- Casto AM, Feldman MW 2011. Genome-wide association study SNPs in the human genome diversity project populations: Does selection affect unlinked SNPs with shared trait associations? PLoS Genet 7: e1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DG 2009. Prediction and interaction in complex disease genetics: Experience in type 1 diabetes. PLoS Genet 5: e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GS, Miller FW, Pandey JP 1999. The role of genetic factors in autoimmune disease: Implications for environmental research. Environ Health Perspect 107 (Suppl 5): 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GS, Bynum ML, Somers EC 2009. Recent insights in the epidemiology of autoimmune diseases: Improved prevalence estimates and understanding of clustering of diseases. J Autoimmun 33: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona E, Dudley JT, Butte AJ 2010. Extreme evolutionary disparities seen in positive selection across seven complex diseases. PLoS One 5: e12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Brown MA 2011. Promise and pitfalls of the Immunochip. Arthritis Res Ther 13: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, Aubin C, Bauer D, Heubach JF, Sandbrink R, et al. 2009. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: A weighted genetic risk score. Lancet Neurol 8: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD, Nutland S, Coleman G, Himsworth M, Hardy M, et al. 2009. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet 41: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Netto MJ, Ward H, Morrison KM, Ramagopalan SV, Dyment DA, DeLuca GC, Handunnetthi L, Sadovnick AD, Ebers GC 2009. Risk alleles for multiple sclerosis in multiplex families. Neurology 72: 1984–1988 [DOI] [PubMed] [Google Scholar]
- De Stefano N, Cocco E, Lai M, Battaglini M, Spissu A, Marchi P, Floris G, Mortilla M, Stromillo ML, Paolillo A, et al. 2006. Imaging brain damage in first-degree relatives of sporadic and familial multiple sclerosis. Ann Neurol 59: 634–639 [DOI] [PubMed] [Google Scholar]
- DeVoss J, Hou Y, Johannes K, Lu W, Liou GI, Rinn J, Chang H, Caspi RR, Fong L, Anderson MS 2006. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med 203: 2727–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. 2006. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314: 1461–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton WW, Pedersen MG, Atladottir HO, Gregory PE, Rose NR, Mortensen PB 2010. The prevalence of 30 ICD-10 autoimmune diseases in Denmark. Immunol Res 47: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas S, Trucco M 1994. The genes influencing the susceptibility to IDDM in humans. J Endocrinol Invest 17: 477–495 [DOI] [PubMed] [Google Scholar]
- Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, Vyse TJ, Rioux JD 2008. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet 4: e1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM 1995. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 81: 935–946 [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4: 330–336 [DOI] [PubMed] [Google Scholar]
- Forabosco P, Bouzigon E, Ng MY, Hermanowski J, Fisher SA, Criswell LA, Lewis CM 2009. Meta-analysis of genome-wide linkage studies across autoimmune diseases. Eur J Hum Genet 17: 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. 2010. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 42: 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HI, Dendrou CA, Healy B, Rainbow DB, Howlett S, Smink LJ, Gregory S, Steward CA, Todd JA, Peterson LB, et al. 2010. Nonobese diabetic congenic strain analysis of autoimmune diabetes reveals genetic complexity of the Idd18 locus and identifies Vav3 as a candidate gene. J Immunol 184: 5075–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourraud PA, McElroy JP, Caillier SJ, Johnson BA, Santaniello A, Hauser SL, Oksenberg JR 2011. Aggregation of multiple sclerosis genetic risk variants in multiple and single case families. Ann Neurol 69: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, et al. 2007. Interleukin 7 receptor α chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet 39: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, et al. 2009. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet 41: 342–347 [DOI] [PubMed] [Google Scholar]
- Guo SW 1998. Inflation of sibling recurrence-risk ratio, due to ascertainment bias and/or overreporting. Am J Hum Genet 63: 252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi S, Andersen O, Rosengren L, Bergstrom T, Wahlstrom J, Nilsson S 2000. Incidence of CSF abnormalities in siblings of multiple sclerosis patients and unrelated controls. J Neurol 247: 616–622 [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Gajdos ZK 2011. Genome-wide association studies: Results from the first few years and potential implications for clinical medicine. Annu Rev Med 62: 11–24 [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science 299: 1057–1061 [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411: 599–603 [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Thomas G, Daly MJ 2009. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet 10: 318–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens AC, van Duijn CM 2008. Genome-based prediction of common diseases: Advances and prospects. Hum Mol Genet 17: R166–R173 [DOI] [PubMed] [Google Scholar]
- Jones JL, Phuah CL, Cox AL, Thompson SA, Ban M, Shawcross J, Walton A, Sawcer SJ, Compston A, Coles AJ 2009. IL-21 drives secondary autoimmunity in patients with multiple sclerosis, following therapeutic lymphocyte depletion with alemtuzumab (Campath-1H). J Clin Invest 119: 2052–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 4: 337–342 [DOI] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol 8: 191–197 [DOI] [PubMed] [Google Scholar]
- Kraft P, Hunter DJ 2009. Genetic risk prediction—Are we there yet? New Engl J Med 360: 1701–1703 [DOI] [PubMed] [Google Scholar]
- Kraft P, Wacholder S, Cornelis MC, Hu FB, Hayes RB, Thomas G, Hoover R, Hunter DJ, Chanock S 2009. Beyond odds ratios—Communicating disease risk based on genetic profiles. Nat Rev Genet 10: 264–269 [DOI] [PubMed] [Google Scholar]
- Kramer S, Schimpl A, Hunig T 1995. Immunopathology of interleukin (IL) 2-deficient mice: Thymus dependence and suppression by thymus-dependent cells with an intact IL-2 gene. J Exp Med 182: 1769–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Nickerson DA 2001. Variation is the spice of life. Nat Genet 27: 234–236 [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA 2005. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J 24: 2968–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC 2003. Aire regulates negative selection of organ-specific T cells. Nat Immunol 4: 350–354 [DOI] [PubMed] [Google Scholar]
- Liston A, Gray DHD, Lesage S, Fletcher AL, Wilson J, Webster KE, Scott HS, Boyd RL, Peltonen L, Goodnow CC 2004a. Gene dosage limiting role of Aire in thymic expression, clonal deletion and organ-specific autoimmunity. J Exp Med 200: 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lesage S, Gray DH, O’Reilly LA, Strasser A, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM, et al. 2004b. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity 21: 817–830 [DOI] [PubMed] [Google Scholar]
- Liston A, Enders A, Siggs OM 2008. Unravelling the association of partial T-cell immunodeficiency and immune dysregulation. Nat Rev Immunol 8: 545–558 [DOI] [PubMed] [Google Scholar]
- Mahdi H, Fisher BA, Kallberg H, Plant D, Malmstrom V, Ronnelid J, Charles P, Ding B, Alfredsson L, Padyukov L, et al. 2009. Specific interaction between genotype, smoking and autoimmunity to citrullinated α-enolase in the etiology of rheumatoid arthritis. Nat Genet 41: 1319–1324 [DOI] [PubMed] [Google Scholar]
- Makaroff L, Linterman M, Liston A 2008. Genetic control of T cell tolerance and susceptibility to autoimmune disease. In Genetic predisposition to disease (ed. Torres S, Marin M) Nova, Hauppauge NY [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC 2005. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: Insights into MHC bias and antigen specificity. Immunity 22: 81–92 [DOI] [PubMed] [Google Scholar]
- McDuffie M 2000. Derivation of diabetes-resistant congenic lines from the nonobese diabetic mouse. Clin Immunol 96: 119–130 [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ 2007. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol 8: 1390–1397 [DOI] [PubMed] [Google Scholar]
- McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, Ippoliti A, Vasiliauskas E, Berel D, Derkowski C, et al. 2010. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet 19: 3468–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda F, Folci M, Baccarelli A, Selmi C 2011. The epigenetics of autoimmunity. Cell Mol Immunol 8: 226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, Amininejad L, Cleynen I, Colombel JF, de Rijk P, Dewit O, et al. 2011. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet 43: 43–47 [DOI] [PubMed] [Google Scholar]
- Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, et al. 1997. Positional cloning of the APECED gene. Nat Genet 17: 393–398 [DOI] [PubMed] [Google Scholar]
- Namjou B, Kothari PH, Kelly JA, Glenn SB, Ojwang JO, Adler A, Alarcon-Riquelme ME, Gallant CJ, Boackle SA, Criswell LA, et al. 2011. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun 12: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S, Walker N, Riches D, Egholm M, Todd JA 2009. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against Type 1 diabetes. Science 324: 387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411: 603–606 [DOI] [PubMed] [Google Scholar]
- Padyukov L, Seielstad M, Ong RT, Ding B, Ronnelid J, Seddighzadeh M, Alfredsson L, Klareskog L 2011. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann Rheum Dis 70: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N 2010. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet 42: 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA 2002. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet 31: 33–36 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. 2007. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39: 596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K 1996. The future of genetic studies of complex human diseases. Science 273: 1516–1517 [DOI] [PubMed] [Google Scholar]
- Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I 1993. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75: 253–261 [DOI] [PubMed] [Google Scholar]
- Sawcer S 2008. The complex genetics of multiple sclerosis: Pitfalls and prospects. Brain 131: 3118–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S, Ban M, Wason J, Dudbridge F 2010. What role for genetics in the prediction of multiple sclerosis? Ann Neurol 67: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdel A, Dai KZ, Lorentzen AR, Flato B, Maslinski S, Thorsby E, Forre O, Spurkland A 2004. Genetic association between juvenile rheumatoid arthritis and polymorphism in the SH2D2A gene. Genes Immun 5: 310–312 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Lusis AJ 2002. The allelic structure of common disease. Hum Mol Genet 11: 2455–2461 [DOI] [PubMed] [Google Scholar]
- Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, et al. 2004. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 53: 3020–3023 [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, et al. 2006. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38: 617–619 [DOI] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, et al. 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, et al. 2010. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surolia I, Pirnie SP, Chellappa V, Taylor KN, Cariappa A, Moya J, Liu H, Bell DW, Driscoll DR, Diederichs S, et al. 2010. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature 466: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJ, Ohashi PS, Griesser H, et al. 1995. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor β. Science 268: 1472–1476 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Zhou YW, Kato M, Mak TW, Nakashima I 1999. Normal regulatory α/β T cells effectively eliminate abnormally activated T cells lacking the interleukin 2 receptor β in vivo. J Exp Med 190: 1561–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert R, Schwendemann J, Kyewski B 2007. Highly variable expression of tissue-restricted self-antigens in human thymus: Implications for self-tolerance and autoimmunity. Eur J Immunol 37: 838–848 [DOI] [PubMed] [Google Scholar]
- The Finnish-German APECED Consortium 1997. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet 17: 399–403 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium 2003. The International HapMap Project. Nature 426: 789–796 [DOI] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium 2005. A high-density screen for linkage in multiple sclerosis. Am J Hum Genet 77: 454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium 2007. Risk alleles for multiple sclerosis identified by a genomewide study. New Engl J Med 357: 851–862 [DOI] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium, Bush WS, Sawcer SJ, De Jager PL, Oksenberg JR, McCauley JL, Pericak-Vance M, Haines JL 2010. Evidence for polygenic susceptibility to multiple sclerosis—The shape of things to come. Am J Hum Genet 86: 621–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium and the Wellcome Trust Case Control Consortium 2 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MHC Sequencing Consortium 1999. Complete sequence and gene map of a human major histocompatibility complex. Nature 401: 921–923 [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3000 shared controls. Nature 464: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39: 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J 2011. The MHC, disease and selection. Immunol Lett 137: 1–8 [DOI] [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. 2003. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423: 506–511 [DOI] [PubMed] [Google Scholar]
- Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C 1997. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 15: 289–292 [DOI] [PubMed] [Google Scholar]
- van der Helm-van Mil AH, Huizinga TW, de Vries RR, Toes RE 2007. Emerging patterns of risk factor make-up enable subclassification of rheumatoid arthritis. Arthritis Rheum 56: 1728–1735 [DOI] [PubMed] [Google Scholar]
- Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, et al. 2005. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 37: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Todd JA 1996. Genetic analysis of autoimmune disease. Cell 85: 311–318 [DOI] [PubMed] [Google Scholar]
- Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, Gohlke H, Wagenpfeil S, Ollert M, Ring J, et al. 2008. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 4: e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger S, Baurecht H, Naumann A, Novak N 2010. Genome-wide association studies on IgE regulation: Are genetics of IgE also genetics of atopic disease? Curr Opin Allergy Clin Immunol 10: 408–417 [DOI] [PubMed] [Google Scholar]
- Wen L, Wong FS, Sherwin R, Mora C 2002. Human DQ8 can substitute for murine I-Ag7 in the selection of diabetogenic T cells restricted to I-Ag7. J Immunol 168: 3635–3640 [DOI] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. 2001. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet 27: 18–20 [DOI] [PubMed] [Google Scholar]
- Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW 1995. Interleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity 3: 521–530 [DOI] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S 2008. CTLA-4 control over Foxp3+ regulatory T cell function. Science 322: 271–275 [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, van Diemen CC, Wijmenga C 2009. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet 10: 43–55 [DOI] [PubMed] [Google Scholar]
- Ziegler AG, Nepom GT 2010. Prediction and pathogenesis in type 1 diabetes. Immunity 32: 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]