The approach to neuroprotection in stroke has relied mainly on exogenously administered drugs derived from research probing the cellular mechanisms of ischemic brain injury. This approach has had limited translational impact. Over 1000 putative neuroprotective agents have been developed, and over 100 have advanced to clinical trials. None, however, have shown clinical efficacy1.
By contrast, a gene-based approach seeks to harness endogenous neuroprotective programs for stroke therapy. Experimentally, ischemic brain injury can be attenuated and stroke outcome improved by gene expression modification in brain. The brain and other organs have highly conserved, endogenous neuroprotective programs, the induction of which reduces ischemic injury. The neuroprotective programs can be induced in the model of ischemic tolerance: brief exposure to sublethal ischemia produces tolerance to a subsequent, severe ischemic challenge. A number of laboratories have taken this approach with differing mechanistic foci which have been reviewed elsewhere2–5. The protection induced by tolerance is substantial, gene-based, and dependent on new protein synthesis. The neuroprotection of tolerance has been demonstrated in experimental cardiopulmonary bypass surgery6 and in stroke in the primate7. A clinical counterpart likely exists in human brain: patients with prodromal transient ischemic attacks have milder strokes8–11. Two small clinical trials have shown potential benefits of preconditioning, and five additional trials are underway12. Thus, there is considerable therapeutic potential in understanding gene expression changes in tolerance and in dissecting the biological mechanisms that regulate them. This is an evolving story deriving mainly from the authors' and our collaborators laboratories.
Individual Genes
Over the last several decades, research has identified numerous genes whose regulation may affect the outcome of stroke. Because cell death following harmful ischemia is mediated by programmed cell death–like mechanisms, many studies of neuroprotection have focused on apoptotic mediators as potential effectors of tolerance to ischemia. The regulation of genes in cell death pathways, particularly those that affect mitochondrial integrity, has been studied extensively. Regulation of genes for caspases, Bcl family members, protein kinases, hypoxia inducible factor (HIF), and apoptosis-inducing factor (AIF)13–16 can alter stroke outcome. For example, the use of antisense to block upregulation of the cell survival protein Bcl-2 during stroke results in a larger infarct, whereas inhibition of the cell death protein Bax reduces stroke volume. Numerous studies have also focused on heat shock proteins, which constitute a highly conserved, gene-based, response to stress. They act as chaperones and have anti-apoptotic and anti-inflammatory properties17, 18. Heat shock proteins are neuroprotective following both exogenous (viral vector gene transfer) and endogenous (transgenic) upregulation. Other highly conserved, widely distributed, gene-based systems with neuroprotective modulatory properties include inflammatory mediators and the toll-like receptor (TLR) system19. Finally, stem cell proliferation also appears to be neuroprotective in stroke20. Paracrine action of factors secreted from these cells is a suggested mechanism of action. Microarray analysis of gene expression in stem cells has identified subpopulations of bone marrow progenitor cells optimal for neuroprotection in stroke21.
Genomics
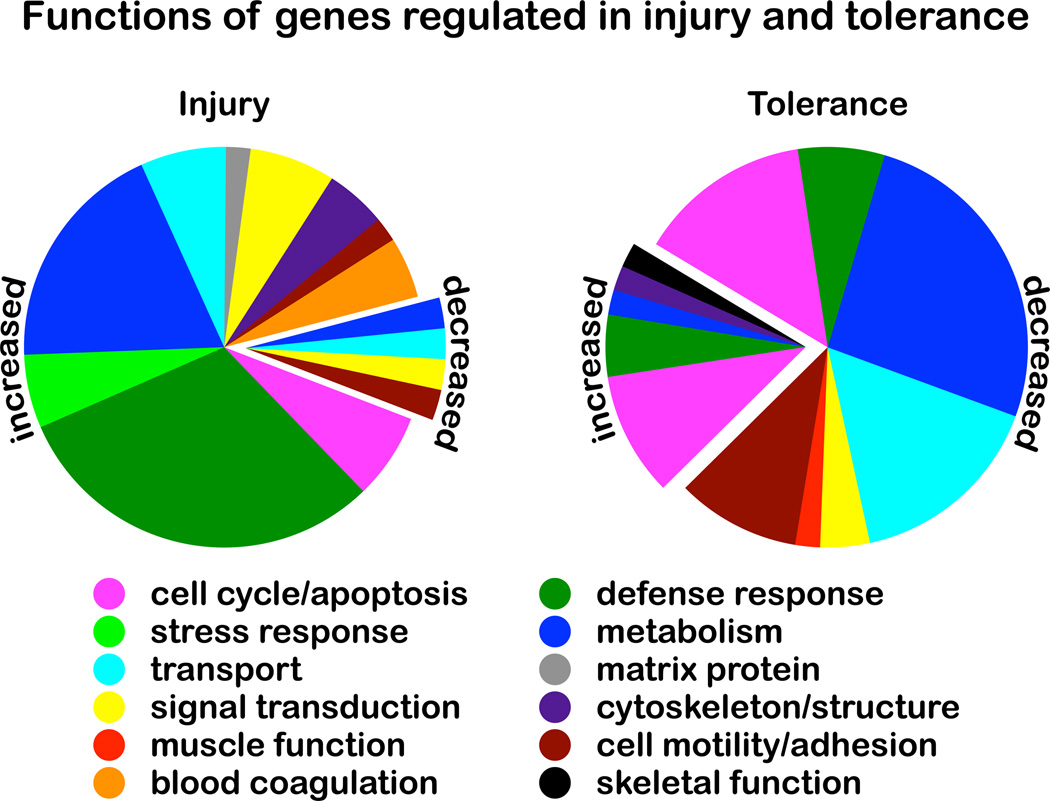
Efforts to identify the molecular effectors of ischemic injury and tolerance relied for many years on a one-gene-at-a-time approach. In the past decade, genomic studies have provided a more complete picture of gene regulation in stroke. The first application of unbiased mRNA screening to the study of endogenous neuroprotection was performed in a mouse model of ischemic tolerance22. The microarray analysis of ischemic tolerance proved a powerful tool for gene discovery and demonstrated that the genomic profile of protection can be determined. The analysis showed that scores to hundreds of genes are regulated after ischemic preconditioning, injury, and tolerance. Additionally, the broad view of gene regulation provided by the microarray analysis offered new insight into mechanisms of stroke neuroprotection. The genes regulated after preconditioning ischemia and the genes regulated after injurious ischemia are remarkably different. Moreover, genes regulated in ischemic tolerance are distinct from those regulated after preconditioning or injurious ischemia. Most notably, regulated genes were predominantly induced in injury but suppressed in tolerance (Figure 1)22.
Figure 1.
Genes whose expression is uniquely increased/decreased in ischemic injury (left) or uniquely increased/decreased in ischemic tolerance (right) are categorized by biological function. In injury (stroke), a majority of regulated genes is expressed at higher levels. By contrast, in tolerance (protection), a majority of regulated genes is expressed at lower levels. Based on data from Stenzel-Poore et al, 2003.
The results from microarray studies of ischemic tolerance underpinned a novel hypothesis about the neuroprotective mechanism: it was proposed that preconditioning reprograms the brain’s response to ischemic challenge, altering the transcriptional response from one that leads to cell death to one that produces a neuroprotected phenotype22. Gene suppresion was hypothesized to be a central neuroprotective feature of tolerance. Among suppressed genes, those that encode ion channels, transporters, and metabolic pathways are particularly affected, reminiscent of changes that allow hibernating animals to survive periods of prolonged oxygen and glucose deprivation23, 24.
The hypothesis that the brain's response to injury can be reprogrammed is supported by genomic profiling in epileptic tolerance. As in ischemic tolerance, a brief noninjurious seizure preconditions the brain so that it is tolerant to subsequent challenge by a prolonged, injurious seizure. Transcriptional changes that occur in the susceptible hippocampal CA3 subfield in mouse have been profiled after seizure preconditioning, epileptic challenge, and epileptic tolerance 25, 26. Similar to ischemic tolerance, preconditioning seizures25, 27, 28 produce a different pattern of gene expression than do injurious seizures29–31. Further, the response to injury is reprogrammed: the set of genes regulated after prolonged seizures differed from the set of genes regulated after prolonged seizure that was precedeced by a preconditioning seizure to produce epileptic tolerance. Consistant with ischemic tolerance, the majority of genes differentially regulated in the tolerant brain were suppressed26. In contrast to ischemic tolerance, genes suppressed in epileptic tolerance encode proteins that participate in calcium signaling and excitatory neurotransmission26. Therefore, the genomic signature of neuroprotection seems to be specific to the stress, as has been previously suggested32. Seizure-preconditioning specifically promotes an anti-excitotoxic phenotype—a phenotype apposite to the inducing stimulus—just as lipopolysaccharide (LPS) preconditioning produces an anti-inflammatory phenotype and preconditioning ischemia produces a hypo-metabolic phenotype32. Of note, these phenotypes are appropriate to the nature of the preconditioning stimulus and not necessarily to the nature of the challenging stimulus.
As an endogenous neuroprotective mechanism, tolerance can be understood as a first insult priming the brain to respond advantageously in the likelihood of a second insult of the same kind. Yet the preconditioned brain appears also to respond advantageously to a second insult of a different kind33, 34. The basis for this is not yet clear. Microarray analyses have made it apparent that the response to any brain challenge is complex, engaging numerous and diverse pathways. There may also be shared neuroprotective mechanisms not detectable at the transcriptional (mRNA) level. Uncovering those mechanisms will advance our understanding of endogenous neuroprotection and facilitate therapeutic intervention.
Transcription factors
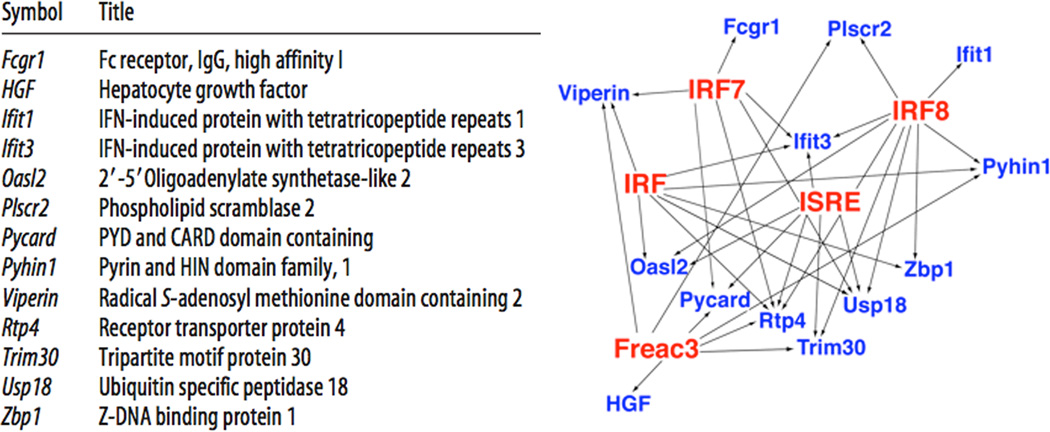
Transcription factors transduce intracellular signaling cascades into genomic and proteomic responses following preconditioning. To identify transcription factors that may coordinate the reprogrammed neuroprotective response, genomic studies compared gene regulation in ischemic tolerance induced by three different preconditioning agents: lipopolysaccharide (LPS, a TLR4 ligand), unmethylated CpGs (a TLR9 ligand), and brief ischemia. The studies identified 13 genes regulated in all three models of ischemic tolerance (Figure 2, left). Bioinformatic analysis of these genes' promoter regions identified transciption response elements for interferon regulatory factors (IRF) (Figure 2, right), suggesting that a set of genes is commonly and coordinately regulated in the neurorprotective response to ischemia. Consistently, preconditioning-induced tolerance to ischemia was abrogated in mice deficient in IRF3 and IRF735. Promoter analysis of genes regulated in all three different models of tolerance supports the view that different preconditioning stimuli activate common neuroprotective mechanisms.
Figure 2.
Analysis of genes regulated in stroke preceded by preconditioning with ischemia, lipopolysaccharide (LPS), or CpGs. Left: Thirteen genes were regulated in all three conditions. Right: Promoter analysis identified five over-represented transcription response elements (TREs; red) that putatively regulate expression of the thirteen genes (blue). Four of the TREs (IRF, IRF7, IRF8, ISRE) associate with interferon signaling. Modified from Stevens et al, 2011 with permission.
microRNAs
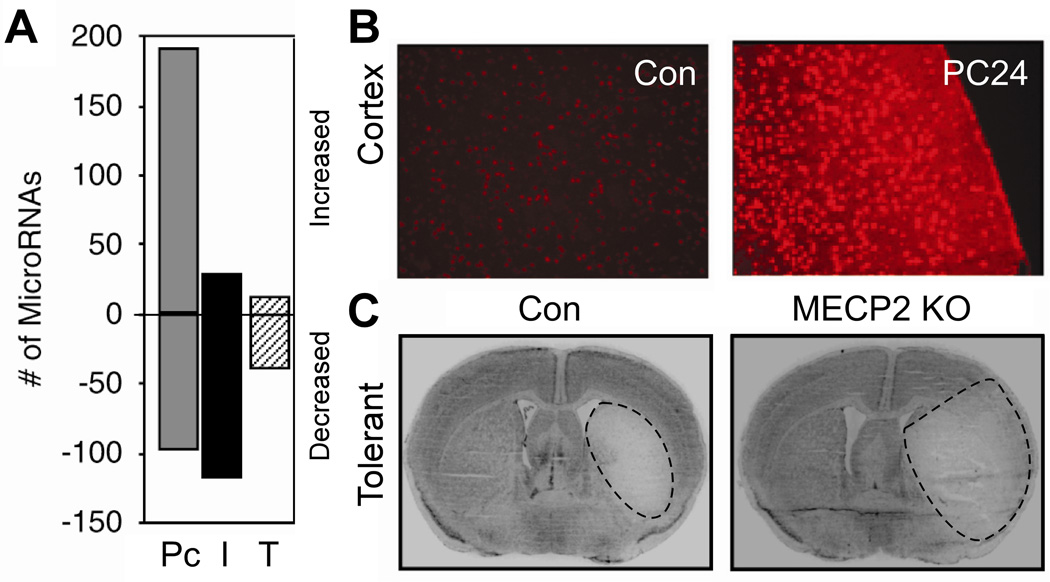
While extrapolation from microarray profiles of tolerant brain has identified a few upstream transcription factors and signaling pathways34 that may coordinate the genomic response in tolerance, the mechanisms at work during the development of the neuroprotected state, between preconditioning and challenge, have not been defined. How does the preconditioning stimulus prepare the brain to respond to subsequent stress in a new, reprogrammed, and protetective manner? Recent work suggests that microRNAs (miRNAs) may play a pivotal role in the molecular response to the preconditioning stimulus36. MiRNAs are short (~22 nucleotide) endogenous, non-coding sequences of RNA that post-transcriptionally regulate gene expression in plants, animals and viruses37–40. Through complimentary base-pairing interactions, miRNAs recognize target mRNA transcripts and direct their incorporation into the RNA-induced silencing complex (RISC), leading to a decrease in protein expression, either through translational repression or mRNA decay (for review 41–43). Recently, miRNA expression was profiled in ischemic preconditioning, ischemic injury, and ischemic tolerance36. Marked regulation of miRNA expression was observed in preconditioned brains, whereas little was observed in ischemic or tolerant brain (Figure 3A)36. Among the most prominent targets of the miRNAs regulated in preconditioned brain was methyl CpG binding protein 2 (MeCP2), a global regulator of gene transcription. Consistent with decreased expression of miRNAs targeting MeCP2, MeCP2 protein was increased in preconditioned brain (Figure 3B). Loss of MeCP2 precluded the induction of tolerance (Figure 3C), thus confirming the importance of this miRNA-mediated pathway36. Accordingly, miRNA profiling of ischemic brain supports the proposition that the preconditioning stimulus regulates miRNAs that target transcription factors and thereby lead to differential gene expression which characterizes tolerant brain. In the case of MeCP2, the effect is to repress transcription.
Figure 3.
MiRNAs in ischemic tolerance. (A) MiRNA screening reveals that the predominant response of miRNAs after preconditioning (P) is upregulation, versus downregulation in ischemia (I) and tolerance (T). (B) MeCP2 (methyl-CpG binding protein) immunostaining shows induction in the cortex 8 h after preconditioning, consistent with decreased expression of MeCP2 miRNA. (C) Infarct is larger in tolerized MeCP2-null mice than in tolerized wildtype (WT) mice. Adapted, with permission, from Lusardi et al, 2010.
Proteomics
Genomic studies of neuroprotection have shown that the responses to ischemia are complex and wide-ranging. Understanding of the molecular mechanisms involved therefore continues to evolve. At the genomic level, the induction of neuroprotection entails a repressive transcriptional response. Identifying the actuators of transcriptional suppression would move us closer to the goal of activating neuroprotection clinically. Recent proteomic studies of ischemic tolerance suggest that these actuators may be epigenetic regulatory proteins44.
The proteome of ischemic tolerance in rodent was characterized using an unbiased, quantitative proteomic approach, followed by biochemical and physiological studies44. The results fit nicely into the concept that the phenotype of neuroprotection is one of transcriptional suppression. Epigenetic silencers, including polycomb group (PcG) proteins and modified histones, are enriched during ischemic tolerance in the brain. Experimentally, changes in PcG protein levels are sufficient to induce or inhibit the tolerant state in neurons. Thus, knockdown of PcG proteins blocks the neuroprotective response, whereas over-expression induces the neuroprotective response. Additional evidence indicates that other epigenetic proteins may also be involved44. A similar biology occurs in ischemic tolerance in retina45.
Polycomb group (PcG) proteins and their antagonists, trithorax group (TrxG) proteins, although known to be present in brain46, had not previously been implicated in neuroprotection. We hypothesize that these proteins, along with their partners, are "master regulators" that switch the mammalian central nervous system neurons from a stress-sensitive (unprotected) to a stress-resistant (protected) state. In Drosophila, where they were originally identified as developmental regulators47, PcG and TrxG proteins alter gene expression by epigenetic means, maintaining over 1000 genes in an active or repressed state48, 49. Studies of PcG proteins in other systems show that they target a wide range of genes, including those regulated in ischemic tolerance, such as potassium channels, which are repressed during tolerance22, 48–51. Recent genome-wide screening of PcG targets in human embryonic fibroblasts identify cellular pathways that PcG proteins may regulate controlling development, differentiation, stem cell biology, and cell fate decisions 48. Notably most of the pathways described are known to be involved in the brain’s response to ischemia. Together, these data suggest that epigenetic regulation is a central mechanism for the induction of ischemic tolerance and that PcG proteins may be the key actuators. An endogenous neuroprotective mechanism mediated by PcG proteins explains many of the genomic and physiologic characteristics of ischemic tolerance in the brain. It is also perhaps a more general regulator of cell fate, as suggested by Suzanne Zukin52.
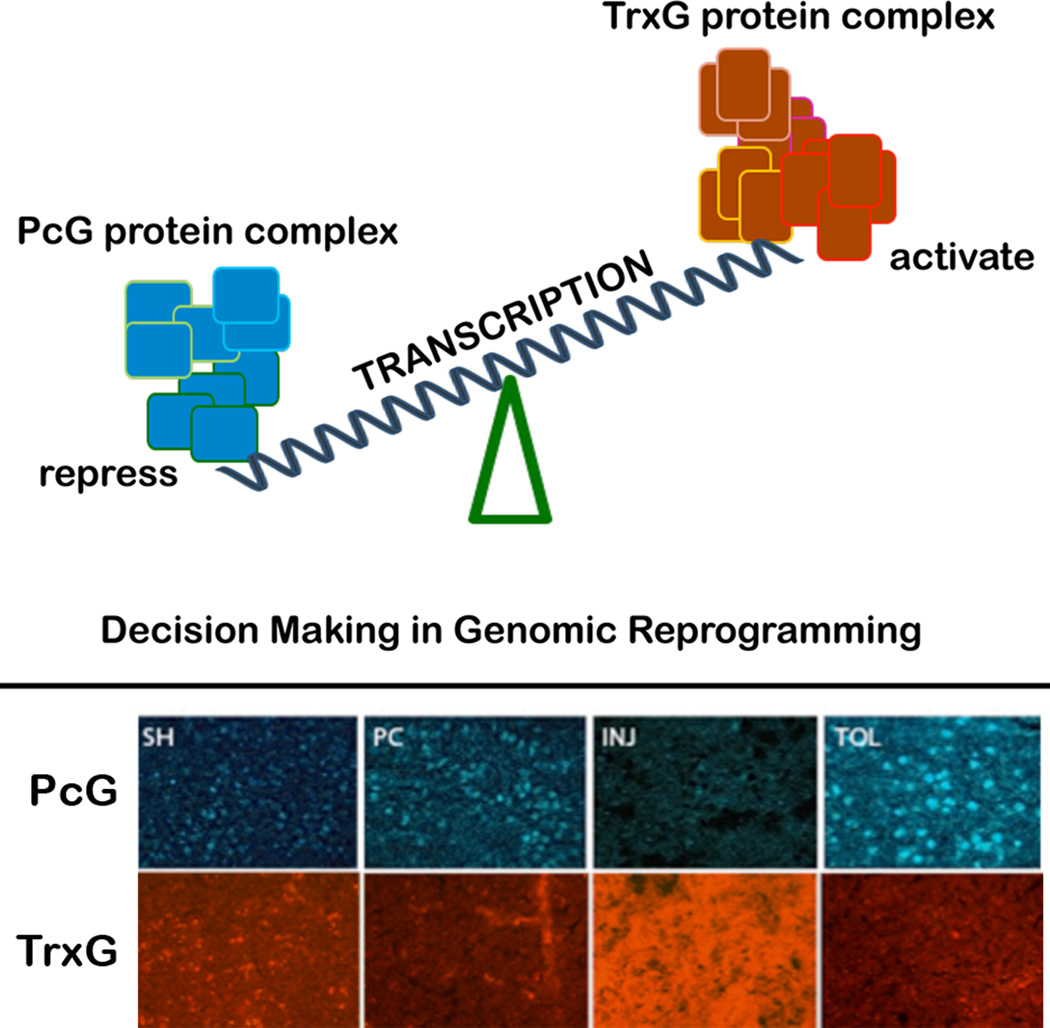
Epigenetic mechanisms control gene expression by remodeling the architecture of chromatin in ways that allow or deny transcription factors access to genes. PcG proteins interact with DNA at polycomb response elements (PRE) to silence the expression of genes, including those that encode electron transporters, endopeptidases, oxidoreductases, and G-protein coupled receptors51, 53, 54. For many genes, silencing by PcG proteins is countered by activation by trithorax group (TrxG) proteins. Figure 4 illustrates a simplified model of epigenetic regulation by PcG and TrxG proteins.
Figure 4.
Top: Proposed bivalent, epigenetic mechanism for regulating the brain's response to ischemia. Increased PcG protein abundance suppresses transcription. Increased TrxG protein abundance activates transcription. Bottom: In tolerance (TOL), PcG proteins increase. In injurious ischemia (INJ), TrxG proteins increase. The balance between PcG and TrxG activity may determine the response to ischemia. Transcriptional suppression and tolerance may be induced increasing PcG proteins, decreasing TrxG proteins, or a combination thereof.
PcG proteins assemble into at least three major complexes, each with a distinct role in epigenetic regulation, that work in concert with one another. The composition of these complexes is dynamic and influential in determining the outcome of biological processes, such as cell fate determination55, 56. Polycomb repressive complex 1 (PRC1), for example, mono-ubiquitinates histones H2A and H2B, whereas PRC2 methylates histone H3. In studies of ischemic tolerance, in rodent brain and neuronal cultures, three PcG proteins - Scmh1, Bmi1, and Ring1B – are robustly upregulated. SCMH1 acts as a link between PRC1 and other proteins57. Bmi1 facilitates monoubiquitination of histone 2A58. Ring1B functions as a ligase in H2a ubiquitination 58. Other proteins that comprise epigenetic regulatory complexes may also be differentially regulated during tolerance. The TrxG protein, ASH1L, is in fact downregulated in ischemic tolerance consistent with the counteracting roles of TrxG and PcG proteins.
Thus, the emerging picture is that PcG and TrxG proteins undergo dynamic regulation during the induction of ischemic tolerance in the brain. This changes the composition of PcG and TrxG complexes, alters the ratio of silencing to activating complexes, and ultimately modulates the expression of target genes. PcG protein levels increase during tolerance, very early after ischemia, suggesting that this response initiates the neuroprotective cascade. The upregulation of PcG proteins during tolerance occurs, at least in part, via a transcription-independent, translational mechanism. Emerging evidence indicates that microRNAs regulate PcG protein expression59, 60.
Conclusion
The discovery that the brain's response to injury can be governed by epigenetic modulation of gene expression offers new insight into mechanisms of brain injury and protection. Specifically, the discovery that PcG /TrxG proteins are active in brain ischemia reveals that an evolutionarily conserved mechanism is active during ischemic stress. This mechanism maintains chromatin in an on or off state, promotes or suppresses gene transcription, and thereby affects cell fate.
Studies of oncogenesis already implicate the PcG/TrxG system as a potent cell fate regulatory system in humans 61. For example, the polycomb group protein EZH2, encoded by the EZH2 gene on chromosome 7q36, functions as a gene repressor. Altered expression of EZH2 occurs in a number of malignances including prostate cancer, where EZH2 knockdown inhibits proliferation of prostate cancer cells 62. In light of this, PcG proteins may be viewed as a prolife signal: turned on transiently, as in tolerance, it promotes cell survival; turned on continuously, as in cancer, it ultimately causes malignancy.
Epigenetic proteins, whose role in neuroprotection was previously unknown, may be “master regulators” of a neuroprotective state in the mammalian brain. Based on our findings that 1) the genomic signature of ischemic tolerance is transcriptional suppression and 2) the proteomic signature of ischemic tolerance is enrichment of epigenetic gene silencers, we submit that widespread changes in gene expression, coordinately modulated by epigenetic regulatory proteins, can modify stroke outcome. In this “omic” view, therapeutic approaches that target a discrete pathway appear of limited value. Thus, in hindsight it is not surprising that clinical trials based on this approach have failed. By contrast, a therapeutic approach that targets the PcG/TrxG system could initiate a comprehensive neuroprotective response in the brain.
Acknowledgments
Sources of funding:
The authors work reviewed here was supported by the National Institute of Neurologic Disorders and Stroke: P01 NS035965(RPS); R01NS059588 (RM); R01NS046560 (AZ); R01NS039016 (DH); and NIH/NCRR/RCMI Grant G12-RR03034 to Morehouse School of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: none
Contributor Information
Roger Simon, Morehouse School of Medicine, rsimon@msm.edu.
Robert Meller, Morehouse School of Medicine, rmeller@msm.edu.
An Zhou, Morehouse School of Medicine, azhou@msm.edu.
David Henshall, Royal College of Surgeons in Ireland, davhenshall@rcsi.ie.
References
- 1.O'Collins VE, Macleod MR, Donnan GA, Horky LL, van der Worp BH, Howells DW. 1,026 experimental treatments in acute stroke. Annals of neurology. 2006;59:467–477. doi: 10.1002/ana.20741. [DOI] [PubMed] [Google Scholar]
- 2.Sandu N, Cornelius J, Filis A, Arasho B, Perez-Pinzon M, Schaller B. Ischemic tolerance in stroke treatment. Expert Rev Cardiovasc Ther. 2009;7:1255–1261. doi: 10.1586/erc.09.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gidday JM. Cerebral preconditioning and ischaemic tolerance. Nat Rev Neurosci. 2006;7:437–448. doi: 10.1038/nrn1927. [DOI] [PubMed] [Google Scholar]
- 4.Dirnagl U, Becker K, Meisel A. Preconditioning and tolerance against cerebral ischaemia: From experimental strategies to clinical use. Lancet Neurol. 2009;8:398–412. doi: 10.1016/S1474-4422(09)70054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durukan A, Tatlisumak T. Preconditioning-induced ischemic tolerance: A window into endogenous gearing for cerebroprotection. Exp Transl Stroke Med. 2010;2:2. doi: 10.1186/2040-7378-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hickey EJ, You X, Kaimaktchiev V, Stenzel-Poore M, Ungerleider RM. Lipopolysaccharide preconditioning induces robust protection against brain injury resulting from deep hypothermic circulatory arrest. J Thorac Cardiovasc Surg. 2007;133:1588–1596. doi: 10.1016/j.jtcvs.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 7.Bahjat FR, Williams-Karnesky RL, Kohama SG, West GA, Doyle KP, Spector MD, et al. Proof of concept: Pharmacological preconditioning with a toll-like receptor agonist protects against cerebrovascular injury in a primate model of stroke. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31:1229–1242. doi: 10.1038/jcbfm.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaller B. Ischemic preconditioning as induction of ischemic tolerance after transient ischemic attacks in human brain: Its clinical relevance. Neurosci Lett. 2005;377:206–211. doi: 10.1016/j.neulet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Weih M, Kallenberg K, Bergk A, Dirnagl U, Harms L, Wernecke KD, et al. Attenuated stroke severity after prodromal tia: A role for ischemic tolerance in the brain? Stroke; a journal of cerebral circulation. 1999;30:1851–1854. doi: 10.1161/01.str.30.9.1851. [DOI] [PubMed] [Google Scholar]
- 10.Wegener S, Gottschalk B, Jovanovic V, Knab R, Fiebach JB, Schellinger PD, et al. Transient ischemic attacks before ischemic stroke: Preconditioning the human brain? A multicenter magnetic resonance imaging study. Stroke; a journal of cerebral circulation. 2004;35:616–621. doi: 10.1161/01.STR.0000115767.17923.6A. [DOI] [PubMed] [Google Scholar]
- 11.Moncayo J, de Freitas GR, Bogousslavsky J, Altieri M, van Melle G. Do transient ischemic attacks have a neuroprotective effect? Neurology. 2000;54:2089–2094. doi: 10.1212/wnl.54.11.2089. [DOI] [PubMed] [Google Scholar]
- 12.Keep RF, Wang MM, Xiang J, Hua Y, Xi G. Is there a place for cerebral preconditioning in the clinic? Transl Stroke Res. 2010;1:4–18. doi: 10.1007/s12975-009-0007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broughton BR, Reutens DC, Sobey CG. Apoptotic mechanisms after cerebral ischemia. Stroke; a journal of cerebral circulation. 2009;40:e331–e339. doi: 10.1161/STROKEAHA.108.531632. [DOI] [PubMed] [Google Scholar]
- 14.Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, et al. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. Embo J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Liang W, Hu X, Zhang W, Stetler RA, Vosler P, et al. Neuroprotection against hypoxic-ischemic brain injury by inhibiting the apoptotic protease activating factor-1 pathway. Stroke; a journal of cerebral circulation. 2010;41:166–172. doi: 10.1161/STROKEAHA.109.561852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobaben S, Grohm J, Seiler A, Conrad M, Plesnila N, Culmsee C. Bid-mediated mitochondrial damage is a key mechanism in glutamate-induced oxidative stress and aif-dependent cell death in immortalized ht-22 hippocampal neurons. Cell Death Differ. 2011;18:282–292. doi: 10.1038/cdd.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan X, Ander BP, Liao IH, Hansen JE, Kim C, Clements D, et al. Recombinant fv-hsp70 protein mediates neuroprotection after focal cerebral ischemia in rats. Stroke; a journal of cerebral circulation. 2010;41:538–543. doi: 10.1161/STROKEAHA.109.572537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuchiya D, Hong S, Matsumori Y, Kayama T, Swanson RA, Dillman WH, et al. Overexpression of rat heat shock protein 70 reduces neuronal injury after transient focal ischemia, transient global ischemia, or kainic acid-induced seizures. Neurosurgery. 2003;53:1179–1187. doi: 10.1227/01.neu.0000090341.38659.cf. discussion 1187–1178. [DOI] [PubMed] [Google Scholar]
- 19.Marsh BJ, Stevens SL, Hunter B, Stenzel-Poore MP. Inflammation and the emerging role of the toll-like receptor system in acute brain ischemia. Stroke; a journal of cerebral circulation. 2009;40:S34–S37. doi: 10.1161/STROKEAHA.108.534917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maysami S, Lan JQ, Minami M, Simon RP. Proliferating progenitor cells: A required cellular element for induction of ischemic tolerance in the brain. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2008;28:1104–1113. doi: 10.1038/jcbfm.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakondi B, Shimada IS, Perry A, Munoz JR, Ylostalo J, Howard AB, et al. Cd133 identifies a human bone marrow stem/progenitor cell sub-population with a repertoire of secreted factors that protect against stroke. Mol Ther. 2009;17:1938–1947. doi: 10.1038/mt.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, et al. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: Similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 23.Storey KB. Biochemistry of natural freeze tolerance in animals: Molecular adaptations and applications to cryopreservation. Biochem Cell Biol. 1990;68:687–698. doi: 10.1139/o90-100. [DOI] [PubMed] [Google Scholar]
- 24.Hochachka PW, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: Molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatazaki S, Bellver-Estelles C, Jimenez-Mateos EM, Meller R, Bonner C, Murphy N, et al. Microarray profile of seizure damage-refractory hippocampal ca3 in a mouse model of epileptic preconditioning. Neuroscience. 2007;150:467–477. doi: 10.1016/j.neuroscience.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez-Mateos EM, Hatazaki S, Johnson MB, Bellver-Estelles C, Mouri G, Bonner C, et al. Hippocampal transcriptome after status epilepticus in mice rendered seizure damage-tolerant by epileptic preconditioning features suppressed calcium and neuronal excitability pathways. Neurobiology of disease. 2008;32:442–453. doi: 10.1016/j.nbd.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Ploski JE, Newton SS, Duman RS. Electroconvulsive seizure-induced gene expression profile of the hippocampus dentate gyrus granule cell layer. J Neurochem. 2006;99:1122–1132. doi: 10.1111/j.1471-4159.2006.04156.x. [DOI] [PubMed] [Google Scholar]
- 28.Borges K, Shaw R, Dingledine R. Gene expression changes after seizure preconditioning in the three major hippocampal cell layers. Neurobiology of disease. 2007;26:66–77. doi: 10.1016/j.nbd.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunsberger JG, Bennett AH, Selvanayagam E, Duman RS, Newton SS. Gene profiling the response to kainic acid induced seizures. Brain Res Mol Brain Res. 2005;141:95–112. doi: 10.1016/j.molbrainres.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Gorter JA, van Vliet EA, Aronica E, Breit T, Rauwerda H, Lopes da Silva FH, et al. Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:11083–11110. doi: 10.1523/JNEUROSCI.2766-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukasiuk K, Kontula L, Pitkanen A. Cdna profiling of epileptogenesis in the rat brain. Eur J Neurosci. 2003;17:271–279. doi: 10.1046/j.1460-9568.2003.02461.x. [DOI] [PubMed] [Google Scholar]
- 32.Stenzel-Poore MP, Stevens SL, King JS, Simon RP. Preconditioning reprograms the response to ischemic injury and primes the emergence of unique endogenous neuroprotective phenotypes: A speculative synthesis. Stroke. 2007;38:680–685. doi: 10.1161/01.STR.0000251444.56487.4c. [DOI] [PubMed] [Google Scholar]
- 33.Plamondon H, Blondeau N, Heurteaux C, Lazdunski M. Mutually protective actions of kainic acid epileptic preconditioning and sublethal global ischemia on hippocampal neuronal death: Involvement of adenosine a1 receptors and k(atp) channels. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1999;19:1296–1308. doi: 10.1097/00004647-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Marsh B, Stevens SL, Packard AE, Gopalan B, Hunter B, Leung PY, et al. Systemic lipopolysaccharide protects the brain from ischemic injury by reprogramming the response of the brain to stroke: A critical role for irf3. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:9839–9849. doi: 10.1523/JNEUROSCI.2496-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens SL, Leung PY, Vartanian KB, Gopalan B, Yang T, Simon RP, et al. Multiple preconditioning paradigms converge on interferon regulatory factor-dependent signaling to promote tolerance to ischemic brain injury. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:8456–8463. doi: 10.1523/JNEUROSCI.0821-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, et al. Ischemic preconditioning regulates expression of micrornas and a predicted target, mecp2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ambros V. The functions of animal micrornas. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 38.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by mirnas: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Shyu AB, Wilkinson MF, van Hoof A. Messenger rna regulation: To translate or to degrade. Embo J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kai ZS, Pasquinelli AE. Microrna assassins: Factors that regulate the disappearance of mirnas. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saugstad JA. Micrornas as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2010;30:1564–1576. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stapels M, Piper C, Yang T, Li M, Stowell C, Xiong ZG, et al. Polycomb group proteins as epigenetic mediators of neuroprotection in ischemic tolerance. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000502. ra15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stowell C, Wang L, Arbogast B, Lan JQ, Cioffi GA, Burgoyne CF, et al. Retinal proteomic changes under different ischemic conditions - implication of an epigenetic regulatory mechanism. Int J Physiol Pathophysiol Pharmacol. 2010;2:148–160. [PMC free article] [PubMed] [Google Scholar]
- 46.Vogel T, Stoykova A, Gruss P. Differential expression of polycomb repression complex 1 (prc1) members in the developing mouse brain reveals multiple complexes. Dev Dyn. 2006;235:2574–2585. doi: 10.1002/dvdy.20876. [DOI] [PubMed] [Google Scholar]
- 47.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: The tale of polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 48.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohler C, Villar CB. Programming of gene expression by polycomb group proteins. Trends Cell Biol. 2008;18:236–243. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Otte AP, Kwaks TH. Gene repression by polycomb group protein complexes: A distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 51.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 52.Zukin RS. Eradicating the mediators of neuronal death with a fine-tooth comb. Sci Signal. 2010;3 doi: 10.1126/scisignal.3125pe20. pe20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, et al. Genome-wide profiling of prc1 and prc2 polycomb chromatin binding in drosophila melanogaster. Nature genetics. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 55.Kerppola TK. Polycomb group complexes--many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 57.Takada Y, Isono K, Shinga J, Turner JM, Kitamura H, Ohara O, et al. Mammalian polycomb scmh1 mediates exclusion of polycomb complexes from the xy body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- 58.Cao R, Tsukada Y, Zhang Y. Role of bmi-1 and ring1a in h2a ubiquitylation and hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Friedman JM, Jones PA, Liang G. The tumor suppressor microrna-101 becomes an epigenetic player by targeting the polycomb group protein ezh2 in cancer. Cell Cycle. 2009;8:2313–2314. doi: 10.4161/cc.8.15.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. Microrna-directed transcriptional gene silencing in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mills AA. Throwing the cancer switch: Reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoon KA, Gil HJ, Han J, Park J, Lee JS. Genetic polymorphisms in the polycomb group gene ezh2 and the risk of lung cancer. J Thorac Oncol. 2010;5:10–16. doi: 10.1097/JTO.0b013e3181c422d9. [DOI] [PubMed] [Google Scholar]