Summary
Aptamers and the SELEX process were discovered over two decades ago. These discoveries have spawned a productive academic and commercial industry. The collective results provide insights into biology, past and present, through an in vitro evolutionary exploration of the nature of nucleic acids and their potential roles in ancient life. Aptamers have helped usher in an RNA renaissance. Here we explore some of the evolution of the aptamer field and the insights it has provided for conceptualizing an RNA world, from its nascence to our current endeavor employing aptamers in human proteomics to discover biomarkers of health and disease.
In vitro techniques for producing nucleotide sequences that bind to particular proteins allow us to explore the role of RNA in ancient life forms, as well as generate new disease biomarkers and therapeutic agents.
1. INTRODUCTION
Aptamers, the output of the SELEX process (Systematic Evolution of Ligands by EXponential enrichment), have now had 20 years to “show their stuff.” They have done so admirably, and in so doing have allowed us to wonder about what else oligonucleotides might be doing that we have yet to discover as we poke around the biosphere. Deeply embedded within the concepts of the RNA World are questions regarding present biology as well as how we got here. A famous Dobzhansky quote echoed often by Carl Woese suffices—“Nothing in biology makes sense except in the light of evolution” (Dobzhansky 1973). Aptamers open our eyes to some of the possibilities.
2. THE HISTORY OF SELEX AND APTAMERS
Let us first provide a little history. Craig Tuerk was finishing his PhD thesis at the University of Colorado, and had taken on the task of more deeply understanding the nature of the “translational operator” within the bacteriophage T4 gene 43 mRNA. A hairpin and the Shine and Dalgarno domain just 5′ to the initiating AUG of the gene 43 mRNA is the RNA motif that is bound by the gene 43 protein (the replicative enzyme encoded by T4) to repress further synthesis of the polymerase when the level of replication is appropriate. Craig decided to mutate completely the hairpin loop within that motif. Those eight nucleotides were the focus of the first SELEX experiment. That first SELEX experiment (Tuerk and Gold 1990) yielded two winning hairpins among the ∼65,000 sequences of length eight—the wild type T4 sequence and another containing four changes (a quadruple “mutation” over eight nucleotides). Those four changes appeared to reduce the loop size from eight nucleotides to four nucleotides, even though the two hairpins bound with the same affinities to the gene 43 protein.
These experiments defined the SELEX process. The resulting ligands were coined “aptamers” (derived from the Greek word aptus; “to fit”) by Andy Ellington and Jack Szostak in independent work that devised the same general strategy (Ellington and Szostak 1990; see also Green et al. 1990). The surprising data on the gene 43 mRNA motif drove us to generalize from the SELEX method to useful (in a commercial sense) single-stranded oligonucleotide shapes that could be identified through SELEX.
At some point shortly after Craig's paper, we expanded the randomized domain from eight nucleotides to 30 or 40 [and, later, at NeXstar, 50 to prove a point (Jellinek et al. 1993)], reasoning that one must access 30 randomized nucleotides or more to provide sufficient length to generate hairpins, G-quartets, bulges, and pseudo-knots. We thought then, and largely think today (but see later), that the helical regions of aptamers provide stable secondary structures that allow loops and other single-stranded regions to “collapse” into whatever three-dimensional shape is most likely. This thinking was influenced by CUUCGG hairpins (Tuerk et al. 1988), data on other common “tetraloops” (Woese et al. 1983), and the extraordinary structures within the loops of tRNAs (Robertus et al. 1974; Suddath et al. 1974). We studied many proteins quickly, as targets, and reached the conclusion that SELEX would yield aptamers to many (if not all) proteins. Before the word “aptamer” became widely used, Craig had chosen the words “nucleic acid antibodies” (meaning antibodies made out of nucleic acids and NOT antibodies to nucleic acids—we are all happy that the word “aptamer” survived).
The history continued with the creation of NeXagen in 1992; NeXagen became, after some biotech stuff, a company called NeXstar. NeXagen and NeXstar were dedicated to the development of aptamers as therapeutic agents, exactly analogous to antibodies or antibody mimics. Many good aptamers were identified, some of which are in clinical development today. The first aptamer taken into the clinic was NX1838 (now called Macugen), a modified RNA aptamer with a low Kd for Vascular Endothelial Growth Factor and an activity that prevented VEGF165 from binding to its high affinity receptors. NX1838 is a VEGF antagonist, and thus an angiogenesis inhibitor (Ruckman et al. 1998). NX1838 was tested against the wet form of age-related macular degeneration (ARMD), and, in the midst of that trial, NeXstar was acquired by Gilead (Gragoudas et al. 2004; Gonzales 2005). Shortly thereafter the therapeutic rights to NX1838 were licensed to Eyetech who finished the clinical development, renamed the compound Macugen, and after FDA approval started selling the drug in about January, 2005 (Doggrell 2005). Macugen would have been a commercial as well as financial success except that Macugen was selected specifically to target the VEGF isoform VEGF165 and does not antagonize isoform VEGF121 because the binding site of Macugen is missing in the shorter protein. Lucentis (and Avastin), two slightly different antibodies aimed at VEGF121 and also VEGF165 beat Macugen in the market based on more rapid and complete clinical response. Although VEGF165 was clearly the predominant and most active isoform, the role of VEGF121 in ARMD was not fully elucidated when Macugen was taken into the clinic (Kaiser 2006). Nevertheless, as concerns emerge that pan-VEGF inhibition is associated with serious cardiovascular and CNS events, it is worth noting that selective inhibition of only VEGF165 may still be a useful option for long-term therapy of ARMD, since some VEGF activity is now known to be required for maintenance of normal blood vessels and retinal neurons (Nishijima et al. 2007; Saint-Geniez et al. 2009).
Several other aptamers are now in clinical development: AS1411 from Antisoma that targets nucleolin for acute myeloid leukemia and renal cell carcinoma; REG1 from Regado that targets Factor IX for coronary artery bypass graft and percutaneous coronary intervention; ARC1779 from Archemix that targets VWF for thrombotic microangiopathies and thrombocytopenic purpura; NU172 from ARCA that targets thrombin for coronary artery bypass graft and percutaneous coronary intervention; E10030 from Ophthotech that targets PDGF-B for ARMD and diabetic retinopathy; ARC1905 from Archemix that targets C5 for ARMD; and NOX-E36 from Noxxon that targets MCP-1 for kidney disease. Several companies including SomaLogic are engaged in developing different versions of aptamers. For example, Archemix and Ophthotech are working on RNA aptamers and NOXXON Pharma is working on spiegelmers, which are mirror-image L-RNA aptamers (Klussmann et al. 1996). Most, but not all of the aptamers in clinical development are modified RNA molecules (modified so as to have slow degradation rates from endogenous human RNases). The present therapeutic market for monoclonal antibodies (with which aptamers would compete, both being aimed largely at extracellular target molecules) is about $35B, and within that growing market there are opportunities for highly specific antagonists. The dominant patent position, staked out in a patent (Gold and Tuerk 1993) approved by the US Patent Office in 1993, will end over the next few years.
3. PROTEOMICS: DRIVING SELEX TO THE BEST POSSIBLE “WINNERS”
In the last few years at NeXstar, one of us became convinced that a major aptamer value was proteomics. We thought extensively about the use of ELISAs for measuring single analytes, and understood the value of using a sandwich of two good monoclonal antibodies to measure a rare protein in plasma, for example. That value, simply stated, is that one can multiply the specificity of each monoclonal for the intended analyte and thus ignore far more abundant proteins toward which the two monoclonal antibodies have higher Kd's. A great ELISA will quantify a nonabundant analyte below 1 pM (and sometimes at 10 fM) in plasma. Although not often stated explicitly, the issue in any assay in complex matrices is noise, not signal, and sandwich assays are intended to reduce noise (Zichi et al. 2008). Nucleic acid biochemists understand this concept deeply—“nested” PCR using contiguous primer pairs is a form of a “sandwich assay” in which specificities can be multiplied.
We were not concerned about the speed of doing SELEX. The SELEX literature contains methods to do SELEX in fewer rounds than we have found to be optimal (Tok and Fischer 2008; Lou et al. 2009), with some methods aiming to find good aptamers in a single round. We doubt that such protocols can work—if the best molecules in a library are present at a frequency of 10−9 to 10−13 (Gold 1995) or even lower, it is very difficult to discard all losers in a single round. It is, however, entirely possible to lose interesting sequences that may initially exist as a single copy, especially in the first round, because starting random libraries of 1014 to 1015 molecules are typically used for practical reasons. Furthermore, the work one does after selection to characterize aptamers is so vast compared to the selections themselves (which we do anyway in 96-well plates with many targets processed in parallel), we see no serious value in speeding up the nonrate limiting piece of the work. The limitation is aptamer quality—the equilibrium and kinetic properties needed to achieve high specificity.
We defined a great aptamer as one that would provide the specificity (in plasma, for example) of a pair of great antibodies. A low Kd was not going to be sufficient if one wants to use a single binding reagent. One can see this through a concocted example. Imagine that albumin in blood is present at 1 mM and that one wants to measure some analyte present at 1 pM. But then imagine that the capture agent (the monoclonal antibody or the aptamer) has a 1 pM Kd (which would be an exceptionally good Kd) for the intended analyte and binds albumin with the horrible affinity of 1 mM (merely a kiss in time). In that situation all measurements of the intended protein would actually measure half intended protein and half albumin. Let us call this the “albumin” problem (and later we will mention the “growth factor” problem, which is even more interesting). One sees immediately why ELISA formats were developed.
Thus we needed a second element of specificity intrinsic to that single capture aptamer, along with equilibrium-based discrimination. That is, we wanted the qualities of a sandwich in a format that used but a single reagent. We examined the original SELEX process exhaustively. The details are published within a patent application (Zichi et al. 2009) as well as papers in press and winding their way through the review process (Keeney et al. 2009; Ostroff et al. 2009). We have learned how to drive aptamer selection to the lowest possible Kd for a given oligonucleotide library, using five-position modified pyrimidines in the libraries. Modified pyrimidines have played an enormous role in the successful enhancement of aptamers. Bruce Eaton's original five-position modifications (Dewey et al. 1995) have been expanded to include both amino acid side-chain adducts (Vaught et al. 2004; Vaught et al. 2010) and more recently adducts that resemble “fragment-based pharmacophores” from the drug industry (Congreve et al. 2008; J. Rohloff, personal communication). We have found many examples of so-called recalcitrant proteins that have yielded lovely aptamers using new oligonucleotide libraries (Zichi et al. 2009).
We also explored second elements of specificity for those modified aptamers because the “albumin” problem is not solved by lower Kds alone, even though we routinely obtain Kds between 10 pM and 100 pM. We have added as a second element of specificity a kinetic component on top of the equilibrium component. Many years ago Hopfield and Ninio independently elaborated the idea of kinetic proofreading (Hopfield 1974; Ninio 1975; Hopfield et al. 1976).
The central idea of kinetic proofreading was primarily concerned with specificity that was enhanced by using ATP or GTP hydrolysis as a method to separate two equilibria events from each other so that one could (almost) use the same binding differentiation twice. We used a version of this thinking: A component of the binding reaction (slow dissociation) can be used after using equilibrium discrimination. We have been able to select aptamers with remarkably slow dissociation rate constants, allowing us to do “kinetic challenges” during SELEX (both by simple dilution and also by incubations with alternative polyanions such as dextran sulfate at high concentration) (Schneider et al. 2009; Zichi et al. 2009).
These new aptamers are so important to the applications we study (biomarker discovery in blood, pathology, and in vivo imaging) that we have renamed them SOMAmers (SOMA; Slow Off-rate Modified Aptamers). The new name helps us distinguish and compare our data with a huge prior literature (Famulok et al. 2007; Mayer 2009).
4. SOMAMER SPECIFICITY
The requirement for our applications, including for the development of “magic bullet” therapeutics, is little to no off-target binding when both equilibrium and kinetic challenges are employed. We have shown that SOMAmers pull down largely the intended analytes from very complex biological matrices. From only these data it is clear that we have solved the “albumin” problem outlined above—in fact, abundant weak-binding proteins in plasma are the IgM's (whose concentrations in plasma are about micromolar, and which are found as multimers, and which have patches of lysines and arginines that bind nucleic acids nonspecifically). Gratifyingly IgMs are largely lost in the pull downs that are a part of the proteomics protocols.
But there are other proteins to fear in biological matrices. Growth factors almost always have heparan sulfate binding sites (even more concentrated patches of lysines and arginines than are present within the IgMs)—those sites are used by growth factors to bind loosely to the external surfaces of cells (which are negatively charged) from which point two-dimensional diffusion on the surface of the cell allows growth factors to find their high affinity receptors (Lieleg et al. 2009). This two-step method for binding was first understood by Peter von Hippel to be the “reduction of the dimensionality of the search” (a phrase he used to describe the diffusion along double-stranded DNA by the lac repressor as it sought its operator) (Von Hippel et al. 1982). Based on the work of Five Prime (Lin et al. 2008) we have an estimate of the number of such secreted proteins in humans, and the number is large: 3400! These proteins represent a SOMAmer-friendly collection of low abundant proteins, but in sum they represent a sink to which SOMAmers or other polyanions might bind. We call this the “growth factor” problem.
The heparan sulfate binding sites of the secreted human proteome have quite different structures from each other because the low affinity binding to cells requires nothing more than weak ionic interactions. That is, the quite different heparan sulfate binding sites on a few thousand human proteins are likely to have randomly disposed positive charges because the target (the outside surface of a cell) has an enormous local concentration of flexible, negative charges. Some of these proteins have significant binding to nontarget aptamers and SOMAmers, but most of those proteins dissociate from noncognate SOMAmers during kinetic challenge. That is, the kinetic challenge step removes the unintended “growth factor” molecules from binding to SOMAmers nonspecifically.
We have published some thoughts about the difficulties with antibodies for biomarker discovery using arrays (Zichi et al. 2008). Our exhaustive attack on the specificity problem has made possible reagents that can be used alone in a “conceptual” sandwich, solving both the albumin and the “growth factor” problems simultaneously.
5. SOMAMER STRUCTURE
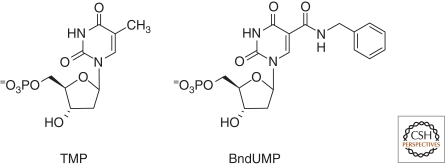
Our studies include an X-ray crystal structure for one SOMAmer bound to its protein target (Janjic and Jarvis, personal communication and manuscript in preparation). The SOMAmer was identified from a library of single-stranded DNAs in which every “T” was substituted with 5-benzyl-dUMP (Fig. 1) (Vaught et al. 2010). The SOMAmer has elements in its structure that have not been observed in single-stranded nucleic acids (which of course do not have access to the hydrophobic benzenes for whatever intramolecular folds might be needed, or for interactions with the target protein). In the structure are elements of benzene—amino acid interactions, benzene stacking on other nucleotides, and even a compact hydrophobic “turn” that is remarkable. Apparently small modifications to standard nucleotide chemistry open up a new world of possible structures, an idea alluded to years ago by Harold B. White III (White 1976). Modification of the pyrimidines of oligonucleotide libraries is a continuing piece of the efforts to make better and better SOMAmers.
Figure 1.
Thymadine-monophosphate (TMP) and the modified nucleotide 5-benzylaminocarbonyl-deoxyuridine-monophosphate (BndUMP).
6. QUANTITATIVE PROBING OF THE PLASMA PROTEOME AT HIGH CONTENT AND LOW LIMITS OF DETECTION
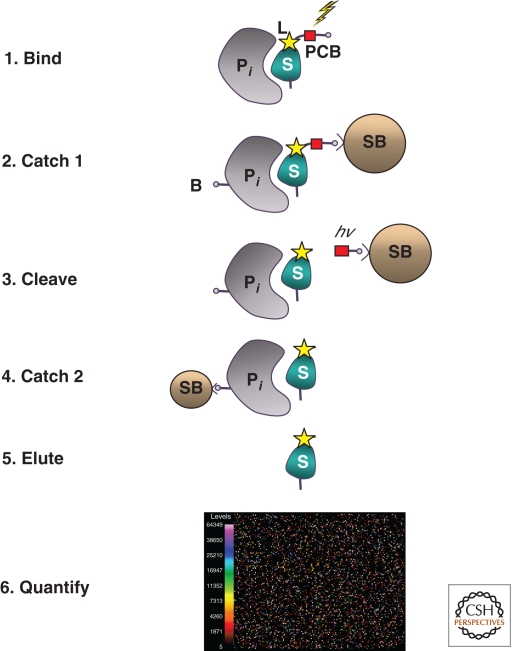
The heart of our work is to find novel biomarkers to use for drug development and for diagnostics. We have devised an assay (Fig. 2) that allows simultaneous measurements of human proteins in plasma or serum (or other matrices) in a manner analogous to mRNA or microRNA profiling from cells or even blood (Derisi et al. 1996; Calin et al. 2004). We have adopted the tools for RNA profiling (and SNP and CNV profiling) for our assay—we measure the SOMAmers themselves after a set of simple biochemical steps that discard the unbound input SOMAmers and allow us to quantify only those SOMAmers that stay bound (through their slow dissociation rate constants) to their cognate proteins. We converted the proteomic exercise required for Biomarker Discovery into a hybridization or QPCR measurement.
Figure 2.
Overview of the SomaLogic proteomics assay. In step 1, the specific protein to be measured (Pi) binds tightly to its cognate SOMAmer binding molecule (S), which includes a photo-cleavable biotin (PCB) and fluorescent label (L) at the 5′ end. In step 2, bound protein-SOMAmer complexes are captured onto streptavidin coated beads (SB) by the photo-cleavable biotin on the SOMAmer. Unbound proteins are washed away. Bound proteins are tagged with NHS-biotin (B). In step 3, the photo-cleavable biotin is cleaved by UV light (hv) and the protein-SOMAmer complexes are released into solution. In step 4, the protein-SOMAmer complexes are captured onto streptavidin coated magnetic beads and the SOMAmer are eluted into solution and recovered for quantification in step 6, hybridization to a custom DNA microarray. Each probe spot contains DNA with sequence complementary to a specific SOMAmer, and the fluorescent intensity of each probe spot is proportional to the amount of SOMAmer recovered, and thus directly proportional to the amount of protein present in the original sample.
This body of work (manuscripts submitted) yields array data that look exactly like the data obtained on a “DNA chip” constructed by, for example, Agilent, Affymetrix, Illumina, or NimbleGen/Roche because we use custom chips that have been printed with the complements of our SOMAmers.
We have found novel biomarkers for a variety of cancers, cardiovascular conditions, degenerative diseases, and more. The content today is “only” >800 SOMAmers (hence we have the capacity to measure >800 human proteins, simultaneously, using only about 15 µl of sample). We easily measure proteins at levels below 1 pM in plasma. Thus we achieved the primary objective we set for ourselves, which was to allow biomarker discovery in a way that could enhance drug development and medicine. We shall see over the next years if the effort we made will be as valuable for patients as we hoped when we started.
7. BACK TO THE RNA WORLD
We appreciate having a chance to tell our molecular biology friends what we have been up to for all these years. What started as an accident (Craig Tuerk's PhD thesis, and the isolation of what we called the major variant but which was in fact the first aptamer) and then became general (the small cottage industry, in universities and companies, doing SELEX) has the chance to be truly useful for healthcare, the fundamental reason we all enjoyed NIH funding over the years.
But of course there are lessons in our work for thinking about evolution. The most important lessons, now supported by a huge published literature (we include the patent literature, a literature that seems a bit obscure to most scientists), are that aptamers (or at least SOMAmers) can be identified for virtually any target molecule, large or small, protein or other.
Data on aptamers support only weakly notions of ubiquitous RNA-networks (e.g., Mattick 2003), which will require more data. Detailed studies of many creatures show that no region of any genome is entirely silent (Nagalakshmi et al. 2008). Although it is tempting to imagine functions for everything we find in a creature, and although it is obvious that evolution grabs noise over time and makes something useful, when we think about fancy networks it is a good idea to remember that noise is real in real things (Thattai and Van Oudenaarden 2001; Raser and O'Shea 2005).
But the aptamer literature is huge and compelling—one can find short single-stranded oligonucleotides that will bind to almost anything; and from binding one can imagine function. An oligonucleotide world seems sensible as a step during evolution, and we always think that early short oligonucleotides as drivers of evolution can be RNA-based, DNA-based, or even modified-oligo-based. Years ago the papers of Harold B. White (White 1976) sensibly posited a lovely idea that it was wrong to assume that the present nucleotides in RNA and DNA were the ones that were (in Woese's and Dobzhansky's way of thinking) the players during early evolution. Perhaps our use of SOMAmers, with many different pyrimidine adducts in our libraries, is analogous to White's idea that a variety of alternative nucleotides existed before simple genetics won so that life could cross Woese's Darwinian Threshold (Woese 2002).
The aptamer literature says “give me 30-40 random nucleotides within an oligo (of whatever chemistry) and binders are there if the number of molecules is large.” The deepest question is why is this true, given the relatively uninteresting chemical qualities of modern nucleotides and the tiny (but with profound effect) pyrimidine adducts we have used. One way of thinking about aptamers and SOMAmers is as analogues of the conotoxins, those wonderful (and frightening) small peptides with remarkable affinities and specificities for various protein targets (Mondal et al. 2005; Halai and Craik 2009). Entropic cost upon binding is a problem solved by using non-mobile participants in a binding pair, and the conotoxins have solved the entropy problem through the use of several disulfide bonds to limit the flexibility of the peptides. Aptamers use base-pairing as the conotoxin disulfide bond equivalents, interactions within the aptamer that reduce flexibility and also provide a chance for the hairpin loops and bulges to settle into energetically favored structures. SOMAmers, with their modified nucleotides, use components more commonly thought to be from the domain of proteins.
8. CONCLUSIONS: WHAT OTHER FUNCTIONS MIGHT OLIGONUCLEOTIDES HAVE?
What have we missed in our studies of present day life? The problem is what to seek.
A huge new area, anticipated by aptamer and ribozyme research (Gold et al. 1997a; Gold et al. 1997b; Winkler et al. 2002), is that of the riboswitches (Winkler et al. 2002; Roth and Breaker 2009). From the aptamer-based capacities for binding to several small molecules, Breaker and colleagues imagined and found that bacteria use aptamers and ribozymes to do feedback regulation. They did have a huge advantage, which we want to mention here.
Bacteria have had more than three billion years of life on earth, which is much more than enough time for every single base pair of every bacterial genome to be tested exhaustively for selective advantages. Imagine that bacteria divided through the ages with a one day generation time, and that they had the present mutation rate of one base pair change per 106 base pairs per replication. Three billion years contain about 1×1012 d, and thus enough time for about 1×1012 bacterial replications. Thus bacteria may have (each) experienced 1×106 mutations per base pair since their beginnings. The word one might use is “hammered”—bacterial genomes have been hammered. Each base pair has been like a slow acting metronome—first a G:C to A:T transition, then (perhaps) a reversion to the original G:C, then a transversion, then…and so on, such that, every single base pair in a bacterium has been changed to something else many many times.
One sees immediately the power of conserved RNA sequences and structures in bacteria. If all bacterial genomes are hammered, what remains in common must be important. This simple thought provoked Carl Woese to make his outstanding contributions (Woese and Fox 1977; Woese 1987, 1998, 2000, 2002). Breaker and his colleagues expanded this idea with their discovery of small conserved RNA sequences that also do something profound—in fact one would argue that the number 1×106 mutations per base pair is the underlying power behind all searches for useful RNA sequences using genomic comparisons in bacteria (Eddy 2005).
Are there additional undiscovered functions for RNAs (or even single-stranded DNAs) that we might imagine? The successful creation of aptamers has made every graduate student audience since 1990 say, at aptamer seminars, “why doesn't the immune system use aptamers instead of antibodies?” Of course this is an example of a question that can only be answered with an evolutionary perspective. If mammals carried the SELEX libraries we use as their information content for an oligonucleotide-based immune system (the genomic-size of a typical SELEX experiment is 1015 times 40 base pairs—that is a lot of genome to use to fight off pathogens), that immune system would require a larger genome than that of any creature we know today (by about a factor of a million). We ought to give credit to the present protein-based immune system for the cleverness with which diversity and binding selectivity is built!
However, somewhere on this planet there might well be a creature who responds with a highly mutable expression cassette of little “aptamers” to bind to and inactivate some invading creature. Experiments have been done to test artificial variations on this theme—so-called decoys with aptamers, expressed inside cells to “immunize” against viruses (Tuerk et al. 1992; Kumar et al. 1997), although no natural examples have been found in the biosphere.
One might also imagine that eukaryotic mRNAs will contain aptamers on their 5′ or 3′ ends (or even internally, using codon choices to co-evolve aptamers) that help localize the proteins expressed by the ribosomes and those ribosomes to specific areas on the inside of cells. It is also just another way to “reduce the dimensionality of the search”—a common problem in biology (Von Hippel et al. 1982). The IRES sequences allow internal translational initiations on polycistronic mRNAs (Kieft et al. 2002; Pfingsten et al. 2006).
One might also imagine that all those proteins we think of as nonspecific nucleic acid binding proteins will nucleate on specific aptamer-like sequences. Aptamers have been selected against many such proteins [ribosomal protein S1 from Escherichia coli was an early example (Tuerk and Gold 1990)]. In principle any protein that is a nonspecific binder to RNA or DNA, single or double stranded, has to prefer some sequence over others. Even a perfect “back-bone binder” will be influenced by the precise interphosphate distances, which will in turn be influenced by the bases themselves. We proposed “genomic SELEX” as a way to get to unexpected biology (Gold et al. 1997a)—we never embarked seriously on that work because of our focus on the medical potential. And so it goes.
Finally, and most strikingly, consider the extraordinary work from Eaton and Feldheim (Gugliotti et al. 2004, 2005; Liu et al. 2006; Feldheim and Eaton 2007). In a paper published in Science, using “catalytic aptamers” made from modified RNA libraries, these authors extended the reach of oligonucleotides to an entirely new realm (Gugliotti et al. 2004). The selected catalytic aptamers were able to recruit metals from solution, to nucleate crystal growth of specific crystal forms, and to do so in a sequence dependent manner. Should this work generalize, one might imagine a future research area of intracellular nanotechnology micro-fabrication catalyzed by oligonucleotides. Perhaps the Eaton-Feldheim work will lead us down the pathway toward an organic-inorganic fusion within modern fabrication and even biology.
Our collective thesis has two components. First, evolutionary time is long, and only in the hammered genomes of bacteria do we see the general proposition that what we have learned from in vitro evolution experiments such as SELEX is common in biology. But some of what we see today in our test tubes will have had that key stochastic accidental moment and will have been frozen and survived. Our tasks include wondering how to find those phenomena without the insights that flow from comparative bacterial genome sequences. But we argue that just because the easier discovery methods are unlikely to work, there are examples of present day oligonucleotide uses that will continue to be uncovered.
Second, modified pyrimidines change the entire reach of SELEX. As long as one can solve the replication problem (which has been done because many DNA polymerases are forgiving with respect to pyrimidine nucleotides modified at their five-positions with rather small adducts), one can change the characteristics and qualities of aptamers toward SOMAmers and so provide high quality reagents for many medical applications.
ACKNOWLEDGMENTS
We thank all of our colleagues who participated in the genesis of aptamer science and building the aptamer biotechnology industry over the past decades. They have contributed immeasurably to this endeavor and made countless insights about aptamers and the nature of biology. We especially thank past and present colleagues at the University of Colorado, NeXstar Pharmaceuticals, and SomaLogic. Finally, we thank the editors for their patient and skilled guidance in the preparation of this manuscript.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. 2004. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A 101: 11755–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, Chessari G, Tisi D, Woodhead AJ 2008. Recent developments in fragment-based drug discovery. J Med Chem 51: 3661–3680 [DOI] [PubMed] [Google Scholar]
- Derisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM 1996. Use of a cdna microarray to analyse gene expression patterns in human cancer. Nat Genet 14: 457–460 [DOI] [PubMed] [Google Scholar]
- Dewey T, Mundt A, Crouch G, Zyniewski M, Eaton B 1995. New uridine derivatives for systematic evolution of rna ligands by exponential enrichment. J Am Chem Soc 117: 8474–8475 [Google Scholar]
- Dobzhansky T 1973. Nothing in biology makes sense except in the light of evolution. Am Bio Teach 35: 125–129 [Google Scholar]
- Doggrell SA 2005. Pegaptanib: The first antiangiogenic agent approved for neovascular macular degeneration. Expert Opin Pharmaco 6: 1421–1423 [DOI] [PubMed] [Google Scholar]
- Eddy SR 2005. A model of the statistical power of comparative genome sequence analysis. PLoS Biol 3: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- Famulok M, Hartig JS, Mayer G 2007. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 107: 3715–3743 [DOI] [PubMed] [Google Scholar]
- Feldheim DL, Eaton BE 2007. Selection of biomolecules capable of mediating the formation of nanocrystals. ACS Nano 1: 154–159 [DOI] [PubMed] [Google Scholar]
- Gold L 1995. Oligonucleotides as research, diagnostic, and therapeutic agents. J Biol Chem 270: 13581–13584 [DOI] [PubMed] [Google Scholar]
- Gold L, Tuerk C 1993. Methods for identifying nucleic acid ligands. US5270163 [Google Scholar]
- Gold L, Brown D, He Y, Shtatland T, Singer BS, Wu Y 1997a. From oligonucleotide shapes to genomic selex: Novel biological regulatory loops. Proc Natl Acad Sci 94: 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L, Singer B, He YY, Brody E 1997b. Selex and the evolution of genomes. Curr Opin Genet Dev 7: 848–851 [DOI] [PubMed] [Google Scholar]
- Gonzales CR 2005. Enhanced efficacy associated with early treatment of neovascular age-related macular degeneration with pegaptanib sodium: An exploratory analysis. Retina 25: 815–827 [DOI] [PubMed] [Google Scholar]
- Gragoudas ES, Adamis AP, Cunningham ET Jr, Feinsod M, Guyer DR 2004. Pegaptanib for neovascular age-related macular degeneration. N Engl J Med 351: 2805–2816 [DOI] [PubMed] [Google Scholar]
- Green R, Ellington AD, Szostak JW 1990. In vitro genetic analysis of the tetrahymena self-splicing intron. Nature 347: 406–408 [DOI] [PubMed] [Google Scholar]
- Gugliotti LA, Feldheim DL, Eaton BE 2004. Rna-mediated metal-metal bond formation in the synthesis of hexagonal palladium nanoparticles. Science 304: 850–852 [DOI] [PubMed] [Google Scholar]
- Gugliotti LA, Feldheim DL, Eaton BE 2005. Rna-mediated control of metal nanoparticle shape. Journal of the Americane Chemical Society 127: 17814–17818 [DOI] [PubMed] [Google Scholar]
- Halai R, Craik DJ 2009. Conotoxins: Natural product drug leads. Nat Prod Rep 26: 526–536 [DOI] [PubMed] [Google Scholar]
- Hopfield JJ 1974. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci 71: 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ, Yamane T, Yue V, Coutts SM 1976. Direct experimental evidence for kinetic proofreading in amino acylation of trnaile. Proc Natl Acad Sci 73: 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinek D, Lynott CK, Rifkin DB, Janjic N 1993. High-affinity RNA ligands to basic fibroblast growth factor inhibit receptor binding. Proc Natl Acad Sci 90: 11227–11231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser PK 2006. Antivascular endothelial growth factor agents and their development: Therapeutic implications in ocular diseases. Am J Ophthalmol 142: 660–668 [DOI] [PubMed] [Google Scholar]
- Keeney T, Kraemer S, Walker JJ, Bock C, Vaught J, Nikrad M, Stewart A, Lollo B, Stanton M, Gold L 2009. Automation of the somalogic proteomics assay: A platform for biomarker discovery. J Assoc Lab Automat 14: 360–366 [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA 2002. Crystal structure of an RNA tertiary domain essential to hcv ires-mediated translation initiation. Nat Struct Biol 9: 370–374 [DOI] [PubMed] [Google Scholar]
- Klussmann S, Nolte A, Bald R, Erdmann VA, Furste JP 1996. Mirror-image RNA that binds d-adenosine. Nat Biotechnol 14: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Kumar PK, Machida K, Urvil PT, Kakiuchi N, Vishnuvardhan D, Shimotohno K, Taira K, Nishikawa S 1997. Isolation of RNA aptamers specific to the ns3 protein of hepatitis c virus from a pool of completely random RNA. Virology 237: 270–282 [DOI] [PubMed] [Google Scholar]
- Lieleg O, Baumgartel RM, Bausch AR 2009. Selective filtering of particles by the extracellular matrix: An electrostatic bandpass. Biophys J 97: 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, et al. 2008. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 320: 807–811 [DOI] [PubMed] [Google Scholar]
- Liu D, Gugliotti LA, Wu T, Dolska M, Tkachenko AG, Shipton MK, Eaton BE, Feldheim DL 2006. Rna-mediated synthesis of palladium nanoparticles on au surfaces. Langmuir 22: 5862–5866 [DOI] [PubMed] [Google Scholar]
- Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT 2009. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci 106: 2989–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS 2003. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25: 930–939 [DOI] [PubMed] [Google Scholar]
- Mayer G 2009. The chemical biology of aptamers. Angew Chem Int Ed Engl 48: 2672–2689 [DOI] [PubMed] [Google Scholar]
- Mondal S, Vijayan R, Shichina K, Babu RM, Ramakumar S 2005. I-superfamily conotoxins: Sequence and structure analysis. In Silico Biol 5: 557–571 [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J 1975. Kinetic amplification of enzyme discrimination. Biochimie 57: 587–595 [DOI] [PubMed] [Google Scholar]
- Nishijima K, Ng YS, Zhong L, Bradley J, Schubert W, Jo N, Akita J, Samuelsson SJ, Robinson GS, Adamis AP, et al. 2007. Vascular endothelial growth factor-a is a survival factor for retinal neurons and a critical neuroprotectant during the adaptive response to ischemic injury. Am J Pathol 171: 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff R, Foreman T, Keeney TR, Stratford S, Walker JJ, Zichi D 2009. The stability of the circulating human proteome to variations in sample collection and handling procedures measured with an aptamer-based proteomics array. Journal of Proteomics. [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS 2006. Structural basis for ribosome recruitment and manipulation by a viral ires rna. Science 314: 1450–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK 2005. Noise in gene expression: Origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A 1974. Structure of yeast phenylalanine tRNA at 3 a resolution. Nature 250: 546–551 [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N 1998. 2′-fluoropyrimidine rna-based aptamers to the 165-amino acid form of vascular endothelial growth factor (vegf165). Inhibition of receptor binding and vegf-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 273: 20556–20567 [DOI] [PubMed] [Google Scholar]
- Saint-Geniez M, Kurihara T, Sekiyama E, Maldonado AE, D'amore PA 2009. An essential role for rpe-derived soluble vegf in the maintenance of the choriocapillaris. Proc Natl Acad Sci 106: 18751–18756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Nieuwlandt D, Eaton B, Stanton M, Gupta S, Kraemer S, Zichi D, Gold L 2009. Multiplexed analyses of test samples. US2009/0042206 [Google Scholar]
- Suddath FL, Quigley GJ, Mcpherson A, Sneden D, Kim JJ, Kim SH, Rich A 1974. Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0angstroms resolution. Nature 248: 20–24 [DOI] [PubMed] [Google Scholar]
- Thattai M, Van Oudenaarden A 2001. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci 98: 8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tok JB, Fischer NO 2008. Single microbead selex for efficient ssdna aptamer generation against botulinum neurotoxin. Chem Commun (Camb) 1883–1885 [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage t4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Tuerk C, Macdougal S, Gold L 1992. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci 89: 6988–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, Stormo G, D'aubenton-Carafa Y, Uhlenbeck OC, Tinoco I Jr, et al. 1988. Cuucgg hairpins: Extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci 85: 1364–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaught JD, Dewey T, Eaton BE 2004. T7 RNA polymerase transcription with 5-position modified utp derivatives. J Am Chem Soc 126: 11231–11237 [DOI] [PubMed] [Google Scholar]
- Vaught J, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK, Eaton B 2010. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc 132: 4141–4151 [DOI] [PubMed] [Google Scholar]
- Von Hippel PH, Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, Stormo GD, Gold L 1982. Autoregulation of gene expression. Quantitative evaluation of the expression and function of the bacteriophage t4 gene 32 (single-stranded DNA binding) protein system. J Mol Biol 162: 795–818 [DOI] [PubMed] [Google Scholar]
- White HB 3rd 1976. Coenzymes as fossils of an earlier metabolic state. J Mol Evol 7: 101–104 [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956 [DOI] [PubMed] [Google Scholar]
- Woese CR 1987. Bacterial evolution. Microbiol Rev 51: 221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR 1998. The universal ancestor. Proc Natl Acad Sci 95: 6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR 2000. Interpreting the universal phylogenetic tree. Proc Natl Acad Sci 97: 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR 2002. On the evolution of cells. Proc Natl Acad Sci 99: 8742–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE 1977. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci 74: 5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Gutell R, Gupta R, Noller HF 1983. Detailed analysis of the higher-order structure of 16s-like ribosomal ribonucleic acids. Microbiol Rev 47: 621–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zichi D, Eaton B, Singer B, Gold L 2008. Proteomics and diagnostics: Let's get specific, again. Curr Opin Chem Biol 12: 78–85 [DOI] [PubMed] [Google Scholar]
- Zichi D, Wilcox SK, Bock C, Schneider DJ, Eaton B, Gold L 2009. Method for generating aptamers with improved off-rates. US2009/0004667 [Google Scholar]