SUMMARY
Genomics methodologies have advanced to the extent that it is now possible to interrogate the gene expression in a single cell but proteomics has traditionally lagged behind and required much greater cellular input and was not quantitative. Coupling protein with gene expression data is essential for understanding how cell behavior is regulated. Advances primarily in mass spectrometry have, however, greatly improved the sensitivity of proteomics methods over the last decade and the outcome of proteomic analyses can now also be quantified. Nevertheless, it is still difficult to obtain sufficient tissue from staged mammalian embryos to combine proteomic and genomic analyses. Recent developments in pluripotent stem cell biology have in part addressed this issue by providing surrogate scalable cell systems in which early developmental events can be modeled. Here we present an overview of current proteomics methodologies and the kind of information this can provide on the biology of human and mouse pluripotent stem cells.
Individual mammalian embryos are not amenable to analysis by classic protein techniques (e.g., Western blotting or mass spectrometry). However, early developmental events can be modeled in scalable pluripotent stem cell systems.
1. WHY STUDY PROTEOMICS IN STEM CELLS AND DEVELOPMENT?
Development from a single fertilized zygote to a complex multicellular organism occurs within a relatively short period of time compared with the total lifetime of the resulting adult individual. This remarkable feat requires the precise orchestration of multiple sequential and parallel events controlling cell specification, division, position, migration and communication. With the discovery of genes encoded in DNA, decades of research assumed that the blueprint for the embryo lay entirely in the regulation of gene transcription; but there is a growing realization that epigenetics and the status of proteins in a cell play equally crucial roles. There are many (up to 50%) changes in protein expression that do not have a corresponding change in mRNA expression (during early differentiation) (Lu et al. 2009). Showing the presence of signaling pathway components in a cell is also not sufficient to assess their importance, because protein modifications of many types can affect the functioning of the protein in the cell. It is essential to know the nature of specific signaling pathways, downstream targets, and inhibitory networks, as well as the kinetics of their activation.
Most embryos, however, do not lend themselves easily to the techniques that are available to protein chemists. Classic Western blotting to identify proteins and their activation status, more contemporary ChIP-chip or ChIP-seq to identify interacting partners, and mass spectrometry (MS) for large-scale protein identification, generally require more cells and tissue than available from the mammalian embryos that are closest in development to humans. In contrast, genomic and gene expression profiles can be generated these days from just a single cell. The gap, however, is beginning to close to the extent that some protein assays can be performed on more limited numbers of embryos directly, although the surrogate model systems offered by pluripotent stem cells from mice and humans, as we describe here, are proving exceptionally informative for events that probably take place in the early embryo.
2. STEM CELLS AND DEVELOPMENTAL POTENCY
Stem cells are defined by (1) their ability to self-renew, and (2) their ability to differentiate into one or more different cell types. At one end of the spectrum are totipotent cells, like the fertilized egg or early blastomeres that can become all cells of the conceptus. At the other end are spermatagonial stem cells that are unipotent and can only differentiate into sperm. Between these extremes are pluripotent cells of the blastocyst stage of embryonic development and the multipotent stem or progenitor cells of specific tissues and organs like the nervous system (neural progenitor cells) and skin. Multipotent stem cells are able to differentiate to different cell types but usually only those that make up the organ or tissue from which they are derived (reviewed in Jaenisch and Young 2008). In adults, they are thought to be the sources of cells for tissue repair.
3. WHAT ARE PLURIPOTENT STEM CELLS?
Research in the 1960s on teratocarcinomas, spontaneous tumors that look like disorganized embryos found in the testes of some strains of mice, eventually led to the discovery in the 1970's that pluripotent stem cells are also present in early mouse embryos. The experiments performed showed that teratocarcinomas can be induced in mice simply by transplanting normal embryos to extrauterine sites. The tumors that formed contained multiple tissue types as well as a stem cell population that could be maintained indefinitely in an undifferentiated state in culture. When injected into syngeneic hosts, say under the kidney capsule, they would form teratocarcinomas, once more containing the same mixture of differentiated cells. These stem cells are known as embryonal carcinoma (EC) cells. They resemble undifferentiated embryonic cells of the mouse blastocyst stage embryo in many respects, expressing the same cell-surface proteins and enzymes (Gokhale and Andrews 2006; Yu and Thomson 2008). Human EC cell lines have also been derived as a similar stem cell population from the spontaneous teratocarcinomas that can occur in young men, it is thought as a result of germ cell development going awry before birth. Research with these cell lines has provided important knowledge about the properties of these tumors and the early differentiation of the stem cells that they contain. It also provided the intellectual framework for the successful derivation and culture of embryonic stem cells (ESCs) directly from embryos without an intervening teratocarcinoma stage both in mice and in humans.
The observation that early embryos can form teratocarcinomas when transplanted to animals led to the hypothesis that intact embryos may contain cells that are, or can become, pluripotent stem cells. A few years later, ESCs were indeed isolated from the inner cell mass of blastocyst-stage embryos (Evans and Kaufman 1981; Martin 1981). These cells were immortal and able to contribute to embryonic development by differentiation when injected into a host blastocyst, just like EC cells. More importantly, however, they could contribute to the germ line, forming gametes. This allowed them to become the most important vehicle for genetic manipulation in mice, the deletion of genes by homologous recombination. Whereas the first mESCs required culture on so-called “feeder” cells, usually rather ill-defined fibroblast cells that can, for instance, be derived from mouse embryos around mid-gestation, to inhibit differentiation, it is now possible to culture them and maintain the undifferentiated state in defined growth media supplemented with either growth factors or inhibitors of signaling pathways that induce differentiation (Ying et al. 2008). Many years after the first isolation of mESC, the first human ESC (hESC) lines were derived (Thomson 1998) following much the same protocol using feeder cells, although in this case, the cell-surface molecular identity of hESC clearly resembled human EC cells more than mESCs, and the growth factor requirements differed between the two species. Whereas mouse embryonic fibroblasts (MEFs) could be replaced by leukemia inhibitory factor (LIF) and bone morphogenetic protein (BMP) for mESCs, this was not the case for hESC, and two other factors are required in addition to an appropriate coating of the plastic dish: a combination of fibroblast growth factor (FGF) and nodal or activin. hESC and mESC did, however, show a similar capacity to differentiate into many cell types in both teratomas in immunocompromised mice and in culture. In addition, the core transcription factors that are now considered the signature of stem cell pluripotency (Loring and Rao 2006) were clearly largely conserved between mice and humans.
Of course, the use of human embryos to derive hESC made them ethically controversial, and several groups addressed the question of whether direct reprogramming of a somatic nucleus was possible, perhaps using signals much like those present in the oocyte, which led to reprogramming of the genome during the generation of the first cloned animal (Campbell et al. 1996). In 2006, however, Yamanaka and colleagues at Kyoto University reported that the introduction of genes encoding four important stem cell transcription factors (Oct4, Sox2, Klf4, and c-Myc) into adult mouse cells by retroviral transduction resulted in reprogramming them into cells with ESC-like properties (Takahashi and Yamanaka 2006). These reprogrammed cells were referred to as “iPSCs,” for induced pluripotent stem cells. In 2007, Yamanaka and the laboratory of Thomson (Takahashi et al. 2007; Yu et al. 2007) described the successful genetic reprogramming of human adult cells into human iPSCs. Because ESCs and iPSCs are pluripotent and appear to model the first differentiation steps in development faithfully, they are frequently used as a surrogate for these processes when large numbers or cell types not accessible from the human embryo are required for study. For all intents and purposes, hESCs and hiPSCs seem to be very similar, although not identical (see, e.g., commentaries Mummery 2011; Panopoulos et al. 2011; Pera 2011). Imprinting and epigenetic memory during reprogramming have been described as being retained during reprogramming so that during early passages, iPSCs may retain a preference to differentiate to the somatic cell type from which it derives, although this may be lost at later passages (Ohi et al. 2011).
4. AIMS OF THIS ARTICLE
In the present article, we will consider what proteomics has added to date to our understanding of pluripotency networks and linked signaling pathways, how cells exit the pluripotent state, and how lineage might be determined during differentiation of stem cells. Genomics provided the genetic tools for reprogramming to pluripotency and direct reprogramming: what can proteomics add? Answering these questions and others like them in stem cells will likely require standardized growth and differentiation protocols for multiple cell lines, much like those available for mESC, and many of these are now being reported. Defined protocols for differentiation are still under development, although exceptionally, the neural lineages now have some robust protocols (Gossrau et al. 2007; Koch et al. 2009; Lee et al. 2011).
To address the contribution of proteomics to understanding pluripotency and differentiation, we divide the text into two main sections:
Current proteomic methods, focusing on challenges and achievements in charting proteome dynamics and posttranslational modifications (PTMs).
Relevance of proteomics to stem cells and early development: We will focus on principles and examples of the regulation, modification, and interaction of proteins rather than provide an extensive literature survey of the cell lines that have been examined by MS-based proteomics.
5. PROTEOMIC APPROACHES: CONTEMPORARY METHODOLOGIES APPLICABLE TO STEM CELL BIOLOGY
5.1. Mass Spectrometry and Its Use
MS has become one of the most powerful tools for protein identification in cell biology. Mass spectrometers are versatile instruments that come in many different types for multiple applications. Nevertheless, they share one basic concept: they all determine the mass of protein products that can then be used to infer identity. Detection of peptides and proteins by MS is enabled by so-called “soft ionization” techniques, such as electrospray (ES) and matrix-assisted laser desorption/ionization (MALDI), which brings these molecules from the solid to the gas phase without destroying them. Protein identification generally does not occur by measuring the mass of the intact protein because its mass alone is usually not distinctive enough to allow a unique assignment. Rather, proteins are digested with proteases such as trypsin, producing peptides that can then be introduced in the mass spectrometer. This has two distinct advantages: first, it produces an assembly of masses (also called a “peptide fingerprint”), which collectively indicate a unique protein. Second, peptides can be fragmented in the mass spectrometer (much more easily than proteins), generating peptide fragments that can be used to deduce a peptide sequence. The latter approach increases the specificity of protein identification tremendously, especially if multiple peptides can be fragmented per protein. It is for this reason that this technique, known as “tandem MS” (where “tandem” denotes the subsequent mass analysis of a peptide and its fragments) now dominates in the field of proteomics. Collectively, a wide range of mass spectrometers are available, each of which can be used for specific research questions (e.g., protein quantification, PTMs, etc.). Table 1 lists the distinguishing features of some of the most current instruments. Table 2 describes their utility in the field of proteomics, along with some examples of how they have been applied in the area of developmental biology.
Table 1.
Characteristics of contemporary mass spectrometers
| MALDI-TOF | MALDI-TOF/TOF | Q-q-TOF | Ion trap | Q-q-Q (triple quad) | FTICR | Orbitrap | |
|---|---|---|---|---|---|---|---|
| Resolving power | 30,000 | 30,000 | 40,000 | <10,000 | <5000 | >500,000 | 30–240,000 |
| Mass accuracy (ppm) | <3 | <3 | <3 | >50 | >100 | <1 | <3 |
| Scan rate | Fast | Fast | Fast | Fast | Moderate | Slow | Fast |
| Dynamic range | + | + | + | + | +++ | + | ++ |
| Sensitivity | + | + | ++ | ++ | +++ | ++ | ++ |
| MS/MS capability | No | Yes | Yes | Yes | Yes | Yes | Yes |
| Identification | + | ++ | +++ | ++ | + | ++ | +++ |
| Quantification | + | + | ++ | + | +++ | ++ | ++ |
| Throughput | + | ++ | ++ | +++ | ++ | + | +++ |
| PTMs | + | ++ | +++ | ++ | ++ | +++ | +++ |
Abbreviations: MALDI, matrix-assisted laser desorption/ionization; TOF, time of flight; MS, mass spectrometry; PTMs, posttranslational modifications.
Table 2.
Mass spectrometers and their use in proteomics research
| Instrument | Application | Literature examples |
|---|---|---|
| MALDI-TOF | Protein identification by mass fingerprinting; analysis purified proteins, single gel bands, or 2D-gel spots | Kurisaki et al. 2005; Han et al. 2011 |
| MALDI-TOF-TOF | Protein identification by mass fingerprinting or peptide fragmentation; analysis of gel bands, 2D-gel spots, or protein mixtures | Yocum et al. 2008; Fathi et al. 2009; Nasrabadi et al. 2010 |
| Q-q-TOF | Protein identification by peptide fragmentation, analysis of complex protein mixtures after coupling to LC; PTM detection; analysis of intact proteins | Williamson et al. 2008; Chaerkady et al. 2009 |
| Ion trap | Protein identification by peptide fragmentation, analysis of complex protein mixtures after coupling to LC; PTM detection | Lopez-Ferrer et al. 2006; Kang et al. 2011 |
| Q-q-Q | Targeted quantification of proteins by multiple reaction monitoring (MRM); PTM detection by neutral loss or precursor ion scanning; top-down proteomics | Yocum et al. 2008; Costenoble et al. 2011 |
| FTICR | Protein identification and quantification; analysis of intact proteins; top-down proteomics; PTM analysis | Van Hoof et al. 2006; Tipton et al. 2011 |
| Orbitrap | Protein identification and quantification, analysis of complex samples after coupling to LC; PTM analysis; top-down proteomics | Kruger et al. 2008; Phanstiel et al. 2008; Van Hoof et al. 2009 |
Literature refers to applications in developmental biology.
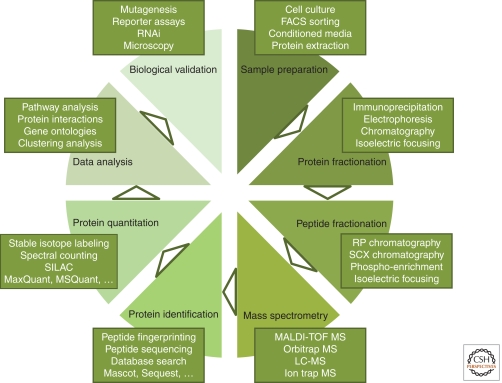
Although MS is one of the key components in proteomics, a number of other developments in accessory techniques have critically contributed to disclosing ever-increasing parts of the proteome. This includes many aspects of proteomic workflows, such as sample preparation, protein and peptide separation, and bioinformatic tools for protein identification, quantification, and data mining (Fig. 1). Some of the challenges that have been met (and that continue to be addressed) are relevant for any proteomic application, whereas others are of particular importance for developmental biology. Therefore, we will briefly mention some of these challenges as well as possible solutions, before describing their application in the field of stem cell biology and mammalian development.
Figure 1.
Workflow for the proteomic analysis of biological samples, from sample preparation to biological validation. The indicated techniques are a nonexhaustive list of examples, which often are used in combination.
6. CHALLENGES IN CHARTING PROTEOMES
If two features stand out distinguishing the proteome from the genome, they are its complexity and dynamic range. Complexity indicates the enormous variation in protein entities that, by far, outnumber the number of genes in the genome. This is caused by the many ways proteins can be processed posttranscriptionally, e.g., by splicing, truncation, or one of several hundred other PTMs. In particular, the decoration of proteins by modifications in a combinatorial way increases the number of protein entities exponentially. For instance, the ∼15 modifications that are known to occur in the tail of Histone 3 alone (Kouzarides 2007) would result in 215 (∼32,000) possible combinations (at least in theory), thereby exceeding the number of genes in mammalian genomes. In turn, this number is dwarfed by proteins containing >100 phosphorylation sites (Gnad et al. 2007), which could produce up to 2100 combinations.
On top of this complexity comes dynamic range, indicating that protein concentration (or copy number) can vary widely among protein species. It is estimated that protein concentration in serum spans 12 orders of magnitude (Anderson and Anderson 2002). For yeast (Picotti et al. 2009) and mammalian cells (Schwanhausser et al. 2011) there is experimental evidence that proteins vary in copy number >6 orders of magnitude. Furthermore, there is no even distribution along this range, because in mammalian cells the 25 most abundant proteins make up 25% of the cellular protein mass, whereas the lower quartile is populated by thousands of proteins (Schwanhausser et al. 2011).
Together, this serves to illustrate that “identifying the human proteome” poses an enormous analytical challenge that will only begin to be tackled in the foreseeable future. This is not to say that meeting this goal only partially cannot be informative―on the contrary. Access to sizable parts of the proteome can be achieved by the implementation of protein and peptide fractionation strategies, or combinations thereof (Fig. 1). Fractionation by protein electrophoresis (Lundby and Olsen 2011), peptide chromatography (Motoyama and Yates 2008), or isoelectric focusing (Krijgsveld et al. 2006) results in reduced complexity per collected fraction, and increased chances to identify more proteins in a subsequent mass spectrometric analysis. Typically, this is the way to uncover low-abundant proteins, which tend to be the ones of highest biological relevance (e.g., kinases and transcription factors). This is the reason why two-dimensional (2D) gel electrophoresis of proteins has largely been replaced by liquid-phase approaches: 2D gels display up to several hundreds of unique (abundant) proteins, whereas a workflow combining protein electrophoresis and peptide chromatography identifies >5000 proteins (Graumann et al. 2008).
This gain in protein number is partly owing to miniaturization of chromatographic columns (∼50 µm diameter) and the introduction of high-pressure liquid chromatography (HPLC) and ultra-high-pressure chromatography (UPLC) pumps capable of stably maintaining low flow rates (∼100 nl/min). On top of this, technical developments in MS have dramatically increased the speed and sensitivity of data acquisition, sequencing 5–10 peptides/sec, or >15,000/h during a chromatographic separation. In combination with fractionation strategies, this results in the identification of many thousands of proteins (Mann and Kelleher 2008). These are not just figures that are of interest to proteomic technologists, but they bear an important message for (stem cell) biologists. Namely, they indicate that relatively low amounts of starting material (105–106 cells) are sufficient to generate proteomic data sets that include all classes of cellular proteins, which are very likely to contain biologically relevant information. It is anticipated that lower cell numbers are within reach (103–104 range), while maintaining high numbers of protein identifications. This means that rare cell populations obtained from tissue culture or fluorescence-activated cell sorting (FACS) are becoming accessible for meaningful proteomic investigation.
7. PHOSPHOPROTEOMICS AND ANALYSIS OF OTHER PTMs
Protein function is often modulated by PTMs. Therefore there is a tremendous interest in identifying PTMs in a biological context. Although many modifications are known to occur in nature, phosphorylation may stand above them all because of its importance in signal transduction and cellular signaling. Even though phosphorylation is widespread, it is often substoichiometric and thus tends to be very low-abundant―and difficult to detect by MS. It is for these reasons that intense efforts have focused on enrichment techniques to isolate phosphopeptides selectively from nonmodified peptides, typically using metal-affinity chromatography (Villen and Gygi 2008; Pinkse et al. 2011). Such approaches have been used to identify many thousands of phosphorylation sites (Dephoure et al. 2008) and have been applied successfully to stem cell research (further described below).
Alternative methods studying PTMs include immunoprecipitation using antibodies, followed by MS. Unfortunately, only a limited number of antibodies are truly modification specific (i.e., disregarding any sequence specificity surrounding the modification). One of the few exceptions is antibodies against phosphorylated tyrosine residues, which have been applied successfully to interrogate specific signaling cascades (Blagoev et al. 2004; Ding et al. 2011). Other modifications have been targeted with moderate success, such as antibodies recognizing acetylated lysines. Nevertheless, recent studies have shown that lysine acetylation modification may be just as widespread as phosphorylation, fulfilling an equally important role in regulating protein function (Yang and Seto 2008); nearly all enzymes in primary metabolism (glycolysis, tricarboxylic acid [TCA] cycle, fatty acid metabolism) can be modified by acetylation, the vast majority of which was shown to be functional (Zhao et al. 2010). This shows that the availability of an analytical tool opens up new ways to study basic cellular processes, even for relatively high-abundant proteins that have been studied for decades. This may be crucial for studying many developmental processes, where cellular differentiation and reprogramming (and tumorigenesis) are accompanied by drastic changes in metabolism (Zhu et al. 2010; Levine and Puzio-Kuter 2011).
8. QUANTITATIVE PROTEOMICS
Another crucial development that has brought proteomic technology considerably closer to cell biology is the ability to quantify protein expression levels. Traditionally, MS has been strong in generating protein inventories; but these lists do not necessarily indicate which proteins are biologically relevant. This is preferably performed in a comparative fashion, contrasting with cells that were isolated from different biological states or that received different treatments. Because MS is not an inherently quantitative method, additional means need to be applied to achieve this goal. One possibility is to take the number of fragmented peptides per protein (“spectral count”) as an approximation for protein abundance. Although this may be useful for some applications, it generally suffers from low sensitivity to detect small differences in protein levels (a less than fivefold change) (Vaudel et al. 2010). A preferred strategy entails the introduction of stable isotopes (e.g., 2H, 13C, 15N, and 18O) into proteins of one sample, that then serve as internal standards for the (unlabeled) proteins in the reciprocal sample (Gouw et al. 2010; Washburn 2011). The principle is based on the premise that labeled and unlabeled proteins/peptides behave identically in the biological experiment and throughout the analytical process, the sole difference being in their mass that can be detected in the mass spectrometer. The relative intensity of a “light” and a “heavy” peak is then a direct measure for their protein abundance in the samples they are derived from.
Technically, proteins can be labeled by stable isotopes in various ways. Among the most powerful methods, cells can be labeled in cell culture in the presence of labeled amino acids (e.g., lysine and/or arginine). This method, known as stable isotope labeling with amino acids in cell culture (SILAC) (Mann 2006), has the advantage that full labeling of all proteins occurs metabolically, i.e., without the need for protein extraction and additional sample handling. Clearly, the prerequisite is that cells of interest can be grown in defined media: the presence of unlabeled amino acids in poorly defined supplements (e.g., serum) will compromise labeling efficiency. The use of dialyzed serum usually circumvents this problem. Another potential caveat is the metabolic conversion of (labeled) arginine into proline, thus diluting the label and complicating downstream data analysis. It should be noted, however, that conversion rates differ greatly between cell types, and therefore testing beforehand is advisable. For highly converting cells, the process can be inhibited by adding an excess of unlabeled proline (Bendall et al. 2008) or by changing the labeling regimen (Van Hoof et al. 2007). Several groups have successfully used SILAC to label ESCs (Van Hoof et al. 2007; Bendall et al. 2008; Graumann et al. 2008; Prokhorova et al. 2009), thus opening the way to use this powerful approach in developmental biology.
If SILAC is not possible or is impractical for a particular cell type, there is a range of possibilities to introduce the isotope label post isolation by chemical means (Bantscheff et al. 2007; Elliott et al. 2009). Without going into great detail here, this includes iTRAQ (Karp et al. 2010), ICPL labeling (Lottspeich and Kellermann 2011), reductive dimethylation (Boersema et al. 2009) and various others. One method is not necessarily better than the other; considerations in choosing either of these methods include: availability of appropriate mass spectrometric infrastructure, price tags of the reagents (some are only commercially available as kits, e.g., iTRAQ), and―pragmatically―experience that is present in the (collaborating) MS laboratory. Availability of appropriate software is becoming less of an issue, because packages accepting various labeling formats are being developed (Cox and Mann 2008; Mortensen et al. 2010).
In summary, it is clear from the above that proteomics has much to teach us about stem cell biology and development, in a highly complementary fashion to genomics, which lacks the ability to accurately chart the behavior and effects of proteins, and is blind to their modifications. However, the application of proteomics has been limited by (1) its requirement for large cellular input (only recently has it become possible to scale up stem cell production to levels appropriate for proteomic analysis, and have proteomics methods been adapted to lower cellular input), (2) its inability to quantify the outcome and compare relative levels of proteomic changes, and (3) limited access for most biologists to high-end proteomic platforms. All of these issues are now being addressed and it is expected that proteomic analysis will reveal much more about stem cell biology in the coming years when complemented with genomic analysis. Yet, the direct application of proteomics to the development of embryos will, for the time being, remain limited to species like zebrafish, fruit flies, and amphibians, in which it is possible to produce large numbers of identically staged embryos or tissues within a reasonable time span and cost. For mice and their embryos as well as for humans, it is likely that yet one higher order of magnitude in sensitivity is required before proteomic analysis, at least by MS, becomes feasible to implement widely in developmental biology and stem cells.
9. PROTEOME BIOLOGY OF STEM CELLS
9.1. What Proteomics Can Teach Us about Cellular Processes
One of the strongest motivations to study cellular processes at the proteome level is the increasing awareness that transcript levels are poor indicators for protein abundance (Schwanhausser et al. 2011). This has been beautifully shown recently for differentiating stem cells, showing that transcription profiles, epigenetic marks, and protein levels were highly divergent (Lu et al. 2009). At the same time, this is one of the few papers describing the dynamics of protein expression during the early phases of differentiation at a relatively large scale. It follows earlier (mostly semiquantitative) studies, mostly studying cellular differentiation. For instance, the first large-scale MS-based study on differentiating hESCs compared the proteomes of undifferentiated hESCs with their derivatives formed after 12 days of undirected “spontaneous” differentiation into a heterogeneous population (Van Hoof et al. 2006). More than 700 of the total of nearly 2300 unique proteins found in both samples were identified as being present in undifferentiated cells only. Of these, 191 were also detected in mouse ESCs in a parallel analysis. Among these were several proteins that at the time were not previously known to be enriched in or specific for ESCs; some examples are proteins like TOP2A, MCM4, KPNA2, and Sall4. Although these numbers represent only a fraction of the actual proteome, the technique as such proved sensitive enough to detect ESC-associated low-abundance transcription factors. These included well-known pluripotency proteins like OCT4 and UTF1 (for review, see Van Hoof et al. 2008). Interestingly, based on their gene ontology, many of the identified proteins were annotated as nuclear, which is to be expected considering the high nucleus-to-cytosol ratio of ESCs. Even though subcellular locations were confirmed for a handful of these proteins using fluorescence microscopy, one of the major advantages of MS-based proteomics over transcriptome analysis, the ability to generate cell compartment-specific data sets, was not implemented in this study.
The nuclear proteomes of hESCs and neural stem cell analogs derived from them were, however, analyzed by a more conventional approach involving isolation of the nuclei by centrifugation before 2D difference gel electrophoresis. The proteins that were differentially expressed were then identified by MS (Barthelery et al. 2009). Although not as comprehensive as an unbiased, discovery-oriented MS-based analysis, this study identified CPSF6 as a novel potential hESC-specific protein.
9.1.1. Membrane Proteomics
In contrast to cytosolic and nuclear proteins, membrane-associated proteins―in particular, transmembrane proteins―are notoriously difficult to purify owing to their generally hydrophobic nature. Therefore, these proteins are likely to be underrepresented in global proteomics analyses. In one study, in an attempt to skew this bias for detection of hydrophilic proteins, samples of hESCs and their tumorigenic counterpart, human EC cells, were enriched for membrane-associated proteins using ultracentrifugation before MS analysis (Dormeyer et al. 2008). The high percentage of commonly expressed proteins confirmed similarities in expression patterns known to exist between hESCs and human EC cells, both of which are pluripotent, divide relatively rapidly, and self-renew by symmetrical division. The relatively few differences in surface proteins that were found might betray signaling pathways that are active specifically in carcinoma cells, shedding light on their tumorigenic behavior as opposed to the benign properties of pluripotent cells in the developing embryo.
9.1.2. Protein Dynamics
The studies described above all used semiquantitative proteomics. However, as mentioned in the previous section, SILAC and iTRAQ are being used increasingly to quantitate the relative differences in protein levels between two samples. The use of SILAC has, however, been challenging because, unfortunately, some hESC lines show a high rate of arginine-to-proline conversion, which compromises the accuracy of SILAC-based quantitation when using heavy stable isotope-containing arginine for protein labeling in vivo. Two approaches have been developed to address this technical issue: inclusion of the “light” forms of the arginine in the “unlabeled” sample (Van Hoof et al. 2007), or lowering the arginine concentration in the medium to a minimum (Bendall et al. 2008). Two independent studies used a very similar quantitative MS approach to compare the proteomes of hESCs and their differentiated derivatives, formed after exposure of the undifferentiated cells to the BMP4-inhibitor noggin, which induces neuronal differentiation (Yocum et al. 2008; Chaerkady et al. 2009). Despite the comparable strategies and analytical methods used, only a small number of proteins were found to be commonly differentially expressed in the two studies. The lack of concordance is not typical for these two studies alone; it appears to be a widespread phenomenon that is often attributed to dissimilarities in differentiation propensities of individual hESC lines, the variety in MS strategies used, and “undersampling,” denoting the property that an MS experiment usually identifies a (random) portion of the sampled proteome.
Combining subcellular fractionation with quantitative MS showed its potential in a search for a cell-surface protein that would allow antibody-based purification of hESC-derived cardiomyocytes (Van Hoof et al. 2010). In this study, directed differentiation was used to induce cardiomyocyte formation from hESCs under SILAC conditions. Although the differentiated population did not consist exclusively of cardiomyocytes, the purified plasma membrane proteome of the cardiomyocyte-enriched populations was compared quantitatively to that of unlabeled undifferentiated hESCs to select differentially expressed surface proteins. In parallel, human fetal heart muscle cells were compared to find surface proteins commonly expressed by primary and stem cell-derived cardiomyocytes. The resulting data sets were then screened for overlapping proteins, identifying EMILIN2 as a candidate that turned out to be suitable for FACS of cardiomyocytes from the heterogeneous pool of hESC-derived differentiated cells. Unfortunately, the protein was not stably associated with the plasma membrane and could only be used to sort fixed but not live cells. The approach nevertheless, was shown to be feasible even for relatively low abundance surface proteins.
9.1.3. Protein–Protein Interactions
An area where MS has been particularly powerful is in the characterization of protein complexes and interactions. Tagging approaches have been designed to capture proteins of interest by affinity purification, followed by proteomic identification of interaction partners. Such studies have been extremely helpful in charting molecular networks, ranging from the local environment of individual proteins to a genome-wide description of protein–protein interaction networks (reviewed in Vermeulen et al. 2008; Wodak et al. 2009; Vidal et al. 2011).
Protein–DNA networks in stem cells have primarily been approached by ChIP-chip and ChIP-seq, taking individual proteins as initial baits, and locating where they bind in the genome. Regions of prime interest include the core pluripotency factors (Oct4, Sox2, and Nanog), in an effort to explain how transcription of these proteins may be modulated by regulatory interactors. Indeed, from these studies it has become apparent that, e.g., enhancer domains of the Oct4 locus interact with a large number of transcriptional regulators, such as Essrb, Tcf3, Zfx, and c-Myc (Young 2011). Recent proteomic screens, taking a more unbiased approach, have expanded these networks considerably by screening for proteins that directly interact with pluripotency factors. This has indicated that Oct4 (Pardo et al. 2010; van den Berg et al. 2010) and Nanog (Wang et al. 2006), either directly or indirectly, interact with an extensive set of proteins, including transcription factors, chromatin remodelers, and components of the basal transcriptional machinery. Many of these interactions have been shown to be functional, indicating that the activity of both Oct4 and Nanog is modulated by a large number of cofactors (Wang et al. 2006; Pardo et al. 2010; van den Berg et al. 2010). This notion also emerged from an elegant study, where affinity purification and lentiviral expression were combined to identify interactions between transcription factors and chromatin remodeling complexes (Mak et al. 2010). These studies are important to position proteins that are pivotal for pluripotency into biochemical context. Importantly, they complement approaches such as ChIP-chip and ChIP-seq, which study genomic localization of individual proteins. The recent protein interaction studies mentioned above have expanded and refined the circuitry controlling pluripotent cell identity, while identifying players potentially contributing to cellular reprogramming.
9.1.4. Protein Phosphorylation Dynamics in Differentiating Human Embryonic Stem Cells
In addition to identifying direct protein interactions in a discovery-oriented manner, MS-based proteomics excels at finding out which signaling pathways are active in a cell, and which become activated or deactivated on external signals. Mapping such dynamic processes goes beyond simply detecting the presence of a protein and determining its subcellular location; it requires the identification of PTMs on these proteins—a task for which MS is exceptionally well suited.
Because phosphorylation is common, and probably one of the earliest PTMs that occur at the onset of differentiation, it was also the first PTM to be investigated in hESCs in a systematic way. An initial screen identified nearly 11,000 phosphorylation sites on more than 4000 proteins in hESCs (Swaney et al. 2009). Among these were several known pluripotency-associated proteins, including the transcription factors OCT4 and SOX2. OCT4 was found to be phosphorylated at serine residue 236 (Ser236), which lies within the DNA-binding domain, implying its involvement in transcriptional activity, whereas multiple phosphorylation sites were found for SOX2 (i.e., Ser246, Ser249, Ser250, and Ser251). However, because these cells were analyzed only in their undifferentiated state, the data set represented a static map of phosphorylated residues of the proteins identified, thereby limiting the interpretation of their biological significance to speculation.
Subsequent studies compared hESCs before and after differentiation to deduce phosphorylation associated with pluripotency and differentiation. Brill et al. (2009) charted the dynamics in phosphorylation that occur in hESCs treated for 4 days with retinoic acid, which induces efficient differentiation into a heterogeneous population of cells. Their semiquantitative analysis indicated that many components of the signaling cascades that are believed to be important for hESC self-renewal are phosphorylated in undifferentiated cells, among which were members of the EGF, VEGF, and PDGF pathways. Indeed, blocking each of the signal transduction pathways individually by inhibiting the activity of receptors at the top of these signaling chains resulted in major morphological changes in the cells in addition to reducing or inducing complete loss of the transcription factors OCT4 and NANOG.
Van Hoof et al. (2009) applied SILAC-based MS to quantify early phosphorylation changes that occur in hESCs at multiple time points (30 min, 1 h, and 4 h) on BMP4-induced differentiation. BMP4 is one of the most rapid inducers of differentiation in hESC (Pera et al. 2004); early derivatives eventually form mesoderm or trophectoderm. The resulting data set, consisting of more than 5000 proteins and 3000 phosphosites, provided quantitative as well as temporal information on the dynamics of phosphorylation events in hESCs during their exit from the pluripotent state and their commitment to a specific lineage. Subjecting the data to an algorithm that links phosphorylation motifs to kinases (Linding et al. 2007), CDK1/2 emerged as a central kinase regulating self-renewal and lineage specification. Furthermore, the largest group identified comprised nucleic acid-binding proteins and transcription factors; both were significantly reduced in differentiating cells, which indicates that these classes of proteins are highly represented in hESCs. Interestingly, the three consecutive serine residues of the transcription factor SOX2 that had been found to be phosphorylated (i.e., Ser249, Ser250, and Ser251) did not show altered phosphorylation levels in the differentiating cells, even though the protein itself was rapidly eliminated after the onset of differentiation. Instead, phosphorylation of these residues was associated with increased binding of the small ubiquitin-related modifier (SUMO) to the proximate lysine residue at position 245 (Lys245), as assessed by mutating the three serine residues to aspartic acids, thereby mimicking the negative charge resulting from phosphorylation. PolySUMOylation of a lysine residue usually targets the modified protein for proteasomal degradation (Gareau and Lima 2010). Interestingly, the combined lysine and serine residues closely match a defined phosphorylation-dependent SUMOylation motif also described for other SOX family members (Hietakangas et al. 2006). Therefore, the unwavering presence of SOX2 in undifferentiated cells, despite continuous phosphorylation and its likely subsequent SUMO-induced degradation in self-renewing hESCs, suggests that the levels of this transcription factor are tightly controlled by a fine balance between translation and degradation. The proposed importance of a consistent level of this core transcription factor in hESCs is in line with that already observed for OCT4: a twofold increase of the latter initiates differentiation into primitive endoderm and mesoderm, whereas a decrease results in the formation of trophectoderm (Niwa et al. 2000).
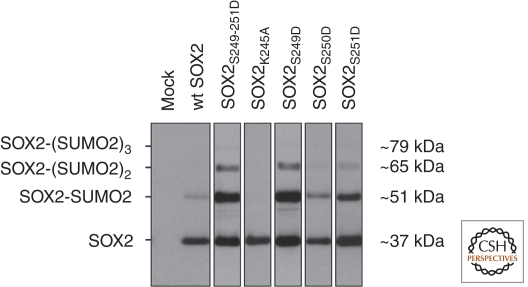
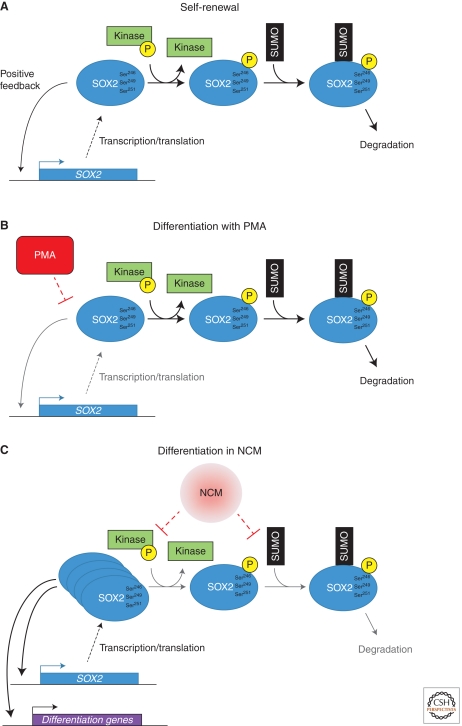
Rigbolt et al. (2011) reported an impressive 6500 different proteins and 23,500 phosphorylation sites identified in differentiating hESCs that were exposed to either nonconditioned medium (NCM) or a diacylglycerol analog (phorbol 12-myristate 13-acetate, PMA). These extremely high numbers of identifications illustrate the rapid progress in size and biological relevance of data sets that can be generated nowadays with MS-based proteomics since the first hESC proteome was profiled five years earlier (Van Hoof et al. 2006). SILAC permitted quantitative cross-comparison between the two conditions in the study of Rigbolt et al. (2011), revealing that, irrespective of the type of differentiation induced, serine residues within basic or acidic amino acid-rich motifs generally became progressively phosphorylated, whereas those with an adjacent proline residue showed reduced phosphorylation. In contrast, a decrease in SOX2 phosphorylation was observed for only Ser246, Ser249, and Ser251 under NCM conditions; phosphorylation ratios remained unchanged on treatment with PMA. Concomitantly, the level of SOX2 protein increased in the cells differentiating in NCM, whereas the level in those differentiated in the presence of PMA decreased. This implies that, when these residues are unphosphorylated, the protein is protected from degradation, whereas phosphorylation does the opposite. This is in agreement with the earlier hypothesis that relates the introduction of an acidic group to the trio of serine residues through phosphorylation to an increase in SUMOylation of Lys245, followed by proteasomal degradation of SOX2 (Van Hoof et al. 2009). Checking the individual serine residues within the triplet independently, we found that mutating Ser249 and Ser251 to aspartic acids increased SUMOylation, whereas mutating Ser250 did not (Fig. 2). Combined, these data fit a model describing regulation of SOX2 protein levels within hESCs and differentiating cells that is dependent on multiple PTMs, i.e., phosphorylation and SUMOylation, the latter of which is causally related to the former (Fig. 3).
Figure 2.
Western blots showing wild-type SOX2 and SOX2 mutants expressed in HeLa cells. HeLa cells that were transfected with 6 × histidine-tagged SUMO2 in combination with either wild-type SOX2 (lane 2) or SOX2 mutants where serine residues 249, 250, and 251 were all (lane 3) or individually (lanes 5–7) mutated to aspartic acid residues, which mimics constitutive phosphorylation of the respective serine residues. SUMO2-SOX2 complexes were purified from the cell lysates with Ni-NTA beads binding the 6 × histidine tag of SUMO2, and then subjected to SDS-PAGE and Western blotting using a SOX2-specific antibody. Wild-type SOX2 (lane 2) is marginally SUMOylated; mutating lysine 245 into alanine (lane 4) shows that this residue is the SUMO target site. Whereas mutation of serine 250 into aspartic acid (lane 6) has no pronounced effect, mutating either all three serine residues simultaneously (lane 3) or serine 249 (lane 5) or 251 (lane 7) individually results in increased polySUMOylation. Mock: negative control of HeLa cells transfected with an empty plasmid (D Van Hoof, J Krijgsveld, and CL Mummery, unpubl.).
Figure 3.
Schematic of the effects of phosphorylation and subsequent SUMOylation of SOX2 in self-renewing and differentiating pluripotent stem cells. The results from the study by Rigbolt et al. (2011) and our own data (Van Hoof et al. 2009; and the unpublished results of Fig. 1) imply that there is a fine balance between the transcription/translation and degradation rates of SOX2 to sustain self-renewal (A). If the transcription/translation feedback loop is disturbed by, e.g., PMA, the continuous phosphorylation, the subsequent SUMOylation, and the eventual degradation of the remaining SOX2 proteins lead to a steady decrease in SOX2 levels, resulting in a shift toward differentiation (B). On the other hand, if the intracellular levels of SOX2 increase owing to a decrease in the degradation rate, the cells will also differentiate as a result of the transcription/translation of differentiation-associated genes (C). The dotted lines indicate that the precise action and targets of these differentiation-inducing conditions are not known. Abbreviations: PMA, phorbol 12-myristate 13-acetate; NCM, non-conditioned medium.
10. FUTURE PERSPECTIVES
Rapid advances in contemporary proteomics methodologies and commercial media preparations for cell expansion and differentiation are now making it possible to scale up cell production at every stage of development. This is independent of whether cells are undifferentiated, at lineage progenitor stages, or are in the process of reprogramming to iPSCs. Genomic and proteomic assays can now be performed simultaneously and with increasing sensitivity on the same samples. Direct genome-wide comparisons of the transcriptome with the proteome will increasingly lead to integrated analyses of stem cells before and during differentiation so that systems biology approaches will be feasible. As importantly, it will be possible to integrate data on the epigenetic status of cells so that their self-renewal can be maintained, not only when the cells are undifferentiated and pluripotent, but also at lineage progenitor stages. Neural progenitors from pluripotent stem cells are at present the only progenitors from pluripotent stem cells that can be expanded robustly in culture. This has a great advantage for future clinical applications, because it does not require repeated return to the undifferentiated cells, which have the capacity for teratomas after transplantation.
Among the specific stem cell questions that still need to be addressed are:
Why do some (pluripotent) stem cells preferentially differentiate into one cell lineage or type above another? A survey of ∼70 hESC lines (Adewumi et al. 2007) showed no obvious differences at the genome/transcriptome level or in multiple other characteristics, when the cells were grown under essentially identical conditions. At what (proteomic) level might this be regulated? Answers to some of these questions might be lurking in the vast amounts of MS data produced these days (Phanstiel et al. 2011).
Recent results show that both mouse and human iPSCs can retain an “epigenetic memory” of the somatic cell that they are derived from, and this only disappears after either secondary reprogramming (in mice) or after prolonged passage (in human). At what level is this regulated? As indicated above, proteomics provide some answers (Phanstiel et al. 2011).
As further refinement takes place and proteomics increases in sensitivity, it may one day become possible to analyze the proteomic status of embryos directly with a limited amount of tissue or cells so that issues of erroneous imprinting in development can be addressed proteome and genome wide. Until that time, stem cells in their various forms will provide a much needed source of new and exciting information on the control of self-renewal and directed differentiation.
ACKNOWLEDGMENTS
Research in the Mummery Laboratory is supported by the Netherlands Proteomics Consortium (NPC: 050-040-250). J.K. is supported by a Vidi grant from the Netherlands Organisation for Scientific Research (NWO).
Footnotes
Editors: Patrick P.L. Tam, W. James Nelson, and Janet Rossant
Additional Perspectives on Mammalian Development available at www.cshperspectives.org
REFERENCES
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. 2007. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25: 803–816 [DOI] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG 2002. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics 1: 845–867 [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B 2007. Quantitative mass spectrometry in proteomics: A critical review. Anal Bioanal Chem 389: 1017–1031 [DOI] [PubMed] [Google Scholar]
- Barthelery M, Jaishankar A, Salli U, Freeman WM, Vrana KE 2009. 2-D DIGE identification of differentially expressed heterogeneous nuclear ribonucleoproteins and transcription factors during neural differentiation of human embryonic stem cells. Proteomics Clin Appl 3: 505–514 [DOI] [PubMed] [Google Scholar]
- Bendall SC, Hughes C, Stewart MH, Doble B, Bhatia M, Lajoie GA 2008. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol Cell Proteomics 7: 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev B, Ong SE, Kratchmarova I, Mann M 2004. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol 22: 1139–1145 [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ 2009. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4: 484–494 [DOI] [PubMed] [Google Scholar]
- Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S 2009. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5: 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, Wilmut I 1996. Sheep cloned by nuclear transfer from a cultured cell line. Nature 380: 64–66 [DOI] [PubMed] [Google Scholar]
- Chaerkady R, Kerr CL, Marimuthu A, Kelkar DS, Kashyap MK, Gucek M, Gearhart JD, Pandey A 2009. Temporal analysis of neural differentiation using quantitative proteomics. J Proteome Res 8: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costenoble R, Picotti P, Reiter L, Stallmach R, Heinemann M, Sauer U, Aebersold R 2011. Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol Syst Biol 7: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP 2008. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding VM, Boersema PJ, Foong LY, Preisinger C, Koh G, Natarajan S, Lee DY, Boekhorst J, Snel B, Lemeer S, et al. 2011. Tyrosine phosphorylation profiling in FGF-2 stimulated human embryonic stem cells. PLoS One 6: e17538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormeyer W, Van Hoof D, Braam SR, Heck AJ, Mummery CL, Krijgsveld J 2008. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J Proteome Res 7: 2936–2951 [DOI] [PubMed] [Google Scholar]
- Elliott MH, Smith DS, Parker CE, Borchers C 2009. Current trends in quantitative proteomics. J Mass Spectrom 44: 1637–1660 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Fathi A, Pakzad M, Taei A, Brink TC, Pirhaji L, Ruiz G, Bordbar MST, Gourabi H, Adjaye J, Baharvand H, et al. 2009. Comparative proteome and transcriptome analyses of embryonic stem cells during embryoid body-based differentiation. Proteomics 9: 4859–4870 [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD 2010. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M 2007. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol 8: R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale PJ, Andrews PW 2006. A prospective on stem cell research. Semin Reprod Med 24: 289–297 [DOI] [PubMed] [Google Scholar]
- Gossrau G, Thiele J, Konang R, Schmandt T, Brustle O 2007. Bone morphogenetic protein-mediated modulation of lineage diversification during neural differentiation of embryonic stem cells. Stem Cells 25: 939–949 [DOI] [PubMed] [Google Scholar]
- Gouw JW, Krijgsveld J, Heck AJ 2010. Quantitative proteomics by metabolic labeling of model organisms. Mol Cell Proteomics 9: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J, Hubner NC, Kim JB, Ko K, Moser M, Kumar C, Cox J, Scholer H, Mann M 2008. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics 7: 672–683 [DOI] [PubMed] [Google Scholar]
- Han I, Jeong SJ, Lee HJ, Koh W, Lee EO, Kim HS, Lee SJ, Chen CY, Jung MH, Kim SH 2011. Proteomic analysis of mesenchymal stem-like cells derived from ovarian teratoma: Potential role of glutathione S-transferase M2 in ovarian teratoma. Proteomics 11: 352–360 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci 103: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SU, Heo S, Lubec G 2011. Mass spectrometric analysis of GABAA receptor subtypes and phosphorylations from mouse hippocampus. Proteomics 11: 2171–2181 [DOI] [PubMed] [Google Scholar]
- Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS 2010. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics 9: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O 2009. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci 106: 3225–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krijgsveld J, Gauci S, Dormeyer W, Heck AJ 2006. In-gel isoelectric focusing of peptides as a tool for improved protein identification. J Proteome Res 5: 1721–1730 [DOI] [PubMed] [Google Scholar]
- Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M 2008. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134: 353–364 [DOI] [PubMed] [Google Scholar]
- Kurisaki A, Hamazaki TS, Okabayashi K, Iida T, Nishine T, Chonan R, Kido H, Tsunasawa S, Nishimura O, Asashima M, et al. 2005. Chromatin-related proteins in pluripotent mouse embryonic stem cells are downregulated after removal of leukemia inhibitory factor. Biochem Biophys Res Commun 335: 667–675 [DOI] [PubMed] [Google Scholar]
- Lee G, Chambers SM, Tomishima MJ, Studer L 2011. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc 5: 688–701 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM 2011. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330: 1340–1344 [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. 2007. Systematic discovery of in vivo phosphorylation networks. Cell 129: 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ferrer D, Ramos-Fernandez A, Martinez-Bartolome S, Garcia-Ruiz P, Vazquez J 2006. Quantitative proteomics using 16O/18O labeling and linear ion trap mass spectrometry. Proteomics 6: S4–S11 [DOI] [PubMed] [Google Scholar]
- Loring JF, Rao MS 2006. Establishing standards for the characterization of human embryonic stem cell lines. Stem Cells 24: 145–150 [DOI] [PubMed] [Google Scholar]
- Lottspeich F, Kellermann J 2011. ICPL labeling strategies for proteome research. Methods Mol Biol 753: 55–64 [DOI] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. 2009. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Olsen JV 2011. GeLCMS for in-depth protein characterization and advanced analysis of proteomes. Methods Mol Biol 753: 143–155 [DOI] [PubMed] [Google Scholar]
- Mak AB, Ni Z, Hewel JA, Chen GI, Zhong G, Karamboulas K, Blakely K, Smiley S, Marcon E, Roudeva D, et al. 2010. A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency. Mol Cell Proteomics 9: 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M 2006. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 7: 952–958 [DOI] [PubMed] [Google Scholar]
- Mann M, Kelleher NL 2008. Precision proteomics: The case for high resolution and high mass accuracy. Proc Natl Acad Sci 105: 18132–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, Cox J, Foster LJ, Heck AJ, Blagoev B, et al. 2010. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res 9: 393–403 [DOI] [PubMed] [Google Scholar]
- Motoyama A, Yates JR III 2008. Multidimensional LC separations in shotgun proteomics. Anal Chem 80: 7187–7193 [DOI] [PubMed] [Google Scholar]
- Mummery C 2011. Induced pluripotent stem cells—A cautionary note. New Engl J Med 364: 2160–2162 [DOI] [PubMed] [Google Scholar]
- Nasrabadi D, Larijani MR, Fathi A, Gourabi H, Dizaj AV, Baharvand H, Salekdeh GH 2010. Nuclear proteome analysis of monkey embryonic stem cells during differentiation. Stem Cell Rev 6: 50–61 [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. 2011. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13: 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Ruiz S, Izpisua Belmonte JC 2011. iPSCs: Induced back to controversy. Cell Stem Cell 8: 347–348 [DOI] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J 2010. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF 2011. Stem cells: The dark side of induced pluripotency. Nature 471: 46–47 [DOI] [PubMed] [Google Scholar]
- Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley E, Ward-van Oostwaard D, Mummery CL 2004. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci 117: 1269–1280 [DOI] [PubMed] [Google Scholar]
- Phanstiel D, Brumbaugh J, Berggren WT, Conard K, Feng X, Levenstein ME, McAlister GC, Thomson JA, Coon JJ 2008. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci 105: 4093–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, et al. 2011. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Methods 8: 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R 2009. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse MW, Lemeer S, Heck AJ 2011. A protocol on the use of titanium dioxide chromatography for phosphoproteomics. Methods Mol Biol 753: 215–228 [DOI] [PubMed] [Google Scholar]
- Prokhorova TA, Rigbolt KT, Johansen PT, Henningsen J, Kratchmarova I, Kassem M, Blagoev B 2009. Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol Cell Proteomics 8: 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4: rs3. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Thomson JA, Coon JJ 2009. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci 106: 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Thomson JA 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Tipton JD, Tran JC, Catherman AD, Ahlf DR, Durbin KR, Kelleher NL 2011. Analysis of intact protein isoforms by mass spectrometry. J Biol Chem 286: 25451–25458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D, Passier R, Ward-Van Oostwaard D, Pinkse MW, Heck AJ, Mummery CL, Krijgsveld J 2006. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics 5: 1261–1273 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Pinkse MW, Oostwaard DW, Mummery CL, Heck AJ, Krijgsveld J 2007. An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat Methods 4: 677–678 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Heck AJ, Krijgsveld J, Mummery CL 2008. Proteomics and human embryonic stem cells. Stem Cell Res 1: 169–182 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J 2009. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5: 214–226 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Dormeyer W, Braam SR, Passier R, Monshouwer-Kloots J, Ward-van Oostwaard D, Heck AJ, Krijgsveld J, Mummery CL 2010. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res 9: 1610–1618 [DOI] [PubMed] [Google Scholar]
- Vaudel M, Sickmann A, Martens L 2010. Peptide and protein quantification: A map of the minefield. Proteomics 10: 650–670 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Hubner NC, Mann M 2008. High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr Opin Biotechnol 19: 331–337 [DOI] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. 2011. Interactome networks and human disease. Cell 144: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villen J, Gygi SP 2008. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3: 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Washburn MP 2011. Driving biochemical discovery with quantitative proteomics. Trends Biochem Sci 36: 170–177 [DOI] [PubMed] [Google Scholar]
- Williamson AJ, Smith DL, Blinco D, Unwin RD, Pearson S, Wilson C, Miller C, Lancashire L, Lacaud G, Kouskoff V, et al. 2008. Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol Cell Proteomics 7: 459–472 [DOI] [PubMed] [Google Scholar]
- Wodak SJ, Pu S, Vlasblom J, Seraphin B 2009. Challenges and rewards of interaction proteomics. Mol Cell Proteomics 8: 3–18 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E 2008. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum AK, Gratsch TE, Leff N, Strahler JR, Hunter CL, Walker AK, Michailidis G, Omenn GS, O’Shea KS, Andrews PC 2008. Coupled global and targeted proteomics of human embryonic stem cells during induced differentiation. Mol Cell Proteomics 7: 750–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA 2011. Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Thomson JA 2008. Pluripotent stem cell lines. Genes Dev 22: 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S 2010. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7: 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]