SUMMARY
Inflammation is triggered when innate immune cells detect infection or tissue injury. Surveillance mechanisms involve pattern recognition receptors (PRRs) on the cell surface and in the cytoplasm. Most PRRs respond to pathogen-associated molecular patterns (PAMPs) or host-derived damage-associated molecular patterns (DAMPs) by triggering activation of NF-κB, AP1, CREB, c/EBP, and IRF transcription factors. Induction of genes encoding enzymes, chemokines, cytokines, adhesion molecules, and regulators of the extracellular matrix promotes the recruitment and activation of leukocytes, which are critical for eliminating foreign particles and host debris. A subset of PRRs activates the protease caspase-1, which causes maturation of the cytokines IL1β and IL18. Cell adhesion molecules and chemokines facilitate leukocyte extravasation from the circulation to the affected site, the chemokines stimulating G-protein-coupled receptors (GPCRs). Binding initiates signals that regulate leukocyte motility and effector functions. Other triggers of inflammation include allergens, which form antibody complexes that stimulate Fc receptors on mast cells. Although the role of inflammation is to resolve infection and injury, increasing evidence indicates that chronic inflammation is a risk factor for cancer.
Pattern recognition receptors (PRRs) signal infection and injury. They detect foreign particles and host debris and stimulate the expression of inflammatory genes.
1. INTRODUCTION
The role of the inflammatory response is to combat infection and tissue injury. Innate immune cells residing in tissues, such as macrophages, fibroblasts, mast cells, and dendritic cells, as well as circulating leukocytes, including monocytes and neutrophils, recognize pathogen invasion or cell damage with intracellular or surface-expressed pattern recognition receptors (PRRs). These receptors detect, either directly or indirectly, pathogen-associated molecular patterns (PAMPs), such as microbial nucleic acids, lipoproteins, and carbohydrates, or damage-associated molecular patterns (DAMPs) released from injured cells. Activated PRRs then oligomerize and assemble large multi-subunit complexes that initiate signaling cascades that trigger the release of factors that promote recruitment of leukocytes to the region.
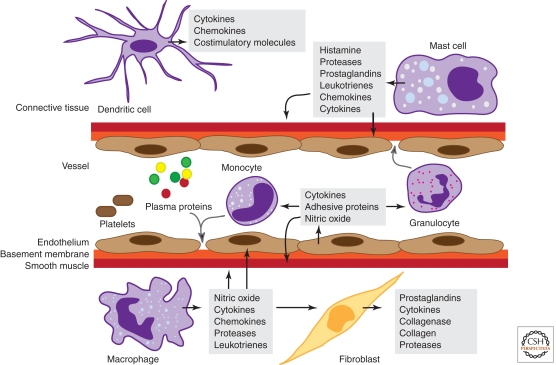
Vascular alterations play an important role in the inflammatory response (Fig. 1). Histamine, prostaglandins, and nitric oxide act on vascular smooth muscle to cause vasodilation, which increases blood flow and brings in circulating leukocytes, whereas inflammatory mediators including histamine and leukotrienes act on endothelial cells to increase vascular permeability and allow plasma proteins and leukocytes to exit the circulation. Cytokines such as tumor necrosis factor (TNF) and interleukin 1 (IL1) promote leukocyte extravasation by increasing the levels of leukocyte adhesion molecules on endothelial cells. Activated innate immune cells at the site of infection or injury, including dendritic cells, macrophages, and neutrophils, remove foreign particles and host debris by phagocytosis, plus they also secrete cytokines that shape the slower, lymphocyte-mediated adaptive immune response.
Figure 1.
Cells and mediators of the inflammatory response. Molecules derived from plasma proteins and cells in response to tissue damage or pathogens mediate inflammation by stimulating vascular changes, plus leukocyte migration and activation. Granulocytes include neutrophils, basophils, and eosinophils.
Below we examine how PRRs signal recognition of infection and injury. We then describe how the ensuing inflammatory response is amplified by the cytokines TNF and IL1β. Next, we discuss the mechanisms that get leukocytes to where they are needed. Finally, we consider inflammatory signaling pathways triggered during allergic or hypersensitivity reactions and the possibility that chronic inflammation promotes tumor development.
2. DAMPs AND PAMPs TRIGGER THE INNATE IMMUNE RESPONSE
DAMPs are endogenous molecules normally found in cells that get released during necrosis and contribute to sterile inflammation. They include ATP, the cytokine IL1α, uric acid, the calcium-binding, cytoplasmic proteins S100A8 and S100A9, and the DNA-binding, nuclear protein HMGB1. Amyloid β fibrils associated with Alzheimer’s disease have also been shown to be pro-inflammatory. PAMPs, in contrast, are pathogen-derived, often essential for microbe survival, and, like DAMPs, structurally diverse. PAMPs include bacterial and viral nucleic acids, fungal β-glucan and α-mannan cell wall components, the bacterial protein flagellin, components of the peptidoglycan bacterial cell wall, and lipopolysaccharide (LPS) from Gram-negative bacteria.
3. TOLL-LIKE RECEPTORS (TLRs)
Members of the TLR family are major PRRs in cells. They are type I transmembrane proteins containing leucine-rich repeats (LRRs) that recognize bacterial and viral PAMPs in the extracellular environment (TLR1, TLR2, TLR4, TLR5, TLR6, and TLR11) or endolysosomes (TLR3, TLR7, TLR8, TLR9, and TLR10). The ligands identified for the different TLRs are listed in Table 1. Signal transduction by TLRs relies on a cytoplasmic Toll/interleukin (IL) 1 receptor (TIR) domain that serves as the docking site for TIR-containing cytoplasmic adaptor proteins (Fig. 2). TIR domains in the receptor for the pro-inflammatory cytokine IL1 function in a similar fashion.
Table 1.
Agonists of mouse and human Toll-like receptors
| TLR | Ligand |
|---|---|
| 1/2 | Triacyl lipopeptides |
| 2/6 | Diacyl lipopeptides |
| 3 | dsRNA |
| 4 | Lipopolysaccharide |
| 5 | Flagellin |
| 7 | ssRNA |
| 8 | ssRNA in humans; unclear in mice |
| 9 | CpG DNA, malarial hemozoin |
| 10a | Unknown |
| 11b | Uropathogenic bacteria, Toxoplasma gondii profilin-like protein |
| 12b | Unknown |
| 13b | Unknown |
aExpressed only in humans.
bExpressed only in mice.
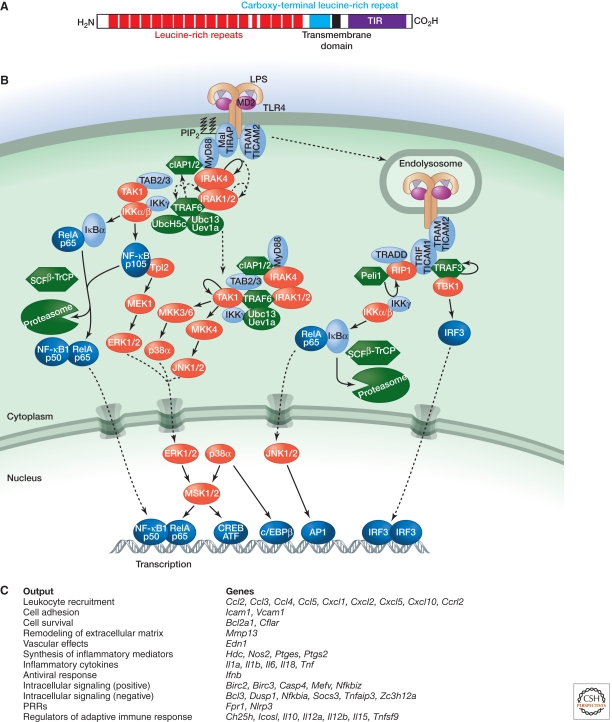
Figure 2.
Signaling by TLR4. (A) Domain structure of human TLR4. (B) Binding of LPS to TLR4 and the coreceptor MD2 triggers interactions between the cytoplasmic TIR domain of TLR4 and TIR-containing adaptor proteins (Mal, MyD88, and TRAM). MyD88 binds IRAK4, which requires its kinase activity to bind the kinases IRAK1 and IRAK2 sequentially. The MyD88–IRAK complex also engages the ubiquitin ligase TRAF6 to make polyubiquitin chains that activate the IKK complex for NF-κB- and ERK-dependent gene transcription. Ubiquitin ligases cIAP1 and cIAP2 recruited to the TLR4 signaling complex regulate translocation of a subset of signaling components to the cytoplasm, where TAK1 activation initiates a MAPK cascade that stimulates gene expression. TLR4 activated at the plasma membrane is endocytosed but can signal within the endosomal compartment via the adaptors TRAM and TRIF. The kinase and ubiquitin ligase combination of RIP1 and Peli1 interacts with TRIF to signal NF-κB activation, whereas TBK1 and TRAF3 stimulate IRF3-dependent transcription. (C) Functional outputs of some of the genes upregulated by TLR4 signaling.
3.1. MyD88-Dependent TAK1 and IKK Activation by TLRs
With the exception of TLR3, all known TLRs engage the adaptor MyD88 either directly (TLR5, TLR7, TLR8, TLR9, TLR10, and TLR11, heterodimeric TLR1-TLR2 and TLR2-TLR6, and the IL1Rs) or in combination with the adaptor TIRAP/Mal (TLR1-TLR2, TLR2-TLR6, and TLR4). MyD88 contains a death domain (DD) in addition to a TIR domain, and this mediates interactions with the DD of the serine/threonine kinase IRAK4 (Lin et al. 2010). Clustering of IRAK4 within the receptor complex probably results in its autophosphorylation. The kinase activity of IRAK4 is required for its DD to bind the DD of the related kinases IRAK1 and IRAK2. MyD88 and the IRAKs nucleate a larger complex, which in the case of TLR4 includes E3 ubiquitin ligases (TRAF6, cIAP1, and cIAP2) and the E2 ubiquitin-conjugating enzyme Ubc13 (Tseng et al. 2010). TRAF6 and Ubc13 catalyze the formation of polyubiquitin chains in which the carboxyl terminus of one ubiquitin forms an isopeptide bond with the ε-amino group of K63 of an adjacent ubiquitin. TRAF6 can build polyubiquitin chains on lysines within itself and IRAK1, and it also promotes K63-linked polyubiquitylation of cIAPs (Skaug et al. 2009; Tseng et al. 2010). These K63-linked chains are thought to recruit the adaptor proteins TAB2 and TAB3, which exist in a complex with the kinase TAK1. The regulatory subunit of the IκB kinase (IKK) complex, known as IKKγ or NEMO, also binds K63-linked polyubiquitin and is recruited to the TLR4 signaling complex (Laplantine et al. 2009; Tseng et al. 2010).
Analyses of knockout mice indicate that both IKK and TAK1 are important for activation of NF-κB transcription factors, whereas only TAK1 is required for activation of the mitogen-activated protein kinases (MAPKs) p38α and JNK (Sato et al. 2005; Shim et al. 2005; Israel 2010). How TAK1 and IKK are activated, however, requires further clarification. Unanchored K63-linked polyubiquitin chains synthesized by TRAF6 and Ubc13 were proposed to activate TAK1 by inducing its autophosphorylation (Xia et al. 2009). Although TAK1 can phosphorylate the activation loops in the kinases IKKβ and MKK6 in vitro, the latter a MAPK kinase upstream of p38α (Skaug et al. 2009), it is not clear whether TAK1 phosphorylates IKKβ in cells. A recent study suggested that TLR4-induced TAK1 autophosphorylation and activation, but not IKK activation, require translocation of the MyD88–TRAF6–Ubc13–cIAP–TAK1–IKKγ signaling complex from TLR4 into the cytosol (Tseng et al. 2010). This translocation depends on TRAF6 and the cIAPs. Studies of gene-targeted mice expressing a kinase-dead version of TAK1 would clarify whether TAK1 requires its kinase activity to activate IKK.
IKK activation, like TAK1 activation, has been linked to polyubiquitin binding, but the chain linkages and the key E2/E3 ubiquitin enzyme combinations appear to differ. Both unanchored polyubiquitin formed by TRAF6 with the E2 UbcH5c (Xia et al. 2009) and linear polyubiquitin conjugated to IKKγ (Rahighi et al. 2009; Tokunaga et al. 2009) have been shown to induce IKK activation. The linear ubiquitin chain assembly complex (LUBAC) joins ubiquitins in a head-to-tail fashion with the carboxy-terminal glycine residue of one ubiquitin linked to the amino terminus of another ubiquitin. E2 enzymes that support LUBAC activity in vitro include UbcH5c and UbcH7 (Gerlach et al. 2011; Ikeda et al. 2011; Tokunaga et al. 2011). Binding of the carboxy-terminal UBAN domain in IKKγ to linear ubiquitin chains may produce conformational changes necessary for IKK activation. The importance of IKKγ binding to polyubiquitin is supported by the identification of mutations in the UBAN domain that impair NF-κB activation and cause immunodeficiency in humans (Table 2) (Döffinger et al. 2001). These studies may explain why NF-κB activation is largely normal in the absence of Ubc13 while MAPK activation is compromised (Yamamoto et al. 2006).
Table 2.
Genes regulating inflammation that are mutated in human disease
| Gene | Protein | Disease |
|---|---|---|
| CIAS1 | NLRP3 | Familial cold autoinflammatory syndrome; Muckle–Wells syndrome; Neonatal-onset multisystem inflammatory disease |
| IKBKG | IKKγ | Anhidrotic ectodermal dysplasia with immunodeficiency |
| NOD2 | NOD2 | Crohn’s inflammatory bowel disease; Blau syndrome characterized by arthritis and uveitis |
| TNFAIP3 | A20 | B-cell lymphomas |
The MAPKs JNK and p38α, like TAK1, are activated downstream from TLR2 and TLR4 in a cIAP-dependent manner. The TAK1-containing signaling complex translocates into the cytosol and recruits the kinase MKK4 to phosphorylate and activate JNK (Tseng et al. 2010). It probably also recruits MKK3 and MKK6 to activate p38α because both kinases associate with TRAF6 in response to LPS (Wan et al. 2009). p38α is required for activation of the transcription factors CREB and c/EBPβ, and it contributes to the induction of several genes, including those encoding chemokines (Cxcl1, Cxcl2), cytokines (IL10, IL12b, IL1a, and IL1b), and regulators of extracellular matrix remodeling (Mmp13) and cell adhesion (Vcam1) (Kang et al. 2008; Kim et al. 2008). JNK regulates the activity of the AP1 transcription factor and stimulates expression of pro-inflammatory mediators such as TNF (Das et al. 2009).
Activation of the IKK complex is required for NF-κB-dependent transcription as well as transcriptional responses downstream from the MAPK ERK. IKKβ substrates include p105, the precursor of the p50 NF-κB1 transcription factor, as well as the IκB proteins that sequester NF-κB transcription factors in the cytosol. Phosphorylation by IKKβ targets these substrates for K48-linked polyubiquitylation by the E3 ubiquitin ligase SCFβ-TrCP and subsequent proteasomal degradation (Kanarek et al. 2010). Degradation of p105, which exists in a complex with the kinase Tpl2, activates a Tpl2–MEK1–ERK kinase cascade that leads to the induction of genes such as Ptgs2 by the CREB/ATF family of transcription factors (Banerjee and Gerondakis 2007). The cyclooxygenase 2 (COX2) enzyme encoded by Ptgs2 is involved in the synthesis of prostaglandins, which are important mediators of pain, inflammation, and fever. IκB degradation allows dimeric NF-κB transcription factors composed largely of RelA (p65) and NF-κB1 (p50) subunits to accumulate in the nucleus and drive expression of a large number of pro-inflammatory genes (Table 3 lists a subset of these genes).
Table 3.
Genes induced by the canonical IKKβ/NF-κB signaling pathway
| Genea | Protein | Function |
|---|---|---|
| Ager | RAGE | PRR belonging to the immunoglobulin superfamily that recognizes multiple ligands including HMGB1 |
| Birc3 | cIAP2 | Ubiquitin ligase that regulates NF-κB activation |
| Casp4 | Caspase-11 | Aspartate-specific cysteine protease implicated in inflammation |
| Ccl2 | MCP1 | Chemokine for monocyte recruitment |
| Ccl3 | MIP1α | Chemokine for leukocyte recruitment |
| Ccl5 | RANTES | Chemokine for monocyte and T-cell recruitment |
| Cd200 | CD200 | Binds CD200R1 and inhibits macrophage activation |
| Cfb | Complement factor B | Serine protease in the alternative complement activation pathway |
| Cflar | c-FLIP | Inhibitor of death receptor-induced apoptosis |
| Csf2 | GM-CSF | Growth factor that promotes differentiation and activation of DCs, macrophages, and neutrophils |
| Cxcl1 | KC | Chemokine for neutrophil recruitment |
| Cxcl2 | MIP2 | Chemokine for neutrophil recruitment |
| F3 | Tissue factor | Coagulation factor |
| Icam1 | ICAM1 | Cell adhesion molecule that interacts with β2 integrins |
| Ifnb1 | IFNβ | Suppressor of virus replication |
| Il1b | IL1β | Cytokine that amplifies the inflammatory response |
| Il6 | IL6 | Pleiotropic cytokine that stimulates fever, production of hepatocyte acute phase proteins, and lymphocyte differentiation |
| Il12b | IL12 p40 | Component of heterodimeric IL12 and IL23, which modulate NK cell and lymphocyte effector functions |
| Mmp9 | MMP9 | Metalloproteinase that degrades extracellular matrix |
| Nfkbia | IκBα | Inhibitor of NF-κB signaling |
| Nfkbib | IκBβ | NF-κB transcriptional coactivator |
| Nos2 | iNOS | Enzyme that makes anti-microbial nitric oxide |
| Sele | E-selectin | Cell adhesion molecule |
| Selp | P-selectin | Cell adhesion molecule |
| Sod2 | MnSOD | Enzyme that converts superoxide to hydrogen peroxide |
| Tnf | TNF | Cytokine that amplifies the inflammatory response |
| Tnfaip3 | A20 | Inhibitor of NF-κB signaling by TLRs and TNF-R1 |
| Vcam1 | VCAM1 | Cell adhesion molecule that interacts with β1 integrin VLA-4 |
aMouse gene nomenclature used.
NF-κB also induces genes that limit the duration and magnitude of the inflammatory response, such as Tnfaip3 and Nfkbia (the latter encodes IκBα and thus forms a negative-feedback loop). These genes prevent the inflammatory response from causing more tissue damage than the initial injury. For example, mice lacking the A20 deubiquitylating enzyme encoded by Tnfaip3 die from unchecked inflammation. A20 is thought to switch off TLR signaling by countering ubiquitylation by TRAF6 or cIAPs (Newton et al. 2008; Skaug et al. 2009; Shembade et al. 2010). Somatic mutation of TNFAIP3 occurs frequently in some human B-cell lymphomas, which suggests that A20 may function as a tumor suppressor, and polymorphisms within the TNFAIP3 locus have been associated with autoimmune disorders, including systemic lupus erythematosus, rheumatoid arthritis, Crohn’s inflammatory bowel disease, and psoriasis (Table 2) (Vereecke et al, 2011). The tumor suppressor gene CYLD, which is mutated in familial cylindromatosis, also encodes a deubiquitylating enzyme that limits NF-κB signaling. CYLD cleaves both linear and K63-linked polyubiquitin efficiently in vitro (Komander et al. 2009), but mice lacking CYLD do not develop the multi-organ inflammation seen in A20-deficient mice, perhaps because CYLD normally is phosphorylated and inactivated by IKK (Reiley et al. 2005). Another NF-κB target gene that suppresses TLR signaling is Cd200, which encodes the membrane glycoprotein ligand for CD200R1 (Mukhopadhyay et al. 2010). TAM (Tyro3, Axl, and Mer) receptor tyrosine kinases are further examples of receptor systems that negatively regulate TLR signaling (Rothlin et al. 2007).
3.2. MyD88-Dependent Type I Interferon (IFN) Induction by TLRs 7 and 9
The MyD88–IRAK4–IRAK1–TRAF6 signaling complex required for pro-inflammatory cytokine production by TLR7 and TLR9 also stimulates synthesis of interferon α (IFNα) and IFNβ in plasmacytoid dendritic cells (pDCs). The induction of type I IFNs and Ifn-related genes is critical to the anti-viral response and also requires TRAF3, IKKα, and the transcription factor IRF7. Phosphorylation of IRF7 by IKKα promotes its dimerization and translocation into the nucleus, where it upregulates expression of type I Ifn genes. Differences have been noted in myeloid DCs1; IRF1 is more important than IRF7, and there does not seem to be a requirement for TRAF3 and IRAK1 (Hoshino et al. 2010; Takeuchi and Akira 2010).
3.3. TRIF-Dependent Signaling by TLRs 3 and 4
TLR4 is endocytosed following ligand binding and, like TLR3 stimulated with dsRNA, transduces signals from within the endosomal compartment. Both receptors recruit the TIR-containing cytoplasmic adaptor TRIF (also known as TICAM1), which in the case of endocytosed TLR4 occurs via the bridging adaptor TRAM (also known as TICAM2). TRIF can activate NF-κB using its RIP homotypic interaction motif (RHIM) to recruit the RHIM-containing kinase RIP1 (Cusson-Hermance et al. 2005), which is, in turn, bound and ubiquitylated by the E3 ubiquitin ligase Peli1 (Chang et al. 2009). K63-linked polyubiquitylation of RIP1 by Peli1 has not been shown, but could be a mechanism for the recruitment of IKKγ and TAK1. Interaction of the DD in RIP1 with the DD in the cytoplasmic adaptor TRADD is important for TRIF-dependent NF-κB and MAPK activation in some cell types (Chen et al. 2008; Ermolaeva et al. 2008; Pobezinskaya et al. 2008). The TRIF-containing complex also induces type I IFN, and this is dependent on TRAF3 plus the kinase TBK1, the latter phosphorylating and activating the transcription factor IRF3 (Takeuchi and Akira 2010). TRAF3 can modify itself with K63-linked polyubiquitin, and this may be important for its interaction with TBK1 and subsequent IRF3 activation.
4. RIG-I-Like Receptors (RLRs)
The RLR family of PRRs, which comprises RIG-I, MDA5, and LGP2, signals the production of pro-inflammatory cytokines and type I IFN in response to viral and bacterial nucleic acids in the cytoplasm (Fig. 3). These cytosolic proteins have a central DExD/H-box helicase domain and a carboxy-terminal regulatory domain (CTD), the latter binding RNA. RIG-I and MDA5 also have two amino-terminal caspase activation and recruitment domains (CARDs), which function as protein–protein interaction motifs. RIG-I recognizes double-stranded RNAs (dsRNAs) that have a 5′ triphosphate and are either viral in origin or generated by RNA polymerase III from microbial DNA templates (Lu et al. 2010; Wang et al. 2010b). MDA5 and LGP2 also bind RNA, but MDA5 and RIG-I have non-redundant functions in sensing certain viruses. LGP2 appears, in most contexts, to act as a positive regulator of signaling by MDA5 and RIG-I (Venkataraman et al. 2007; Satoh et al. 2010).
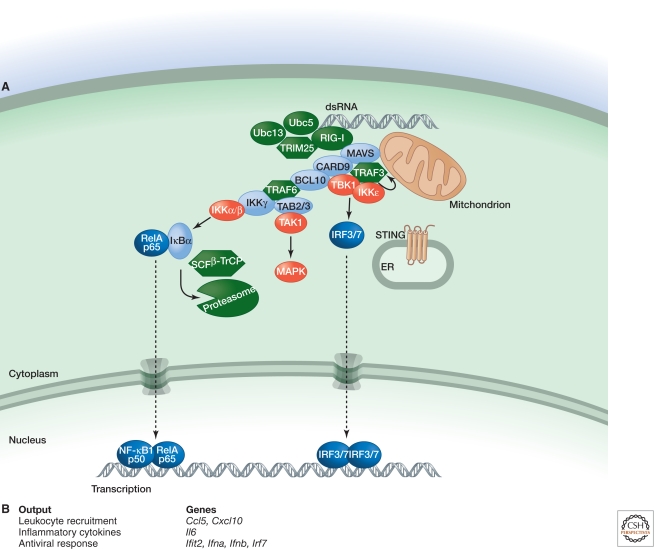
Figure 3.
Signaling by RIG-I. (A) RIG-I binding to dsRNA that has a 5′ triphosphate and polyubiquitin, the latter generated by the ubiquitin ligase TRIM25 and E2 ubiquitin-conjugating enzymes Ubc5 and Ubc13, promotes RIG-I binding to mitochondrial MAVS. Subsequently, a larger complex containing the adaptor proteins CARD9 and BCL10 is assembled for MAPK and NF-κB activation. TRAF3, the kinases TBK1 and IKKε, and ER-resident protein STING are required for activation of transcription factors IRF3 and IRF7. (B) Functional outputs of some of the genes upregulated by MAVS signaling.
Binding of RIG-I to RNA causes a conformational change (Jiang et al. 2011) that enables its CARDs to bind K63-linked polyubiquitin generated by the E3 ubiquitin ligase TRIM25. In an ill-defined manner, this interaction causes RIG-I to engage the mitochondrial CARD-containing protein MAVS (also called IPS-1, VISA, and Cardif) (Zeng et al. 2010). Subsequently, MAVS forms large aggregates (Hou et al. 2011) that stimulate MAPK activation and transcription induced by IRF3, IRF7, and NF-κB. Many of the components found downstream from TRIF in TLR signaling also are engaged by MAVS. For example, TRAF3 plus the kinases TBK1 and IKKε mediate IRF3/7 activation and induction of type I Ifn genes (Takeuchi and Akira 2010). RIG-I, but not MDA5, also requires the transmembrane protein STING to induce IFN (Ishikawa and Barber 2008). STING is located in the endoplasmic reticulum (ER), but its precise role in IFN induction by dsRNA requires further study. Experiments with fibroblasts from gene-targeted mice also implicate TRAF6, IKKγ, and the DD-containing proteins TRADD, RIP1, and FADD in IFN induction (Balachandran et al. 2004; Zhao et al. 2007; Michallet et al. 2008; Yoshida et al. 2008). Note that loss of TRADD, RIP1, or FADD produces a defect less severe than does MAVS or TBK1 deficiency. The extent of IRF3 phosphorylation and dimerization has not been determined in cells lacking TRADD, RIP1, or FADD; it remains possible that these proteins, like TRAF6, contribute to type I Ifn gene expression by activating NF-κB (Wang et al. 2010a). In DCs, MAVS engages the CARD-containing adaptors CARD9 and BCL10 to activate NF-κB (Poeck et al. 2010). BCL10 engages TRAF6 in lymphocytes to activate NF-κB (Sun et al. 2004b), and a similar pathway may operate downstream from MAVS. A role for FADD, TRADD, and RIP1 in MAVS signaling by DCs has not been examined.
5. NOD-LIKE RECEPTORS (NLRs)
Members of the Nod-like receptor (NLR) family of cytosolic PRRs are best known for their ability to signal NF-κB activation (NOD1 and NOD2) or secretion of the pro-inflammatory cytokines IL1β and IL18 (NLRP1/NALP1, NLRP3/NALP3/cryopyrin, and NLRC4/Ipaf) (Fig. 4). These proteins typically contain a CARD or pyrin domain at the amino terminus, a central nucleotide-binding oligomerization NACHT domain, and carboxy-terminal LRRs. NOD2 and NLRP3 have received considerable attention because their mutation is linked to inflammatory disease. NOD2 mutations are associated with Crohn’s inflammatory bowel disease and Blau syndrome, whereas mutations in the CIAS1 gene encoding NLRP3 are associated with familial cold autoinflammatory syndrome, Muckle–Wells syndrome, and neonatal-onset multisystem inflammatory disease (Table 2).
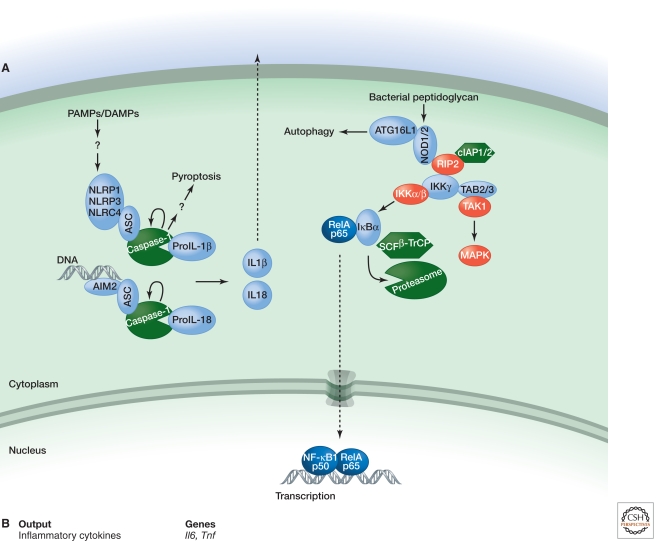
Figure 4.
Signaling by NLRs. (A) NLRP1, NLRP3, and NLRC4 respond to diverse PAMPS and DAMPs by engaging the caspase-1 adaptor protein ASC, whereas AIM2 binds ASC in response to cytoplasmic dsDNA. Activation of caspase-1 within each inflammasome complex results in processing of pro-IL1β and pro-IL18 and secretion of their biologically active forms. Caspase-1 activation also triggers a rapid form of cell death termed pyroptosis. NOD1 and NOD2 sense different components of bacterial peptidoglycan and stimulate either the autophagy machinery or gene transcription via NF-κB and MAPK activation. The latter outcome requires interaction of NOD1 or NOD2 with the kinase RIP2, which may be ubiquitylated by cIAPs in order to recruit TAB2/3 and IKKγ for TAK1 and IKK activation. (B) Functional outputs of some of the genes upregulated by NOD1 or NOD2 signaling.
NOD1 and NOD2 are sensors of different bacterial peptidoglycan components, but they both interact with the CARD-containing kinase RIP2 to activate MAPK and NF-κB signaling (Park et al. 2007). cIAPs are proposed to bind and ubiquitylate RIP2, and K63-linked polyubiquitylation of RIP2 recruits TAK1 for IKK and MAPK activation (Yang et al. 2007; Hitotsumatsu et al. 2008; Bertrand et al. 2009). NOD1 and NOD2 also have been shown to stimulate autophagy2 independently of RIP2 (Travassos et al. 2010).
Distinct PAMPS and DAMPs trigger NLRP1, NLRP3, and NLRC4 to nucleate signaling complexes termed “inflammasomes” (Table 4). The CARD- and PYRIN-domain-containing adaptor ASC is a critical inflammasome component, binding the CARD in the zymogen form of the aspartate-specific cysteine protease caspase-1 (Mariathasan et al. 2004). The proximity of caspase-1 zymogens within the inflammasome complex is believed to facilitate their autocatalytic activation. Caspase-1 substrates include pro-IL18 and pro-IL1β, the latter being upregulated transcriptionally by MyD88-dependent TLR signaling. Inflammasome activation also results in an extremely rapid form of cell death termed “pyroptosis.” The suicide of infected macrophages by pyroptosis is important for bacterial clearance (Miao et al. 2010), but the critical substrates of caspase-1 in this process still have to be determined.
Table 4.
Stimuli detected by inflammasome sensors that activate caspase-1
| Sensor | Known stimuli |
|---|---|
| AIM2 | Cytoplasmic DNA |
| NLRP1 | Bacillus anthracis lethal toxin |
| NLRP3 | ATP |
| Ionophore nigericin | |
| Marine toxin maitotoxin | |
| Crystals such as monosodium urate, calcium pyrophosphate dihydrate, silica, and asbestos | |
| Fungi such as Candida albicans | |
| Bacteria such as Staphylococcus aureus, Neisseria gonorrhoeae, Salmonella typhimurium, and Listeria monocytogenes | |
| Viruses such as sendai, adenovirus, and influenza | |
| NLRC4 | Bacterial flagellin |
| Certain bacterial type III secretion systems |
Intriguingly, immunofluorescence microscopy of endogenous inflammasome components in mouse macrophages infected with Salmonella typhimurium suggests that inflammasome assembly occurs at a single focus within a cell (Broz et al. 2010). One question that continues to vex the field is how NLRP1, NLRP3, and NLRC4 sense PAMPS and DAMPs, because direct binding has not been shown. The diversity of entities that trigger NLRP3-dependent caspase-1 activation suggests that NLRP3 might respond to a particular stress-activated signaling pathway. Both potassium efflux and the generation of reactive oxygen species (ROS) have been proposed as critical events upstream of NLRP3 activation, but the precise nature of NLR activation remains obscure.
ASC-dependent caspase-1 activation is also triggered in response to cytoplasmic DNA that appears during an infection or after tissue injury. The responsible PRR is not an NLR but the IFN-induced protein AIM2, which has a HIN200 domain for DNA binding and a pyrin domain to engage ASC. Cytoplasmic DNA also triggers type I IFN production, but this requires neither AIM2 nor TLRs. STING is a critical signaling component in this pathway, but whether any one DNA receptor is essential for its activation is unclear (Hornung and Latz 2010).
6. THE PRO-INFLAMMATORY CYTOKINE TUMOR NECROSIS FACTOR (TNF)
Induction of the cytokines IL1β and TNF by PRRs serves to amplify the inflammatory response because they too promote NF-κB and MAPK activation. Binding of IL1β to IL1R triggers MyD88-dependent signaling (Muzio et al. 1997), whereas TNF mediates most of its pro-inflammatory effects by binding to TNF receptor I (TNF-RI) (Peschon et al. 1998).
6.1. NF-κB and MAPK Activation by TNF
TNF-RI (also called TNFRSF1A) is a type I transmembrane protein that has cysteine-rich extracellular domains (CRDs) for TNF binding. Its cytoplasmic tail contains a DD that recruits the DD-containing adaptor TRADD and kinase RIP1 (Fig. 5). TRADD facilitates binding of RIP1 to TNF-R1 and recruits TRAF2, which is an adaptor for the ubiquitin ligases cIAP1 and cIAP2 (Chen et al. 2008; Ermolaeva et al. 2008; Pobezinskaya et al. 2008). Analyses of cells lacking cIAPs, RIP1, or TRAF2 indicate that all three contribute to NF-κB and MAPK activation, but the details of how they do so continue to be unraveled. Ubiquitylation of RIP1 by the cIAPs and E2 UbcH5 is believed to be important for recruitment of NEMO and TAK1, and subsequently for IKK activation (Varfolomeev et al. 2008; Xu et al. 2009), although TRAF2 ubiquitin ligase activity has been invoked recently as well (Alvarez et al. 2010). In addition, in some cell types, ubiquitylation of TRAF2 or cIAPs rather than RIP1 may support IKK activation (Li et al. 2009; Wong et al. 2010). Recruitment of LUBAC components sharpin, HOIL-1, and HOIP to the TNF-RI complex is linked to cIAP ubiquitin ligase activity, and linear ubiquitylation of RIP1 and NEMO may stabilize the TNF-RI signaling complex (Haas et al. 2009; Gerlach et al. 2011; Ikeda et al. 2011; Tokunaga et al. 2011). Underscoring the importance of sharpin in TNF signaling, mutation of Sharpin in mice causes chronic proliferative dermatitis that is rescued by TNF deficiency (Gerlach et al. 2011).
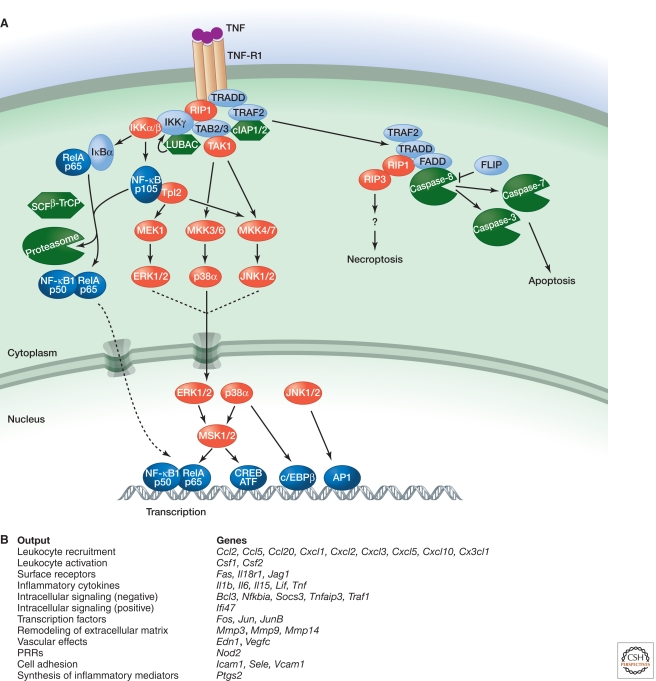
Figure 5.
Signaling by TNF-R1. (A) Binding of TNF to TNF-R1 causes the cytoplasmic death domain (DD) in TNF-R1 to bind the DD-containing proteins TRADD and RIP1. TRADD also binds TRAF2, which serves as an adaptor for the ubiquitin ligases cIAP1 and cIAP2. Ubiquitylation of RIP1, and potentially other components of the complex, recruits IKKγ and TAK1 for NF-κB and MAPK activation. Recruitment of LUBAC for linear ubiquitylation of IKKγ may stabilize the signaling complex. Translocation of TRADD, TRAF2, and RIP1 to the cytoplasm nucleates a second complex that contains the adaptor protein FADD and caspase-8. If c-FLIP levels are low, activation of caspase-8 and its substrates caspase-3 and caspase-7 causes apoptotic cell death. Inhibition of protein synthesis and caspases, as might occur in a virus-infected cell, promotes necroptotic cell death that is dependent on the kinase activities of RIP1 and RIP3. (B) Functional outputs of some of the genes upregulated by TNF signaling.
IKKβ activation by TNF triggers not only NF-κB transcription but also the Tpl2–MEK1–ERK kinase cascade activated by TLRs (see above). In fibroblasts, but not macrophages or B cells, Tpl2 activation by TNF has been linked to activation of the MKK4–JNK pathway as well. In addition, activation of the kinase MSK1 by ERK may enhance NF-κB transcriptional activity through phosphorylation of RelA (Banerjee and Gerondakis 2007). Genetic studies indicate that TNF-induced JNK activation is mediated largely by upstream kinases TAK1 and MKK7, whereas p38 activation requires TAK1 and MKK3/MKK6 (Brancho et al. 2003; Sato et al. 2005; Shim et al. 2005).
6.2. Apoptosis and Necroptosis Induction by TNF
The TNF-RI-associated signaling complex formed in response to TNF is transient as TRADD, RIP1, and TRAF2 shift to the cytoplasm to form what is referred to as complex II (Micheau and Tschopp 2003). Here, the DD in TRADD binds the DD in the adaptor FADD, whereas the death effector domain (DED) in FADD interacts with the DEDs in the prodomain of caspase-8 or its catalytically inactive homolog c-FLIP. If c-FLIP levels are low, then stable, active dimers of caspase-8 are formed. Caspase-8 substrates include caspase-3 and caspase-7, which are activated by proteolytic processing and cleave vital cellular proteins to cause apoptotic cell death (Green 2012). Because expression of c-FLIP is driven by NF-κB (Micheau et al. 2001) and many viruses have evolved strategies to inhibit NF-κB activation (Shisler and Jin 2004; Taylor et al. 2009), apoptosis in the absence of c-FLIP constitutes a host defense mechanism against infection. Certain viruses encode their own FLIPs, but the infected host cell may yet prevail in its attempt to die because TNF activates a form of cell death called necroptosis when protein synthesis and caspases are inhibited. This death is dependent on the kinase activity of RIP1 and the related kinase RIP3 (Cho et al. 2009; He et al. 2009; Zhang et al. 2009). These kinases interact through their RHIMs, but it is not clear what triggers their kinase activity or what substrates are phosphorylated in order to kill the cell.
7. SELECTINS AND INTEGRINS
One important output of LPS, TNF, and IL1β signaling in endothelial cells is increased surface expression of transmembrane proteins involved in cell adhesion, such as P-selectin, E-selectin, ICAM1, and VCAM1. All four are induced by NF-κB in mice, whereas upregulation of human P-selectin relies on fusion of secretory granules with the plasma membrane. Glycoproteins including CD44, P-selectin glycoprotein ligand 1 (PSGL1), and E-selectin ligand (ESL1) on leukocytes interact with the selectins to mediate leukocyte rolling along vessel walls. Engagement of either PSGL1 or CD44 triggers a signaling pathway that causes leukocyte β2 integrins LFA1 and MAC1 expressed on the cell surface to adopt a more extended conformation that increases their affinity for endothelial ICAM1. Interactions between the β2 integrins and ICAM1 then slow leukocyte rolling further. β2 integrin activation via this “inside-out” signaling (Devreotes and Horwitz 2012) requires the membrane-associated Src family kinases Fgr, Hck, and Lyn. These tyrosine kinases probably phosphorylate cytoplasmic immunoreceptor tyrosine-based activation motifs (ITAMs) in the transmembrane adaptors DAP12 and FcRγ to create docking sites for the tandem SH2 domains in the tyrosine kinase Syk (Cantrell 2012), but note that they can also phosphorylate integrin β subunits. Studies with neutrophils from gene-targeted mice indicate that sequential activation of Syk and the kinase Btk is essential for slow rolling on E-selectin and ICAM1 (Zarbock et al. 2008; Yago et al. 2010).
Further β2 integrin activation needed for leukocyte arrest before migration across the endothelial cell barrier is mediated by chemokines and chemoattractants immobilized on the endothelial cell surface. These factors engage G-protein-coupled receptors (GPCRs) on the leukocyte surface (see below). Note that integrin engagement also elicits “outside-in” signaling in leukocytes, and, similarly to Fc receptor signaling (see below), this activates leukocyte effector functions (Lowell 2011). Clustering of ICAM1 on endothelial cells also triggers signals that facilitate leukocyte migration across the endothelium into the surrounding tissue. Phosphorylation of VE-cadherin appears to loosen adherens junctions, whereas activation of myosin light chain kinase (MLCK) mediates endothelial cell contraction (Muller 2011).
8. G-PROTEIN-COUPLED RECEPTORS (GPCRs)
Lipid-based inflammatory mediators such as prostaglandins, leukotrienes, and platelet-activating factor; vasoactive amines such as histamine and serotonin; complement fragments C3b, C3a, and C5a; chemokines; proteases; and bacterial or mitochondrial formylated peptides all activate signaling by GPCRs linked to heterotrimeric G-proteins composed of α, β, and γ subunits. Following ligand binding, or cleavage in the case of protease-activated receptors (PARs), Gα and Gβγ interact with ion channels or enzymes such as adenylyl cyclase, phospholipase C (PLC), and phosphoinositide 3-kinase (PI3K). In addition, active GPCRs are phosphorylated by GPCR kinases (GRKs) to stimulate binding of arrestins, adaptors that stimulate GPCR endocytosis as well as MAPK activation. To highlight some of the pathways engaged by GPCRs during inflammation, below we focus on signaling by the chemoattractants C5a and the prototypical formylated peptide formyl-Met-Leu-Phe (fMLP) (Fig. 6).
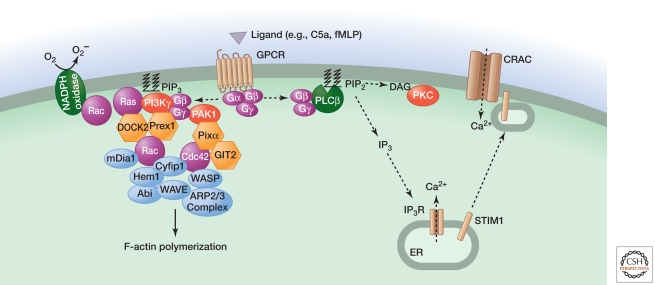
Figure 6.
Signaling by GPCRs activated by chemoattractants C5a and fMLP. C5a or fMLP binding to their respective GPCRs triggers dissociation of Gαi-GTP from Gβγ, the latter interacting with PLCβ, the p101 regulatory subunit of PI3Kγ, and PAK1. Activated PLCβ generates the second messengers IP3 and DAG to elevate intracellular calcium and activate PKC, respectively. These outcomes regulate JNK activation, vesicle exocytosis, and superoxide production by the NADPH oxidase. PIP3 generated by PI3Kγ, whose activation also involves the GTPase Ras, stimulates GEFs (DOCK2 and Prex1) that activate Rac GTPases. PAK1 interacts with the GEF PIXα for activation of another Rho family GTPase called Cdc42. Rac1, Rac2, and Cdc42 together regulate chemotaxis by coordinating alterations to the actin cytoskeleton via mDia1 and the ARP2/3 complex. Rac2 is also an essential component of the NADPH oxidase. The signaling components regulating gene transcription are less defined.
8.1. C5a Receptor (C5aR) and Formyl Peptide Receptors (FPRs)
Complement protein C5a is produced by complement plasma proteases activated by IgM- and IgG-containing antibody complexes (the classical pathway), pathogens coated with host mannose-binding lectin or C-reactive protein (the lectin pathway), or pathogens in isolation (the alternative pathway). C5a can also be generated by non-complement proteases such as thrombin and kallikrein, which are components of the clotting system activated in response to endothelial cell injury. C5a and formyl peptides, the latter of bacterial or mitochondrial origin, stimulate leukocyte chemotaxis, degranulation, superoxide production for microbe killing, and, as mentioned above, activation of integrins for cell adhesion. Similarly to TNF, C5a stimulates endothelial cells to increase expression of cytokines, chemokines, and cell adhesion molecules such as E-selectin, ICAM1, and VCAM1 (Albrecht et al. 2004).
C5a and fMLP activate predominantly pertussis toxin-sensitive Gi proteins. The Gβγ dimer that is released activates several enzymes, including PLCβ (Camps et al. 1992). Hydrolysis of phosphatidylinositol 4,5-bisphosphate in the plasma membrane by PLC yields the second messengers inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) (Bootman 2012). Binding of IP3 to its receptor causes depletion of calcium stores within the ER and relocation of the calcium-binding type I transmembrane protein STIM1 from the ER to structures near the plasma membrane (Brechard et al. 2009). STIM1 then activates the plasma membrane calcium-release-activated calcium channel (CRAC) to cause an influx of calcium into the cell. Elevated intracellular calcium together with protein kinase C (PKC) activation by DAG is important for vesicle exocytosis, superoxide production by the NADPH oxidase, and JNK activation (Li et al. 2000). Calcium stimulates lysosome exocytosis by activating the synaptotagmin regulator of vesicle fusion SYT7 (Colvin et al. 2010).
GTP-bound Ras, activated by a mechanism that is unclear, and Gβγ dimers activate PI3Kγ by binding to its p101 regulatory subunit and p110γ catalytic subunit, respectively. Phosphatidylinositol 3,4,5-trisphosphate (PIP3) produced by PI3Kγ activates Rac guanine-nucleotide exchange factors (GEFs), such as Prex1 and DOCK2 (Welch et al. 2002; Kunisaki et al. 2006), and contributes to superoxide production and chemokinesis3 (Suire et al. 2006; Ferguson et al. 2007; Nishio et al. 2007). The GTPase RhoG also has a role in superoxide production but is dispensable for neutrophil migration (Condliffe et al. 2006). The GTPase Rac2 appears to be essential for the assembly of filamentous actin (F-actin) and the NADPH oxidase, whereas Rac1 localizes F-actin to the leading edge of the cell facing the chemoattractant (Sun et al. 2004a). This asymmetrical polymerization of F-actin drives membrane protusions in the direction of migration. Active Rac is thought to exert its effect on the actin cytoskeleton by interacting with the adaptor Cyfip1 (also called Sra1), which, in combination with several proteins, stimulates the ARP2/3 actin nucleation complex (Devreotes and Horwitz 2012).
The GEF PIXα also is required for F-actin assembly at the leading edge in C5a-stimulated neutrophils. It is recruited to Gβγ via the kinase PAK1 and appears to function by interacting with the GTPase Cdc42 and the GTPase-activating (GAP) protein GIT2 (Li et al. 2003; Mazaki et al. 2006). Activated Cdc42 interacts with the adaptor WASP to engage the ARP2/3 actin nucleation complex. WASP appears to work in concert with the actin-nucleating protein mDia1, because neutrophils lacking both WASP and mDia1 show a profound defect in chemotaxis (Shi et al. 2009).
9. Fc RECEPTORS
Repeated exposure to a polyvalent foreign substance can elicit an inflammatory response called a hypersensitivity reaction if the host makes antibodies against the substance. Immune complexes containing the antigen and IgG or IgM antibodies activate complement proteases, culminating in the generation of C3a and C5a, which signal leukocyte recruitment and activation (see above); the opsonin C3b, which coats and promotes phagocytosis of bacteria; and the membrane attack complex for bacterial cell lysis (C5b-9). In addition, complexes containing IgG or IgE antibodies engage Fc receptors on leukocytes. Members of the Fc receptor family are type I transmembrane proteins (with the exception of human GPI-anchored FcγRIIIB) that produce activating (human FcγRI, FcγRIIA, FcγRIIC, FcγRIIIA, FcγRIIIB, and FcεRI) or inhibitory (human FcγRIIB) signals. Mast cells expressing the high-affinity receptor for IgE, FcεRI, play a central role in allergic reactions. FcεRI engagement causes intracellular granules to fuse with the plasma membrane such that preformed inflammatory mediators including histamine, serotonin, and proteases are released into the extracellular environment. Activated mast cells also secrete pro-inflammatory prostaglandins, leukotrienes, and cytokines, but these are synthesized de novo.
9.1. FcεRI
FcεRI is an αβγ2 heterotetramer. Its α-chain contains extracellular Ig-like domains for binding the heavy-chain constant region of IgE, whereas the β-chain and a γ-chain homodimer transduce signals via cytoplasmic ITAMs (Fig. 7) (Cantrell 2012). IgE-induced clustering of FcεRI promotes activation of Src family kinases Lyn and Fyn. Lyn substrates include both positive and negative regulators of mast cell activation, which fine-tune the magnitude and duration of the response. Lyn stimulates activation by phosphorylating the FcRγ ITAM, which recruits the SH2 domains in Syk. Subsequent Syk-dependent phosphorylation of the transmembrane adaptors LAT1 and LAT2 recruits additional SH2-containing signaling components, such as PLCγ and the adaptors Grb2 and Gads. SH3 domains in Grb2 and Gads bind proline-rich regions in additional proteins such as the adaptors SLP76 and Gab2. SLP76 interacts with the Rho/Rac GEF VAV1, which contributes to PLCγ and JNK activation. Fyn-dependent phosphorylation of Gab2 recruits the SH2-containing p85 regulatory subunit of PI3Kδ. PIP3 produced by PI3Kδ retains proteins containing plextrin homology (PH) domains at the plasma membrane, such as PLCγ, Gab2, Akt, and Btk. The kinase Btk phosphorylates and enhances the activity of PLCγ (Alvarez-Errico et al. 2009).
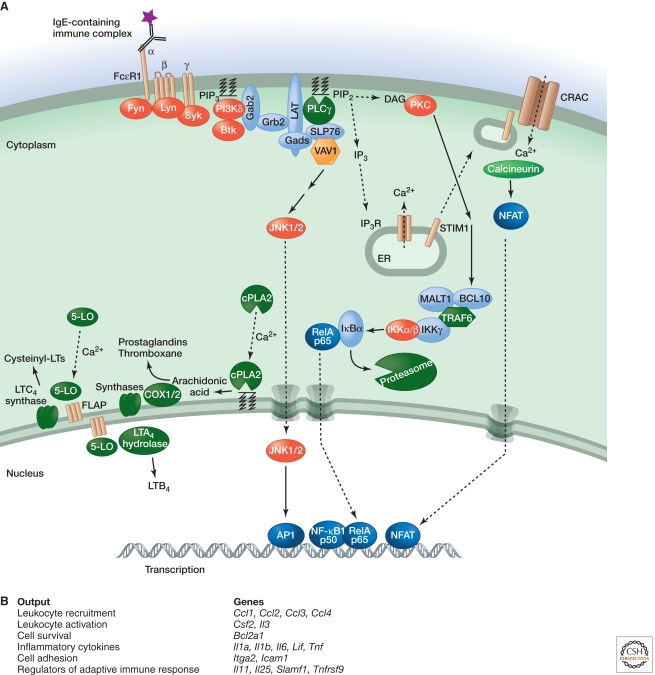
Figure 7.
Signaling by FcεRI. (A) Binding of the Fc region of antigen-bound IgE to FcεRI activates the Src family kinases Lyn and Fyn. Tyrosine phosphorylation of the FcRγ ITAM recruits the tyrosine kinase Syk, which is required for phosphorylation of LAT transmembrane adaptor proteins. Phosphorylated LAT1 binds PLCγ and the adaptors Gads and Grb2. Gads recruits the adaptor SLP76, which regulates activation of PLCγ and the GEF Vav1. Grb2 binds Gab2, which is phosphorylated by Fyn and binds the p85 regulatory subunit of PI3Kδ. PIP3 generated by PI3Kδ retains signaling components such as Gab2, PLCγ, and Btk at the plasma membrane. IP3 generated by PLCγ depletes ER calcium stores, which causes a STIM1-dependent influx of calcium that promotes mast cell degranulation. Elevated intracellular calcium also activates the phosphatase calcineurin, stimulates NFAT-dependent gene expression, and triggers the translocation of cPLA2 and 5-lipoxygenase (5-LO) to the nuclear envelope, cytoplasmic lipid bodies, or ER. cPLA2 releases arachidonic acid from membrane phospholipids. COX enzymes and downstream synthases metabolize arachidonic acid into prostaglandins and thromboxane, whereas leukotriene (LT) synthesis from arachidonic acid involves five-lipoxygenase-activating protein (FLAP), 5-LO, and downstream LTC4 synthase or LTA4 hydrolase. DAG generated by PLCγ activates PKC, which is important for IKK activation via MALT1, BCl10, and TRAF6, as well as subsequent NF-κB-dependent gene transcription. IKKβ has also been implicated in mast cell degranulation independent of NF-κB activation. (B) Functional outputs of some of the genes upregulated by FcεRI signaling.
PLC-γ signaling triggers STIM1-dependent calcium influx (Bootman 2012), which is essential for normal mast cell degranulation, leukotriene synthesis, and activation of NFAT transcription factors via the calcium-dependent phosphatase calcineurin (Baba et al. 2008; Vig et al. 2008). NFAT promotes expression of the cytokines TNF and IL13. Eicosanoids including leukotrienes and prostaglandins are derived from arachidonic acid, which is liberated from phospholipids by cytosolic phospholipase A2 in response to elevated intracellular calcium and MAPK activation (Fujishima et al. 1999).
PKC activation by DAG is required for degranulation and activation of NF-κB, the latter contributing to the induction of TNF and IL6. BCL10, MALT1 (also called paracaspase), and TRAF6 regulate NF-κB activation but are dispensable for degranulation (Klemm et al. 2006; Chen et al. 2007; Yang et al. 2008), whereas IKKβ is required for both functions (Suzuki and Verma 2008). The mechanism by which IKKβ is activated for degranulation remains unclear, but, once activated, IKKβ appears to promote exocytosis by phosphorylating the SNARE receptor SNAP23.
9.2. Fcγ Receptors
Activating Fcγ receptors, in common with FcεRI, contain cytoplasmic ITAMs and stimulate Src family kinases plus Syk. The downstream signaling events that promote phagocytosis, degranulation, cytokine production, and superoxide production are less well defined but probably involve many of the components engaged by FcεRI. Superoxide production by the NADPH oxidase that assembles on phagosomal membranes requires Vav-mediated activation of Rac GTPases, the putative Rac adaptor CAPRI, and PLCγ. Depending on the cell type and context, CAPRI, VAV, and Rac also contribute to remodeling of the actin cytoskeleton for phagocytosis, along with PI3K and its adaptor Gab2 (Gu et al. 2003; Zhang et al. 2005; Hall et al. 2006; Utomo et al. 2006; Jakus et al. 2009). FcγRIIB is unique amongst Fc receptors in that it suppresses ITAM signaling through an immunoreceptor tyrosine-based inhibitory motif (ITIM). Lyn-dependent ITIM phosphorylation recruits the SH2-containing inositol 5′-phosphatase (SHIP), which hydrolyzes PIP3 and thereby limits recruitment of PH-domain-containing proteins such as PLC-γ, Btk, and VAV (Lowell 2011).
10. INFLAMMATION AS A RISK FACTOR FOR CANCER
Bacteria and viruses that establish persistent infections are linked to certain cancers. For example, Helicobacter pylori bacteria increase the risk of gastric cancer and MALT lymphoma, whereas Hepatitis B and Hepatitis C viruses increase the risk of hepatocellular carcinoma. Similarly, pancreatitis is a risk factor for pancreatic ductal adenocarcinoma, and this has been modeled successfully in mice expressing oncogenic K-Ras in adult pancreatic acinar cells (Guerra et al. 2011). Studies with gene-targeted mice have confirmed that inflammatory signaling pathways promote tumor development. For example, deletion of FcRγ suppresses squamous cell carcinoma driven by keratinocyte-specific expression of the human papilloma virus oncogene HPV16 (Andreu et al. 2010). Similarly, IL6 deficiency in hematopoietic cells suppresses colon tumors that develop in response to procarcinogen azoxymethane plus colitis-inducing dextran sulfate sodium (DSS) (Grivennikov et al. 2009). IL6 enhances proliferation and survival of intestinal epithelial cells through activation of the transcription factor STAT3.
In another mouse model, the ability of tobacco smoke to promote lung tumors driven by oncogenic K-Ras is reduced by IKKβ deletion in myeloid cells (Takahashi et al. 2010). Finally, IL6 or TNF-R1 deficiency protects obese mice from hepatocellular carcinomas that form in response to the pro-carcinogen diethylnitrosamine by limiting lipid accumulation and inflammatory infiltrates in the liver (Park et al. 2010). Note, however, that in all of these tumor models, inflammation alone is insufficient for tumor development, which implies that carcinogen-induced mutations are needed.
11. CONCLUDING REMARKS
Many of the major players in inflammatory signaling have been identified, but the importance and complexity of posttranslational modifications such as ubiquitylation in these pathways continue to be unraveled. Binding of TLRs, TNF and IL1 receptors, GPCRs, integrins, selectins, and Fc receptors to their ligands triggers the formation of multi-subunit signaling complexes, but it remains to be seen how diverse inflammatory stimuli can activate intracellular PRRs such as NLRP3 and NLRC4. An attractive hypothesis is that posttranslational modifications to NLR family members are key to their activation. Once activated, both surface and intracellular PRRs stimulate transcription of inflammatory genes; TLRs, RLRs, and some NLRs (e.g., NOD1 and NOD2) engage common downstream signaling pathways to stimulate transcription factors such as NF-κB, AP1, CREB, and c/EBPβ, whereas caspase-1-activating PRRs (e.g., AIM2, NLRP3, and NLRC4) stimulate similar pathways indirectly via the secretion of IL1β and IL18. Going forward, it will be important to understand how innate immune cells exposed to multiple inflammatory mediators and stimuli in vivo integrate signals from diverse receptors, because this will offer insight into what critical components might be targeted for therapeutic benefit in inflammatory disorders.
DCs are very heterogeneous. pDCs acquire DC morphology and secrete large amounts of IFN during virus infections. They can be distinguished from other DC subsets by their cell surface markers. The myeloid DCs referenced were derived in vitro from bone marrow cells with granulocyte/macrophage colony-stimulating factor (GM-CSF).
Autophagy is the process by which cytoplasmic components, including organelles and invading bacteria, are sequestered inside double-membrane vesicles and then delivered to the lysosome for degradation.
Chemokinesis refers to random cell migration, whereas chemotaxis is directed cell migration along a chemical gradient.
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Albrecht EA, Chinnaiyan AM, Varambally S, Kumar-Sinha C, Barrette TR, Sarma JV, Ward PA 2004. C5a-induced gene expression in human umbilical vein endothelial cells. Am J Pathol 164: 849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465: 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Errico D, Lessmann E, Rivera J 2009. Adapters in the organization of mast cell signaling. Immunol Rev 232: 195–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu P, Johansson M, Affara NI, Pucci F, Tan T, Junankar S, Korets L, Lam J, Tawfik D, DeNardo DG, et al. 2010. FcRγ activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell 17: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Nishida K, Fujii Y, Hirano T, Hikida M, Kurosaki T 2008. Essential function for the calcium sensor STIM1 in mast cell activation and anaphylactic responses. Nat Immunol 9: 81–88 [DOI] [PubMed] [Google Scholar]
- Balachandran S, Thomas E, Barber GN 2004. A FADD-dependent innate immune mechanism in mammalian cells. Nature 432: 401–405 [DOI] [PubMed] [Google Scholar]
- Banerjee A, Gerondakis S 2007. Coordinating TLR-activated signaling pathways in cells of the immune system. Immunol Cell Biol 85: 420–424 [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M 2009. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30: 789–801 [DOI] [PubMed] [Google Scholar]
- *.Bootman M 2012. Calcium signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancho D, Tanaka N, Jaeschke A, Ventura JJ, Kelkar N, Tanaka Y, Kyuuma M, Takeshita T, Flavell RA, Davis RJ 2003. Mechanism of p38 MAP kinase activation in vivo. Genes Dev 17: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechard S, Plancon S, Melchior C, Tschirhart EJ 2009. STIM1 but not STIM2 is an essential regulator of Ca2+ influx-mediated NADPH oxidase activity in neutrophil-like HL-60 cells. Biochem Pharmacol 78: 504–513 [DOI] [PubMed] [Google Scholar]
- Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med 207: 1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Carozzi A, Schnabel P, Scheer A, Parker PJ, Gierschik P 1992. Isozyme-selective stimulation of phospholipase C-β2 by G protein β γ-subunits. Nature 360: 684–686 [DOI] [PubMed] [Google Scholar]
- *.Cantrell D 2012. Lymphocyte activation and behavior. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a006056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Jin W, Sun SC 2009. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol 10: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pappu BP, Zeng H, Xue L, Morris SW, Lin X, Wen R, Wang D 2007. B cell lymphoma 10 is essential for FcεR-mediated degranulation and IL-6 production in mast cells. J Immunol 178: 49–57 [DOI] [PubMed] [Google Scholar]
- Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, Huang HL, Pike KA, Hao Z, Su YW, et al. 2008. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc Natl Acad Sci 105: 12429–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK 2009. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137: 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin RA, Means TK, Diefenbach TJ, Moita LF, Friday RP, Sever S, Campanella GS, Abrazinski T, Manice LA, Moita C, et al. 2010. Synaptotagmin-mediated vesicle fusion regulates cell migration. Nat Immunol 11: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe AM, Webb LM, Ferguson GJ, Davidson K, Turner M, Vigorito E, Manifava M, Chilvers ER, Stephens LR, Hawkins PT 2006. RhoG regulates the neutrophil NADPH oxidase. J Immunol 176: 5314–5320 [DOI] [PubMed] [Google Scholar]
- Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA 2005. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem 280: 36560–36566 [DOI] [PubMed] [Google Scholar]
- Das M, Sabio G, Jiang F, Rincon M, Flavell RA, Davis RJ 2009. Induction of hepatitis by JNK-mediated expression of TNF-α. Cell 136: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Devreotes P, Horwitz R 2012. Cell migration and chemotaxis. Cell death. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, Bodemer C, Kenwrick S, Dupuis-Girod S, Blanche S, et al. 2001. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat Genet 27: 277–285 [DOI] [PubMed] [Google Scholar]
- Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, Tschopp J, Pasparakis M 2008. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol 9: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, Andrews S, Crabbe T, Finan P, Jones G, Jackson S, et al. 2007. PI(3)Kγ has an important context-dependent role in neutrophil chemokinesis. Nat Cell Biol 9: 86–91 [DOI] [PubMed] [Google Scholar]
- Fujishima H, Sanchez Mejia RO, Bingham CO III, Lam BK, Sapirstein A, Bonventre JV, Austen KF, Arm JP 1999. Cytosolic phospholipase A2 is essential for both the immediate and the delayed phases of eicosanoid generation in mouse bone marrow-derived mast cells. Proc Natl Acad Sci 96: 4803–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, et al. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471: 591–596 [DOI] [PubMed] [Google Scholar]
- *.Green D 2012. Cell death. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a006080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Botelho RJ, Yu M, Grinstein S, Neel BG 2003. Critical role for scaffolding adapter Gab2 in FcγR-mediated phagocytosis. J Cell Biol 161: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernández-Porras I, Cañamero M, Rodriguez-Justo M, Serrano M, Barbacid M 2011. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 19: 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, et al. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36: 831–844 [DOI] [PubMed] [Google Scholar]
- Hall AB, Gakidis MA, Glogauer M, Wilsbacher JL, Gao S, Swat W, Brugge JS 2006. Requirements for Vav guanine nucleotide exchange factors and Rho GTPases in FcγR- and complement-mediated phagocytosis. Immunity 24: 305–316 [DOI] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137: 1100–1111 [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Advincula R, Malynn BA, Werts C, et al. 2008. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity 28: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Latz E 2010. Intracellular DNA recognition. Nat Rev Immunol 10: 123–130 [DOI] [PubMed] [Google Scholar]
- Hoshino K, Sasaki I, Sugiyama T, Yano T, Yamazaki C, Yasui T, Kikutani H, Kaisho T 2010. Critical role of IκB kinase α in TLR7/9-induced type I IFN production by conventional dendritic cells. J Immunol 184: 3341–3345 [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang Q, Chen ZJ 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skånland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel A 2010. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb Perspect Biol 2: a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakus Z, Simon E, Frommhold D, Sperandio M, Mocsai A 2009. Critical role of phospholipase Cγ2 in integrin and Fc receptor-mediated neutrophil functions and the effector phase of autoimmune arthritis. J Exp Med 206: 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang CQ, Gale M, Patel SS, Marcotrigiano J 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanarek N, London N, Schueler-Furman O, Ben-Neriah Y 2010. Ubiquitination and degradation of the inhibitors of NF-κB. Cold Spring Harb Perspect Biol 2: a000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YJ, Chen J, Otsuka M, Mols J, Ren S, Wang Y, Han J 2008. Macrophage deletion of p38α partially impairs lipopolysaccharide-induced cellular activation. J Immunol 180: 5075–5082 [DOI] [PubMed] [Google Scholar]
- Kim C, Sano Y, Todorova K, Carlson BA, Arpa L, Celada A, Lawrence T, Otsu K, Brissette JL, Arthur JS, et al. 2008. The kinase p38α serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat Immunol 9: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm S, Gutermuth J, Hultner L, Sparwasser T, Behrendt H, Peschel C, Mak TW, Jakob T, Ruland J 2006. The Bcl10–Malt1 complex segregates FcεRI-mediated nuclear factor κB activation and cytokine production from mast cell degranulation. J Exp Med 203: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D 2009. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y, Nishikimi A, Tanaka Y, Takii R, Noda M, Inayoshi A, Watanabe K, Sanematsu F, Sasazuki T, Sasaki T, et al. 2006. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J Cell Biol 174: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A 2009. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J 28: 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Dorf ME 2009. PKC phosphorylation of TRAF2 mediates IKKα/β recruitment and K63-linked polyubiquitination. Mol Cell 33: 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y, Smrcka AV, Wu G, Li L, Liu M, Huang CK, Wu D 2003. Directional sensing requires Gβ γ-mediated PAK1 and PIX α-dependent activation of Cdc42. Cell 114: 215–227 [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D 2000. Roles of PLC-β2 and -β3 and PI3Kγ in chemoattractant-mediated signal transduction. Science 287: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H 2010. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature 465: 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell CA 2011. Src-family and Syk kinases in activating and inhibitory pathways in innate immune cells: Signaling cross talk. Cold Spring Harb Perspect Biol 3: a002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P 2010. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18: 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430: 213–218 [DOI] [PubMed] [Google Scholar]
- Mazaki Y, Hashimoto S, Tsujimura T, Morishige M, Hashimoto A, Aritake K, Yamada A, Nam JM, Kiyonari H, Nakao K, et al. 2006. Neutrophil direction sensing and superoxide production linked by the GTPase-activating protein GIT2. Nat Immunol 7: 724–731 [DOI] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11: 1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, Konig M, et al. 2008. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 28: 651–661 [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114: 181–190 [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J 2001. NF-κB signals induce the expression of c-FLIP. Mol Cell Biol 21: 5299–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Plüddemann A, Hoe JC, Williams KJ, Varin A, Makepeace K, Aknin M, Bowdish DME, Smale ST, Barclay AN, et al. 2010. Immune inhibitory ligand CD200 induction by TLRs and NLRs limits macrophage activation to protect the host from meningococcal septicemia. Cell Host Microbe 16: 236–247 [DOI] [PubMed] [Google Scholar]
- Muller WA 2011. Mechanisms of leukocyte transendothelial migration. Annu Rev Pathol 28: 323–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Ni J, Feng P, Dixit VM 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278: 1612–1615 [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- Nishio M, Watanabe K, Sasaki J, Taya C, Takasuga S, Iizuka R, Balla T, Yamazaki M, Watanabe H, Itoh R, et al. 2007. Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat Cell Biol 9: 36–44 [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Nunez G 2007. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol 178: 2380–2386 [DOI] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M 2010. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Torrance DS, Stocking KL, Glaccum MB, Otten C, Willis CR, Charrier K, Morrissey PJ, Ware CB, Mohler KM 1998. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J Immunol 160: 943–952 [PubMed] [Google Scholar]
- Pobezinskaya YL, Kim YS, Choksi S, Morgan MJ, Li T, Liu C, Liu Z 2008. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol 9: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, et al. 2010. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nat Immunol 11: 63–69 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Wu X, Granger E, Sun SC 2005. Regulation of the deubiquitinating enzyme CYLD by IκB kinase γ-dependent phosphorylation. Mol Cell Biol 25: 3886–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G 2007. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131: 1124–1136 [DOI] [PubMed] [Google Scholar]
- Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, Akira S 2005. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol 6: 1087–1095 [DOI] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci 107: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Ma A, Harhaj EW 2010. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327: 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang J, Mullin M, Dong B, Alberts AS, Siminovitch KA 2009. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol 182: 3837–3845 [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, et al. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev 19: 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisler JL, Jin XL 2004. The vaccinia virus K1L gene product inhibits host NF-κB activation by preventing IκBα degradation. J Virol 78: 3553–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ 2009. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem 78: 769–796 [DOI] [PubMed] [Google Scholar]
- Suire S, Condliffe AM, Ferguson GJ, Ellson CD, Guillou H, Davidson K, Welch H, Coadwell J, Turner M, Chilvers ER, et al. 2006. Gβγs and the Ras binding domain of p110γ are both important regulators of PI(3)Kγ signalling in neutrophils. Nat Cell Biol 8: 1303–1309 [DOI] [PubMed] [Google Scholar]
- Sun CX, Downey GP, Zhu F, Koh AL, Thang H, Glogauer M 2004a. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood 104: 3758–3765 [DOI] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ 2004b. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell 14: 289–301 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Verma IM 2008. Phosphorylation of SNAP-23 by IκB kinase 2 regulates mast cell degranulation. Cell 134: 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Ogata H, Nishigaki R, Broide DH, Karin M 2010. Tobacco smoke promotes lung tumorigenesis by triggering IKKβ- and JNK1-dependent inflammation. Cancer Cell 17: 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- Taylor SL, Frias-Staheli N, Garcia-Sastre A, Schmaljohn CS 2009. Hantaan virus nucleocapsid protein binds to importin α proteins and inhibits tumor necrosis factor α-induced activation of nuclear factor κB. J Virol 83: 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. 2009. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K 2011. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471: 633–636 [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. 2010. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 11: 55–62 [DOI] [PubMed] [Google Scholar]
- Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M 2010. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol 11: 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utomo A, Cullere X, Glogauer M, Swat W, Mayadas TN 2006. Vav proteins in neutrophils are required for FcγR-mediated signaling to Rac GTPases and nicotinamide adenine dinucleotide phosphate oxidase component p40(phox). J Immunol 177: 6388–6397 [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D 2008. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor α (TNFα)-induced NF-κB activation. J Biol Chem 283: 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178: 6444–6455 [DOI] [PubMed] [Google Scholar]
- Vereecke L, Beyaert R, van Loo G 2011. Genetic relationships between A20/TNFAIP3, chronic inflammation, and autoimmune disease. Biochem Soc Trans 39: 1086–1091 [DOI] [PubMed] [Google Scholar]
- Vig M, DeHaven WI, Bird GS, Billingsley JM, Wang H, Rao PE, Hutchings AB, Jouvin MH, Putney JW, Kinet JP 2008. Defective mast cell effector functions in mice lacking the CRACM1 pore subunit of store-operated calcium release-activated calcium channels. Nat Immunol 9: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Xiao H, Affolter J, Kim TW, Bulek K, Chaudhuri S, Carlson D, Hamilton T, Mazumder B, Stark GR, et al. 2009. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J Biol Chem 284: 10367–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, Garcia-Sastre A, Balachandran S, Beg AA 2010a. NF-κB RelA subunit is crucial for early IFN-β expression and resistance to RNA virus replication. J Immunol 185: 1720–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. 2010b. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR 2002. P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108: 809–821 [DOI] [PubMed] [Google Scholar]
- Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J 2010. RIPK1 is not essential for TNFR1-induced activation of NF-κB. Cell Death Differ 17: 482–487 [DOI] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ 2009. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol Cell 36: 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yago T, Shao B, Miner JJ, Yao L, Klopocki AG, Maeda K, Coggeshall KM, McEver RP 2010. E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin αLβ2-mediated slow leukocyte rolling. Blood 116: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, et al. 2006. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol 7: 962–970 [DOI] [PubMed] [Google Scholar]
- Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA 2007. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem 282: 36223–36229 [DOI] [PubMed] [Google Scholar]
- Yang YJ, Chen W, Carrigan SO, Chen WM, Roth K, Akiyama T, Inoue J, Marshall JS, Berman JN, Lin TJ 2008. TRAF6 specifically contributes to FcεRI-mediated cytokine production but not mast cell degranulation. J Biol Chem 283: 32110–32118 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Takaesu G, Yoshida H, Okamoto F, Yoshioka T, Choi Y, Akira S, Kawai T, Yoshimura A, Kobayashi T 2008. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J Biol Chem 283: 36211–36220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbock A, Abram CL, Hundt M, Altman A, Lowell CA, Ley K 2008. PSGL-1 engagement by E-selectin signals through Src kinase Fgr and ITAM adapters DAP12 and FcRγ to induce slow leukocyte rolling. J Exp Med 205: 2339–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Guo J, Dzhagalov I, He YW 2005. An essential function for the calcium-promoted Ras inactivator in Fcγ receptor-mediated phagocytosis. Nat Immunol 6: 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J 2009. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325: 332–336 [DOI] [PubMed] [Google Scholar]
- Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R 2007. The NEMO adaptor bridges the nuclear factor-κB and interferon regulatory factor signaling pathways. Nat Immunol 8: 592–600 [DOI] [PubMed] [Google Scholar]