Abstract
Post-transcriptional events which regulate mRNA biogenesis are fundamental to the control of gene expression. A nascent mRNA is therefore steered through multimeric RNA–protein complexes that mediate its capping, splicing, polyadenylation, nuclear export, and ultimately its translation. Kaposi’s sarcoma-associated herpesvirus (KSHV) mRNA transport and accumulation protein, or ORF57, is a functionally conserved protein found in all herpesviruses which plays a pivotal role in enhancing viral gene expression at a post-transcriptional level. As such, ORF57 has been implicated in multiple steps of RNA biogenesis, including augmenting viral splicing, protecting viral RNAs from degradation to enhancing viral mRNA nuclear export and translation. In this review, we highlight the multiple roles of KSHV ORF57 in regulating the post-transcriptional events which are fundamental to the control of virus gene expression.
Keywords: KSHV, ORF57, mRNA export, mRNA stability
Kaposi’s Sarcoma-Associated Herpesvirus
Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV) or human herpesvirus 8 is a gamma-2 herpesvirus associated with multiple AIDS-related malignancies (Chang et al., 1994), including KS, a highly vascular tumor of endothelial lymphatic origin (Ganem, 2006), and two lymphoproliferative disorders; primary effusion lymphoma and multicentric Castleman’s disease (Chang et al., 1994; Cesarman et al., 1995; Soulier et al., 1995). In sub-Saharan Africa, widespread HIV infection has turned KS into an epidemic disease and KS is now amongst the most common of all diagnosed malignancies (Parkin et al., 2008). Moreover, solid-organ transplant recipients are at an increased risk of developing KS due to a pre-existing infection or recipients being infected by donors (Piselli et al., 2009).
Like other herpesviruses, KSHV has two distinct forms of infection, latency, and lytic replication (Ganem, 2006). However, in contrast to other oncogenic herpesviruses, where latent gene expression plays a prominent role in tumorigenesis, lytic replication plays an important part in the tumorigenicity, pathogenesis, and spread of KSHV infection (Cai et al., 2010). Specifically, lytic replication appears to be a necessary antecedent step in KS development from the primary target of viral infection, the B lymphocyte reservoir, to endothelial cells where tumors are observed. Moreover, lytic gene expression potentially contributes to the development of KS through the expression of lytic viral proteins which mediate paracrine secretion of growth and angiogenic factors that are essential for tumor growth and development (Ballon et al., 2011; Bottero et al., 2011). In addition, they sustain the population of latently infected cells that would otherwise be reduced due to the poor persistence of the KSHV episome during spindle cell division (Grundhoff and Ganem, 2004). Therefore, it is essential to study the molecular mechanisms which regulate lytic replication to fully understand KSHV pathogenesis. This in turn may lead to novel therapeutic interventions which could become an important strategy for the treatment of KSHV-associated diseases.
Post-transcriptional events which regulate mRNA biogenesis are fundamental to the control of gene expression. As a consequence, cells have evolved a “gene expression production line” that encompasses the routing of a nascent transcript through multimeric mRNA–protein complexes that mediate its splicing, polyadenylation, nuclear export, and translation (Hastings and Krainer, 2001; Proudfoot, 2011; Rodríguez-Navarro and Hurt, 2011). These pathways are particularly important for herpesviruses which replicate in the host cell nucleus and express numerous lytic intronless mRNAs. Due to the reliance of herpesviruses on the host cell machinery for efficient processing of their mRNAs, an immediate issue arises concerning the mechanism by which the viral intronless mRNAs are efficiently exported from the nucleus, given that the majority of cellular bulk mRNA nuclear export is intimately linked, and dependent upon, splicing (Luo and Reed, 1999; Valencia et al., 2008).
To circumvent the problem associated with efficient intronless viral mRNA nuclear export KSHV encodes a multifunctional protein, the mRNA transport and accumulation (MTA) protein, or ORF57, which functions in many aspects of RNA processing (Malik et al., 2004b). Many of these properties are also conserved in ORF57 homologs throughout herpesviruses including HSV-1 ICP27; HSV-2 UL54; EBV SM/EB2; CMV UL69, VZV ORF4, HHV-6 U42, HHV-7 U42, HVS ORF57 (Swaminathan, 2005; Boyne and Whitehouse, 2006a; Boyne et al., 2008a; Sandri-Goldin, 2008; Toth and Stamminger, 2008; Ote et al., 2009). The most widely characterized of these is the herpes simplex (HSV-1) homolog ICP27, which has been used as a prototype for herpes virus infection (Sandri-Goldin, 2008). Functionally, ORF57 and its homologs regulate viral and cellular RNA processing, which results in accumulation of target genes and may also contribute to host cell shut-off (Hardwicke and Sandri-Goldin, 1994; Hardy and Sandri-Goldin, 1994; Ruvolo et al., 1998; Whitehouse et al., 1998). However, ORF57 homologs are evolutionarily diverged, with little sequence conservation and have adapted into different roles in the respective viruses. Therefore this review focuses on KSHV ORF57 and its role in the lytic replication cycle and mRNA processing.
The KSHV ORF57 Protein
ORF57 gene and protein structure
Kaposi’s sarcoma-associated herpesvirus lytic gene expression is initiated by the immediate early protein the replication and transcription activator, RTA, which is necessary and sufficient for complete reactivation into the lytic cascade (Lukac et al., 1998, 1999). RTA functions as a viral transcription factor to initiate transcription of various viral genes by multiple mechanisms (Dourmishev et al., 2003). Interestingly, although RTA has been shown to be able to recognize specific RTA response elements in promoters, it activates the ORF57 promoter indirectly via an interaction with RBP-Jκ (Liang et al., 2002; Chang et al., 2005). Additional cellular cofactors including HMGB1 are also thought to be necessary for efficient RTA-mediated activation of the ORF57 promoter (Song et al., 2004; Harrison and Whitehouse, 2008).
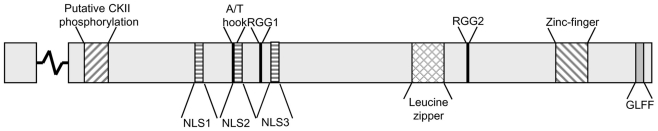
ORF57 is transcribed as a monocistronic pre-mRNA, containing a small intron comprising 109 nucleotides. It is interesting to note that both RTA and ORF57, as immediate early proteins contain introns enabling efficient mRNA processing prior to ORF57 action on numerous delayed early and late intronless transcripts. The ORF57 gene encodes a 455 aa protein, with only the first 16 aa translated from the first exon. Currently there are no solved structures for KSHV ORF57 or any of its homologs, but computer predictions indicate that the N-terminal region is mainly unstructured, whereas the C-terminal domain forms primarily alpha helices (Taylor et al., 2011). Nevertheless, several functional regions have been identified (Figure 1).
Figure 1.
Putative motifs within the KSHV ORF57 protein. KSHV ORF57 is a 455 amino acid protein with an exon junction within the codon for aa 17. The functional motifs comprise a putative CKII phosphorylation site (region 18–43); three nuclear localization signals (NLS1,2,3; regions 101–107, 121–131, 143–152); an A/T hook (region 119–131); two arginine–glycine–glycine motifs (RGG1,2; regions 138–140, 372–374); a leucine zipper (region 343–364); a zinc finger domain (region 423–432); and a hydrophobic glycine–leucine–phenylalanine–phenylalanine motif (GLFF; region 448–451). The putative phosphorylation sites were predicted using GPS V2.1 (Xue et al., 2008).
Three nuclear localization signals (NLS) are located in the N-terminal region of ORF57. Each NLS consists of a stretch of basic residues and each individual NLS is sufficient to localize ORF57 into the nucleus. Mutation of any two of these NLS is sufficient to severely inhibit ORF57 function (Majerciak et al., 2006). Furthermore, it has been shown that these NLS also confer nucleolar localization (Figure 2). Nucleolar trafficking is thought to be essential for KSHV ORF57 function, as well as the related HVS ORF57 (Boyne and Whitehouse, 2006b, 2009), however as yet the role of the nucleolus in ORF57 function is yet to be determined.
Figure 2.
Cellular localization of KSHV ORF57. BCBL-1 cells were reactivated with TPA to express the lytic KSHV genes. Cells were fixed and permeabilized and proteins immunostained using an ORF57-specific antibody and a monoclonal antibody to C23 and FITC and Texas Red secondary antibodies. ORF57 can be seen localizing in the nucleus, as well as the nuclear speckles and colocalizing in the nucleolus with C23.
ORF57 binds to both viral and cellular target RNAs, for example cellular IL-6 and viral vIL-6 have been shown to interact with ORF57 using crosslinking immunoprecipitation (CLIP) assays (Kang et al., 2011). Two RGG motifs, which are putative RNA binding sites are found in ORF57. However, deletion of the first RGG motif has no effect on ORF57 RNA binding, while deletion of the second C-terminal RGG motif results in a lack of RNA binding and consequently inefficient nuclear export of a viral target RNA. Although, this could be due to a deficiency in RNA binding or binding of cellular interacting proteins (Nekorchuk et al., 2007). However, more recent evidence using site-directed mutants of the RGG motifs suggests to defects in homodimerization and intranuclear trafficking of the RGG2 mutant (Taylor et al., 2011). Interestingly, the N-terminal domain is also thought to be sufficient to bind RNA (Majerciak et al., 2006). A similar observation has also been seen in the highly conserved Herpesvirus saimiri ORF57 protein (Goodwin et al., 1999). Further work is necessary to fully elucidate the ORF57 RNA binding domain or indeed domains.
ORF57 also contains an uncharacterized AT-Hook motif. AT-Hook motifs are generally found in DNA binding proteins and are also found in a series of transcriptional cofactors (Aravind and Landsman, 1998). ORF57 has been shown to bind directly to DNA, with the AT-Hook domain implicated in this interaction (Palmeri et al., 2007). EMSA-based analysis shows a deletion of the AT-Hook region lead to a loss of ORF57 activation of the polyadenylated nuclear RNA (PAN) promoter and diminished ability of ORF57 to bind DNA. It is intriguing to note that ORF57 has been described to act as a cofactor for RTA-mediated transactivation (Kirshner et al., 2000), which will be discussed in more detail later. Additionally, a putative leucine zipper domain is located in the C-terminal domain of ORF57, however the function of this region is still unclear. Although, a leucine rich region in the ORF57 homolog ICP27 from HSV-1, has been shown to contain a functional nuclear export signal (Sandri-Goldin, 1998). Together with the NLS, this motif might therefore be responsible for the ability of ORF57 to shuttle between nucleus and cytoplasm (Bello et al., 1999).
Finally, the C-terminal domain of ORF57 also contains a zinc finger domain and a GLFF motif. The zinc finger domain is conserved throughout ORF57 homologs whereas the GLFF motif is restricted to gamma-herpesviruses (Goodwin et al., 2000). The functional relevance of the zinc finger domain of KSHV ORF57 is not fully characterized, however work on HSV-1 ICP27 highlighted a potential role in dimerization and binding of the nuclear export factor TAP (Hernandez and Sandri-Goldin, 2010, 2011). The GLFF motif plays a role in transactivation as well as repression properties of the HVS ORF57 protein and SM protein in EBV, however, again no role has been shown in KSHV to date (Goodwin et al., 2000; Ruvolo et al., 2004).
Caspase-7 Cleavage of ORF57
An interesting recent study presented a hypothesis for a possible cellular mechanism against lytic KSHV virus infection (Majerciak et al., 2010). The authors showed that infection with KSHV or reactivation into the lytic cycle induces the caspase-8 mediated apoptosis pathway. This leads to the expression of caspase-7 which was shown to cleave ORF57 33 amino acids from the N-terminus. This cleaved form of ORF57 has been shown to be deficient in its functional roles of viral mRNA maturation and processing. Furthermore, cells expressing caspase-7 showed little lytic reactivation and underwent caspase-induced apoptosis, whereas cells expressing full-length ORF57 appeared to show diminished activity of caspase-7 and were able to enter into a full lytic cycle. The mechanisms underlying this delicate balancing act are yet to be defined, but understanding how caspase-7 is suppressed in a number of lytically active cells, and how caspase-8 is induced leading to apoptosis in the remainder of cells presents an interesting scenario for a potential antiviral mechanism against KSHV. It is, however, possible that this cleavage is simply a neutral event, or perhaps even beneficial to the virus. For example, KSHV could utilize this as a post-translational mechanism to control ORF57 activity. Alternatively, it maybe possible that the virus could utilize this cleavage event to prevent full lytic reactivation in the majority of cells maintaining the large latent pool of infected cells.
The Multifunctional Roles of ORF57 in mRNA Biogenesis
The role of ORF57 in splicing
The mechanism of pre-mRNA splicing and the function of ORF57 and its homologs is closely linked. For example, HSV-1 ICP27 effectively inhibits splicing to reduce host cell gene expression, thereby eliciting a host cell shut-off (Smith et al., 2005). This is mediated by an interaction between ICP27 and SAP145, an essential pre-mRNA splicing factor (Bryant et al., 2001). No such effect has been reported for KSHV ORF57 to date, as KSHV encodes a number of spliced transcripts. On the contrary, KSHV ORF57 is able to enhance splicing of a number of viral genes (Majerciak et al., 2008). This enhancement effect on splicing is not limited to KSHV genes as a similar enhancement has been observed on non-viral mRNA constructs. ORF57 was able to enhance splicing of reporter constructs which are normally only poorly spliced due to a large exon prior to the spliced intron. ORF57 forms a complex with the spliceosome, as demonstrated by association with the small spliceosomal RNAs (U1, U2, U4, U5, and U6), as well as the splicing factors SF2/ASF and U2AF (Majerciak et al., 2008). Interestingly, association of ORF57 with unspliced pre-mRNA could only be observed in the presence of nuclear extract and not purified ORF57, indicating that the association with pre-mRNA is indirect and ORF57 is acting as a modulator of normal splicing rather than directly recruiting splicing factors to the mRNA. A similar effect has since also been reported for the EBV homolog of ORF57, SM, which acts as an alternative splicing factor, influencing the choice of splice site (Verma and Swaminathan, 2008). To this end, SM has been reported to interact directly with SRp20 (Verma et al., 2010) however it remains to be determined whether the same is true for KSHV ORF57.
ORF57 functions in intronless viral mRNA export
Spicing of cellular mRNAs is intrinsically linked to a number of upstream and downstream processes, including transcription, 3′ end formation, mRNA export, RNA stability, and translation (Moore and Proudfoot, 2009). This link is due to the recruitment of numerous proteins to an mRNA during the splicing process. One such complex of proteins is the human transport/export (hTREX) complex which comprises multiple proteins including UAP56, Aly, the multi-protein THO complex, and the recently discovered Tex1 and CIP29 (Masuda et al., 2005; Dufu et al., 2010). hTREX is deposited onto the 5′ end of the mRNA in a splicing-dependent manner through an interaction between Aly and the cap-binding complex (Cheng et al., 2006). Aly then recruits the remainder of the hTREX complex before interacting with the nuclear export receptor protein, TAP. The handover from Aly to TAP is promoted by arginine methylation of Aly within its TAP and RNA binding domains (Hung et al., 2010). Methylation of these sites reduces the RNA binding affinity of Aly and allows TAP to efficiently displace Aly during the export process. TAP then interacts with nucleoporins to facilitate the transport of the mRNP through the nuclear pore (Kohler and Hurt, 2007). However, whilst understanding of the export of intron-containing genes has increased significantly over recent years, the mechanisms underlying intronless cellular mRNA export are poorly understood. This is of particular interest with regards to ORF57 as numerous KSHV mRNAs are intronless. However, a recent study has identified that some intronless cellular mRNAs can also be exported by hTREX and TAP, increasing our understanding of how intronless mRNAs are processed (Lei et al., 2011).
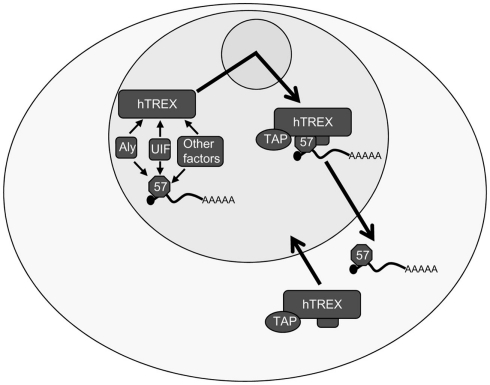
A pivotal function of ORF57 in viral mRNA metabolism is its role in nuclear export of viral mRNA. To efficiently export viral intronless mRNAs from the nucleus ORF57 is able to recruit the entire hTREX complex to intronless viral mRNAs (Boyne et al., 2008b; Tunnicliffe et al., 2010). ORF57 is able to bind directly to the RNA and recruit the export adapter, Aly to the 5′ end of the RNA, bypassing the requirement for splicing-dependent recruitment of Aly. Aly then recruits the remainder of the hTREX complex as well as TAP to form an export competent viral RNP which then allows efficient translocation through the nuclear pore complex. This function is also conserved in the HVS ORF57 homolog (Colgan et al., 2009).
However, a long-standing conundrum with this model has been the role of the exporter adapter Aly. Surprisingly, depletion studies in both mammalian and viral systems show that Aly is dispensable for mRNA export. The explanation for this appears to lie in an apparent redundancy in the eukaryotic hTREX components; specifically due to the discovery of a second export adapter protein, UAP56 interacting factor (UIF; Hautbergue et al., 2009). UIF binds to the mRNA in a splicing-independent reaction and is recruited to the mRNA via a direct interaction with the histone chaperone FACT. UIF can then interact with TAP to facilitate delivery of the mRNA to the nuclear pore. To this end, ORF57 is now known to interact directly with both Aly and UIF and to recruit the entire hTREX complex through these interactions (Jackson et al., 2011), as well as interacting with other cellular factors such as RBM15 and OTT3 (Majerciak et al., 2011; Figure 3). While our understanding of the proteins involved in the export of intronless KSHV mRNAs has improved, the exact mechanism remains elusive. For example, it has been demonstrated that ORF57 interacts directly with UIF and Aly, but it is not known whether these interactions occur simultaneously or independent of one another. siRNA depletion of both Aly and UIF is sufficient to severely impair ORF57-mediated mRNA export. However, depletion of Aly and UIF individually also causes a slight decrease in the level of mRNA export, perhaps suggesting that both proteins are required simultaneously for the efficient export KSHV mRNAs.
Figure 3.
Kaposi’s sarcoma-associated herpesvirus ORF57-mediated intronless viral mRNA nuclear export. ORF57 binds to intronless viral mRNA in the nucleus and interacts directly with the export adapters Aly and UIF. Via the interaction with one or both of these export adapters ORF57 can then recruit the remainder of the hTREX complex, including the proteins UAP56 and the THO complex as well as the export receptor, TAP. ORF57 shuttles this complex through the nucleolus prior to the mRNA being exported into the cytoplasm in a TAP-dependent mechanism. The hTREX components are then recycled back into the nucleus while ORF57 remains bound to the viral mRNA for involvement in further downstream mechanisms.
The role of ORF57 in mRNA stability
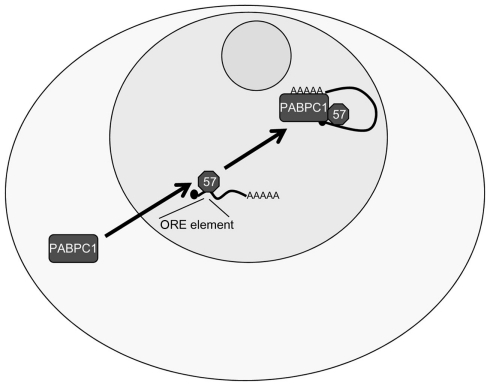
Another major role of KSHV ORF57 is to enhance RNA stability. Multiple studies have shown that a series of KSHV mRNAs are stabilized by ORF57 (e.g., ORF47, ORF59, and PAN), whereas others show no dependence on ORF57 (e.g., GCR and K5), suggesting that a specific RNA motif may be recognized by ORF57 to enhance the stability of target mRNAs (Kirshner et al., 2000; Boyne et al., 2008b). Recent analysis has now shed light on a possible ORF57 response element (ORE), although to date information is mainly limited onto the KSHV PAN. An ORE has been identified in the 5′ end of PAN and further analysis showed that the transfer of this element confers ORF57 responsiveness to an intronless reporter construct (Sei and Conrad, 2011). Deletion and mutational mapping have demonstrated that a nine nucleotide long core sequence is essential for the binding of ORF57 to PAN. A similar response element was independently confirmed, however the suggested minimal element of nine nucleotides is one position shifted compared to the sequence identified by Sei and Conrad (Massimelli et al., 2011). The binding of ORF57 to PAN via this RNA sequence is believed to enhance PAN RNA stability. In addition, two further proteins have been shown to interact with the ORF57 binding sequence, namely PABPC1 and EIB-AP5 (Massimelli et al., 2011). As the PABPC1 binding to the ORE is ORF57-dependent and specific to this binding site, it has been suggested that PABPC1 recruitment might be involved in stabilizing the PAN RNA in an ORF57-dependent manner (Massimelli et al., 2011; Figure 4).
Figure 4.
Kaposi’s sarcoma-associated herpesvirus ORF57 stabilizes PAN RNA in the nucleus. ORF57 binds to the ORF57 response element (ORE) in the 5′ of the KSHV PAN RNA, stabilizing PAN, and increasing expression. Additionally, ORF57 is able to redistribute PABPC1 to the nucleus, as well as PABPC1 being able to bind to the ORE, possibly explaining the stabilizing ability of the ORF57–PABPC1 interaction on PAN RNA.
An additional ORE has been identified within the KSHV vIL-6 mRNA (Kang et al., 2011). This sequence however only shares a 4-bp core (GGAU) with the PAN minimal ORE. Interestingly, investigations of ORF57 binding to this ORE highlighted a mechanism by which ORF57 stabilizes and protects vIL-6 from miRNA-mediated degradation. ORF57 protects the mRNA from degradation by a miRNA by a competitive binding mechanism. Moreover, the cellular IL-6 is also protected by ORF57 binding from the action of a second miRNA. How the other ORF57 target mRNAs are recognized remains still unclear, but considering the pace of recent findings, a better understanding of the underlying mechanism of ORF57 RNA binding and RNA stability seems closer than ever.
ORF57 functions to enhance the translation of viral transcripts
The multistep processes of mRNA biogenesis and maturation are not limited to the mechanisms that take place in the nucleus. A major protein complex deposited onto cellular mRNA during splicing is the exon junction complex (EJC; Bono and Gehring, 2011). The EJC is deposited ∼20 nts upstream of every exon–exon junction of spliced mRNA. The EJC functions in nonsense mediated decay targeting aberrant mRNAs which contain premature stop codons (Chang et al., 2007). In addition, the EJC plays a critical role in enhancing the translation of cellular mRNA transcripts, although the mechanisms by which it achieves this process have only been recently elucidated. It is believed that the EJC interacts with multiple translational enhancement proteins. For example, the EJC associated SKAR is able to recruit the 40S ribosomal subunit S6 protein kinase 1 (S6K1) to newly synthesized mRNA. This recruitment of S6K1 leads to an mTORC1 signaling cascade that results in enhancement of the pioneer round of translation (Ma et al., 2008). Additionally, the cellular protein PYM is able to bind to the EJC proteins Y14 and Magoh. PYM is then able to interact with the 48S preinitiation complex through a direct interaction with the small ribosomal subunit. This recruitment of the 48S preinitiation complex by PYM then acts to enhance the pioneer round of translation of cellular mRNAs (Diem et al., 2007).
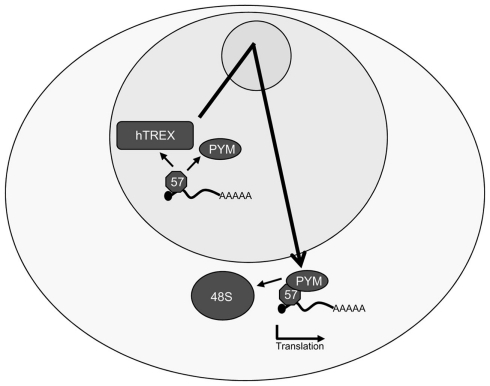
Analysis of the complexes recruited to a viral mRNA by ORF57 has shown that ORF57 only recruits hTREX components to viral intronless mRNAs, not EJC components, which leads to the intriguing question of how the viral transcripts are efficiently translated, when lacking an EJC. The answer lies in that ORF57 is able to enhance translation of viral transcripts itself. To this end, ORF57 interacts directly with PYM in vitro and in vivo, bypassing the need for PYM to interact with the EJC, and recruit the 48S preinitiation complex to intronless KSHV mRNAs (Figure 5; Boyne et al., 2010a,b). The functional importance of the PYM–ORF57 interaction was confirmed through in vivo use of transdominant mutants of PYM lacking the C- and N-terminal domains that are essential for the interaction of PYM with both the EJC and the 48S preinitiation complex. Importantly, these transdominant PYM mutants are still able to interact with ORF57 and expression in vivo alongside ORF57 dramatically reduced expression levels of late KSHV proteins and, concurrently KSHV virion production highlighting the importance of the KSHV ORF57–PYM interaction for enhancement of KSHV mRNA translation.
Figure 5.
Kaposi’s sarcoma-associated herpesvirus ORF57 enhances the translation of intronless viral mRNAs. As well as interacting with the hTREX complex in the nucleus, ORF57 also binds directly to the cellular protein PYM and recruits it to intronless KSHV mRNAs. ORF57 then shuttles through the nucleolus in an mRNP complex with the hTREX components, the intronless viral mRNA, PYM, and possibly other factors before exporting the mRNA. PYM recruits the 48S preinitiation complex to the viral mRNA, enhancing the pioneer round of translation in an ORF57-dependent manner.
To date no other ORF57 homolog has been shown to interact with PYM to enhance translation by a similar method, although translational enhancement is not something that is unique to KSHV ORF57. The ICP27 protein of HSV-1 has been shown to enhance the translation of several late proteins including VP16 and ICP5 (Fontaine-Rodriguez and Knipe, 2008), although the mechanism of this enhancement has not yet been defined. Interestingly, ICP27 does not affect the translation of all HSV-1 proteins as protein levels of the viral glycoprotein gD are not affected by the presence of ICP27, suggesting that whatever translational enhancement effect ICP27 is having, it is not a global effect on all mRNAs. Similarly, the SM protein of EBV has been shown to enhance translation of intronless viral transcripts. Moreover, insertion of an intron into intronless EBV viral transcripts negated the requirement for SM for efficient export and translation (Ricci et al., 2009). One possible explanation for this is that SM functions in a similar manner to KSHV ORF57 and recruits translational enhancement proteins. Therefore inserting an artificial intron allows the formation of an EJC which can recruit PYM independently of SM thereby enhancing the translation of viral transcripts.
A possible role for ORF57 in transcriptional enhancement
The roles of ORF57 are not limited to post-transcriptional processes; ORF57 also interacts with the KSHV transcriptional activator, RTA (Malik et al., 2004a). RTA can transactivate a number of KSHV and cellular promoters by binding directly to promoter regions containing an RTA responsive element (RRE) or interact with other transcriptional control proteins (Dourmishev et al., 2003). Alternatively, RTA can target transcriptional repressors for degradation through the ubiquitin proteasome pathway through its E3 ubiquitin ligase activity (Yu et al., 2005; Gould et al., 2009). Importantly, ORF57 has been shown to interact directly with RTA through its N-terminal region and through this interaction synergistically transactivates a number of viral promoters, including its own promoter as well as promoters for PAN/nut-1, Kaposin, ori-Lyt (L), K-bZIP, and TK (Kirshner et al., 2000; Malik et al., 2004a; Palmeri et al., 2007).
The A/T hook domain in the ORF57 N-terminus has been shown to confer a DNA binding ability on ORF57, although deletion of this domain did not completely abrogate DNA binding (Palmeri et al., 2007). Moreover, ORF57 was able to transactivate the PAN/nut-1 promoter irrespective of whether there was an intact A/T hook domain. Furthermore, ORF57 has also been shown to have a low transactivation effect on other viral promoters, such as Kaposin and TK in the absence of RTA (Kirshner et al., 2000). However, transactivation by ORF57 in the context of a lytic infection appears to be dependent on the ORF57–RTA interaction (Malik et al., 2004a). Additionally, transactivation by the ORF57–RTA complex appears to be promoter-, transcript-, and cell line-specific (Palmeri et al., 2007). Spontaneous KSHV reactivation in lytic cells is an inefficient process that is limited by the expression of RTA. It is interesting to note that ORF57 is able to enhance the expression of RTA in vivo, and one possibility is that activation of the RTA promoter by the ORF57–RTA complex is one mechanism by which KSHV overcomes the initial hurdle of inefficient reactivation.
Conclusion and Future Prospects
A number of recent studies have highlighted that ORF57 and its homologs are highly multifunctional with major roles in transcriptional activation, splicing, RNA stability, RNA nuclear export, and translational enhancement. The key function of this family of proteins, conserved amongst all α-, β-, and γ-herpesviruses, is the enhancement of viral mRNA transcript accumulation, although different homologs may have alternative ways they perform this function, as well as additional roles they play within the viral lifecycle.
A major question yet to be answered is how the multifunctional aspects of ORF57 are controlled. As all ORF57 homologs are designated as S/R proteins it seems highly likely that they are heavily posttranslationally modified, perhaps by multiple modifications. The ORF57 homolog of CMV, UL69, has been shown to be phosphorylated by CDKs, and colocalizes with CDK9 (Rechter et al., 2009). Inhibiting CDK activity in cell culture affects the nuclear localization of UL69 and causes it to form aggregates within the nucleus. Moreover, KSHV ORF57 is known to interact with the major cellular kinase, casein kinase II, and has been shown to be phosphorylated (Malik and Clements, 2004). This phosphorylation regulates the interaction between ORF57 and the cellular protein hnRNP K, a multifunctional protein involved in regulating gene expression. However, ORF57 has multiple protein partners, both cellular and viral, the majority of which presumably have their interaction controlled by some form of posttranslational modification. Further analysis of which residues of ORF57 are phosphorylated, and what effect this has on the various protein–protein interactions, as well as export and import of ORF57, is therefore essential to understanding how the multiple roles of ORF57 are controlled. Moreover, arginine methylation is known to affect protein import and export within the cell as well as protein–protein interactions. The HSV-1 ICP27 protein has recently been shown to be methylated on three residues within its RGG box (Souki and Sandri-Goldin, 2009; Souki et al., 2009), and that this methylation affected the interaction between ICP27 and the cellular proteins Aly and SRPK1, as well as regulating its export; although import of ICP27 into the nucleus is not affected by methylation. It is therefore important to test whether KSHV ORF57 is methylated in a similar way within either RGG1 or RGG2, and what effect any potential methylation has on its function. Additionally, a protein-wide screen of which, if any, ORF57 residues are methylated could lead to an understanding of how various ORF57 interactions and functions are coordinated.
It is clear from recent data from a number of independent research groups that ORF57 binds specifically to PAN RNA in conjunction with PABPC1 to stabilize PAN and increase its expression (Borah et al., 2011; Kumar et al., 2011; Massimelli et al., 2011; Sei and Conrad, 2011). However, one major unanswered question relating to this is why does ORF57 stabilize PAN? It would be interesting to see in the context of the virus what effect a PAN lacking the MRE would have on virus replication. Presumably, as PAN constitutes ∼80% of all polyadenylated RNA in a KSHV lytically active cell, the binding to PAN is an important aspect of ORF57 function. Furthermore, the understanding of ORF57 RNA binding is currently very limited with little awareness of how ORF57 is able to distinguish between cellular and viral transcripts, as well as the mechanistic way in which ORF57 recognizes and binds to the RNA. Answering these questions is vital to continue our understanding of the role of ORF57 in viral mRNA biogenesis.
Current understanding of the mechanisms involved in both viral and cellular mRNA biogenesis is still in its infancy. The function of some of the key proteins in mRNA export, such as the THO complex, is still poorly understood, as is the way that mRNA is stabilized both by cellular and viral mechanisms. Elucidation of the functions that ORF57 has and the roles it plays in various aspects of mRNA biogenesis will not only improve our understanding of viral mechanisms, but it will act as a model for the wider field of RNA biogenesis and processing. Moreover, from a clinical perspective a major difference between KSHV and the other human oncogenic herpesviruses is that KSHV requires reactivation and lytic expression for the majority of its tumorigenic properties. For this reason, studying the switch between latency and lytic replication is essential for both fully understanding the mechanism of KSHV tumorigenesis and also for developing treatments to prevent the onset or spread of KSHV-associated malignancies. Therefore, the study and understanding of the immediate early proteins that control this latent–lytic switch, including ORF57, is paramount for the future development of novel KSHV therapeutics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank both past and present members of the Whitehouse laboratory and collaborating groups for their contributions. Work discussed herein that was produced by the Whitehouse laboratory was supported by grants from the Biotechnology and Biological Sciences Research Council, Wellcome Trust, Yorkshire Cancer Research, and Leverhulme Trust. Adrian Whitehouse is the recipient of a BBSRC Research Development Fellowship.
References
- Aravind L., Landsman D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26, 4413–4421 10.1093/nar/26.18.4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon G., Chen K., Perez R., Tam W., Cesarman E. (2011). Kaposi sarcoma herpesvirus (KSHV) vFLIP oncoprotein induces B cell transdifferentiation and tumorigenesis in mice. J. Clin. Invest. 121, 1141–1153 10.1172/JCI44417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello L. J., Davison A. J., Glenn M. A., Whitehouse A., Rethmeier N., Schulz T. F., Barklie Clements J. (1999). The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80(Pt 12), 3207–3215 [DOI] [PubMed] [Google Scholar]
- Bono F., Gehring N. H. (2011). Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 8, 24–29 10.4161/rna.8.1.13618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S., Darricarrère N., Darnell A., Myoung J., Steitz J. A. (2011). A Viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog. 7, e1002300. 10.1371/journal.ppat.1002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottero V., Kerur N., Sadagopan S., Patel K., Sharma-Walia N., Chandran B. (2011). Phosphorylation and polyubiquitination of transforming growth factor β-activated kinase 1 are necessary for activation of NF-κB by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. J. Virol. 85, 1980–1993 10.1128/JVI.01911-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J. R., Colgan K. J., Whitehouse A. (2008a). Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front. Biosci. 13, 2928–2938 10.2741/2898 [DOI] [PubMed] [Google Scholar]
- Boyne J. R., Colgan K. J., Whitehouse A. (2008b). Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4, e1000194. 10.1371/journal.ppat.1000194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J. R., Jackson B. R., Taylor A., Macnab S. A., Whitehouse A. (2010a). Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 29, 1851–1864 10.1038/emboj.2010.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J. R., Jackson B. R., Whitehouse A. (2010b). ORF57: master regulator of KSHV mRNA biogenesis. Cell Cycle 9, 2702–2703 10.4161/cc.9.14.12627 [DOI] [PubMed] [Google Scholar]
- Boyne J. R., Whitehouse A. (2006a). Gamma-2 herpes virus post-transcriptional gene regulation. Clin. Microbiol. Infect. 12, 110–117 10.1111/j.1469-0691.2005.01317.x [DOI] [PubMed] [Google Scholar]
- Boyne J. R., Whitehouse A. (2006b). Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 15190–15195 10.1073/pnas.0604890103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J. R., Whitehouse A. (2009). Nucleolar disruption impairs Kaposi’s sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 583, 3549–3556 10.1016/j.febslet.2009.10.040 [DOI] [PubMed] [Google Scholar]
- Bryant H. E., Wadd S. E., Lamond A. I., Silverstein S. J., Clements J. B. (2001). Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 75, 4376–4385 10.1128/JVI.75.9.4376-4385.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Verma S. C., Lu J., Robertson E. S. (2010). Molecular biology of Kaposi’s sarcoma-associated herpesvirus and related oncogenesis. J. Adv. Virus Res. 78, 87–142 10.1016/B978-0-12-385032-4.00003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E., Chang Y., Moore P. S., Said J. W., Knowles D. M. (1995). Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332, 1186–1191 10.1056/NEJM199505043321802 [DOI] [PubMed] [Google Scholar]
- Chang P. J., Shedd D., Miller G. (2005). Two subclasses of Kaposi’s sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 79, 8750–8763 10.1128/JVI.79.13.8182-8188.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Cesarman E., Pessin M. S., Lee F., Culpepper J., Knowles D. M., Moore P. S. (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- Chang Y.-F., Imam J. S., Wilkinson M. F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.-S., Hsu J. L., Dias A., Reed R. (2006). Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127, 1389. 10.1016/j.cell.2006.10.044 [DOI] [PubMed] [Google Scholar]
- Colgan K. J., Boyne J. R., Whitehouse A. (2009). Uncoupling of hTREX demonstrates that UAP56 and hTHO-complex recruitment onto herpesvirus saimiri intronless transcripts is required for replication. J. Gen. Virol. 90, 1455–1460 10.1099/vir.0.007476-0 [DOI] [PubMed] [Google Scholar]
- Diem M. D., Chan C. C., Younis I., Dreyfuss G. (2007). PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 14, 1173–1179 10.1038/nsmb1321 [DOI] [PubMed] [Google Scholar]
- Dourmishev L. A., Dourmishev A. L., Palmeri D., Schwartz R. A., Lukac D. M. (2003). Molecular genetics of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67, 175–212 10.1128/MMBR.67.2.175-212.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufu K., Livingstone M. J., Seebacher J., Gygi S. P., Wilson S. A., Reed R. (2010). ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 24, 2043–2053 10.1101/gad.1898610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine-Rodriguez E. C., Knipe D. M. (2008). Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 82, 3538–3545 10.1128/JVI.02395-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. (2006). KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu. Rev. Pathol. 1, 273. 10.1146/annurev.pathol.1.110304.100133 [DOI] [PubMed] [Google Scholar]
- Goodwin D. J., Hall K. T., Giles M. S., Calderwood M. A., Markham A. F., Whitehouse A. (2000). The carboxy terminus of the herpesvirus saimiri ORF 57 gene contains domains that are required for transactivation and transrepression. J. Gen. Virol. 81, 2253–2265 [DOI] [PubMed] [Google Scholar]
- Goodwin D. J., Hall K. T., Stevenson A. J., Markham A. F., Whitehouse A. (1999). The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 73, 10519–10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould F., Harrison S. M., Hewitt E. W., Whitehouse A. (2009). Kaposi’s sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J. Virol. 83, 6727–6738 10.1128/JVI.00351-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A., Ganem D. (2004). Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 113, 124–136 10.1172/JCI200417803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwicke M. A., Sandri-Goldin R. M. (1994). The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68, 4797–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Sandri-Goldin R. M. (1994). Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68, 7790–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. M., Whitehouse A. (2008). Kaposi’s sarcoma-associated herpesvirus (KSHV) Rta and cellular HMGB1 proteins synergistically transactivate the KSHV ORF50 promoter. FEBS Lett. 582, 3080–3084 10.1016/j.febslet.2008.04.055 [DOI] [PubMed] [Google Scholar]
- Hastings M. L., Krainer A. R. (2001). Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13, 302. 10.1016/S0955-0674(00)00212-X [DOI] [PubMed] [Google Scholar]
- Hautbergue G. M., Hung M. L., Walsh M. J., Snijders A. P., Chang C. T., Jones R., Ponting C. P., Dickman M. J., Wilson S. A. (2009). UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 19, 1918–1924 10.1016/j.cub.2009.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez F. P., Sandri-Goldin R. M. (2010). Herpes simplex virus 1 regulatory protein ICP27 undergoes a head-to-tail intramolecular interaction. J. Virol. 84, 4124–4135 10.1128/JVI.02319-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez F. P., Sandri-Goldin R. M. (2011). Head-to-tail intramolecular interaction of herpes simplex virus type 1 regulatory protein ICP27 is important for its interaction with cellular mRNA export receptor TAP/NXF1. MBio 1, e00268-10. 10.1128/mBio.00268-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M. L., Hautbergue G. M., Snijders A. P., Dickman M. J., Wilson S. A. (2010). Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res. 38, 3351–3361 10.1093/nar/gkq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B. R., Boyne J. R., Noerenberg M., Taylor A., Hautbergue G. M., Walsh M. J., Wheat R., Blackbourn D. J., Wilson S. A., Whitehouse A. (2011). An interaction between KSHV ORF57 and UIF Provides mRNA-adaptor redundancy in herpesvirus intronless mRNA export. PLoS Pathog. 7, e1002138. 10.1371/journal.ppat.1002138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. G., Pripuzova N., Majerciak V., Kruhlak M., Le S. Y., Zheng Z. M. (2011). Kaposi sarcoma-associated herpesvirus ORF57 promotes escape of viral and human IL6 from microRNA-mediated suppression. J. Virol. 85, 2620–2630 10.1128/JVI.00720-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner J. R., Lukac D. M., Chang J., Ganem D. (2000). Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74, 3586–3597 10.1128/JVI.74.8.3586-3597.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Hurt E. (2007). Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Kumar G. R., Shum L., Glaunsinger B. A. (2011). Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol. Cell. Biol. 31, 3113–3125 10.1128/MCB.01342-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Dias A. P., Reed R. (2011). Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl. Acad. Sci. U.S.A. 108, 17985–17990 10.1073/pnas.1113076108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Chang J., Lynch S. J., Lukac D. M., Ganem D. (2002). The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16, 1977–1989 10.1101/gad.996502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac D. M., Kirshner J. R., Ganem D. (1999). Transcriptional activation by the product of open reading frame 50 of Kaposi’s sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73, 9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac D. M., Renne R., Kirshner J. R., Ganem D. (1998). Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252, 304–312 10.1006/viro.1998.9486 [DOI] [PubMed] [Google Scholar]
- Luo M. J., Reed R. (1999). Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. U.S.A. 96, 14937–14942 10.1073/pnas.96.13.7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. M., Yoon S. O., Richardson C. J., Julich K., Blenis J. (2008). SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133, 303–313 10.1016/j.cell.2008.02.031 [DOI] [PubMed] [Google Scholar]
- Majerciak V., Kruhlak M., Dagur P. K., Mccoy J. P., Jr., Zheng Z. M. (2010). Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J. Biol. Chem. 285, 11297–11307 10.1074/jbc.M109.068221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V., Uranishi H., Kruhlak M., Pilkington G. R., Massimelli M. J., Bear J., Pavlakis G. N., Felber B. K., Zheng Z.-M. (2011). Kaposi’s sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J. Virol. 85, 1528–1540 10.1128/JVI.01709-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V., Yamanegi K., Allemand E., Kruhlak M., Krainer A. R., Zheng Z. M. (2008). Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 82, 2792–2801 10.1128/JVI.01856-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V., Yamanegi K., Nie S. H., Zheng Z. M. (2006). Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281, 28365–28378 10.1074/jbc.M603095200 [DOI] [PubMed] [Google Scholar]
- Malik P., Blackbourn D. J., Cheng M. F., Hayward G. S., Clements J. B. (2004a). Functional co-operation between the Kaposi’s sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85, 2155–2166 10.1099/vir.0.79784-0 [DOI] [PubMed] [Google Scholar]
- Malik P., Blackbourn D. J., Clements J. B. (2004b). The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279, 33001–33011 10.1074/jbc.M313008200 [DOI] [PubMed] [Google Scholar]
- Malik P., Clements J. B. (2004). Protein kinase CK2 phosphorylation regulates the interaction of Kaposi’s sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 32, 5553–5569 10.1093/nar/gkh876 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Massimelli M. J., Kang J.-G., Majerciak V., Le S.-Y., Liewehr D., Steinberg S., Zheng Z. M. (2011). Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 7, 1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. (2005). Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19, 1512–1517 10.1101/gad.1302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., Proudfoot N. J. (2009). Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Nekorchuk M., Han Z., Hsieh T. T., Swaminathan S. (2007). Kaposi’s sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81, 9990–9998 10.1128/JVI.00896-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ote I., Lebrun M., Vandevenne P., Bontems S., Medina-Palazon C., Manet E., Piette J., Sadzot-Delvaux C. (2009). Varicella-zoster virus IE4 protein interacts with SR proteins and exports mRNAs through the TAP/NXF1 pathway. PLoS ONE 4, e7882. 10.1371/journal.pone.0007882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D., Spadavecchia S., Carroll K. D., Lukac D. M. (2007). Promoter- and cell-specific transcriptional transactivation by the Kaposi’s sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 81, 13299–13314 10.1128/JVI.00732-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin D. M., Sitas F., Chirenje M., Stein L., Abratt R., Wabinga H. (2008). Part I: cancer in indigenous Africans – burden, distribution, and trends. Lancet Oncol. 9, 683–692 10.1016/S1470-2045(08)70175-X [DOI] [PubMed] [Google Scholar]
- Piselli P., Busnach G., Citterio F., Frigerio M., Arbustini E., Burra P., Pinna A. D., Bresadola V., Ettorre G. M., Baccarani U., Buda A., Lauro A., Zanus G., Cimaglia C., Spagnoletti G., Lenardon A., Agozzino M., Gambato M., Zanfi C., Miglioresi L., Di Gioia P., Mei L., Ippolito G., Serraino D. (2009). Risk of Kaposi sarcoma after solid-organ transplantation: multicenter study in 4767 recipients in Italy, 1970–2006. Transplant. Proc. 41, 1227–1230 10.1016/j.transproceed.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. (2011). Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechter S., Scott G. M., Eickhoff J., Zielke K., Auerochs S., Müller R., Stamminger T., Rawlinson W. D., Marschall M. (2009). Cyclin-dependent kinases phosphorylate the cytomegalovirus RNA export protein pUL69 and modulate its nuclear localization and activity. J. Biol. Chem. 284, 8605–8613 10.1074/jbc.M805693200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci E. P., Mure F., Gruffat H., Decimo D., Medina-Palazon C., Ohlmann T., Manet E. (2009). Translation of intronless RNAs is strongly stimulated by the Epstein–Barr virus mRNA export factor EB2. Nucleic Acids Res. 37, 4932–4943 10.1093/nar/gkp497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S., Hurt E. (2011). Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 23, 302–309 10.1016/j.ceb.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Ruvolo V., Sun L., Howard K., Sung S., Delecluse H. J., Hammerschmidt W., Swaminathan S. (2004). Functional analysis of Epstein-Barr virus SM protein: identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J. Virol. 78, 340–352 10.1128/JVI.78.1.340-352.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo V., Wang E., Boyle S., Swaminathan S. (1998). The Epstein–Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. U.S.A. 95, 8852–8857 10.1073/pnas.95.15.8852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R. M. (1998). ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12, 868–879 10.1101/gad.12.6.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R. M. (2008). The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13, 5241–5256 10.2741/3078 [DOI] [PubMed] [Google Scholar]
- Sei E., Conrad N. K. (2011). Delineation of a core RNA element required for Kaposi’s sarcoma-associated herpesvirus ORF57 binding and activity. Virology 419, 107–116 10.1016/j.virol.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. W. P., Malik P., Clements J. B. (2005). The herpes simplex virus ICP27 protein: a multifunctional post-transcriptional regulator of gene expression. Biochem. Soc. Trans. 33, 499–501 10.1042/BST0330737 [DOI] [PubMed] [Google Scholar]
- Song M. J., Hwang S., Wong W., Round J., Martinez-Guzman D., Turpaz Y., Liang J., Wong B., Johnson R. C., Carey M., Sun R. (2004). The DNA architectural protein HMGB1 facilitates RTA-mediated viral gene expression in gamma-2 herpesviruses. J. Virol. 78, 12940–12950 10.1128/JVI.78.6.3072-3082.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souki S. K., Gershon P. D., Sandri-Goldin R. M. (2009). Arginine methylation of the ICP27 RGG box regulates ICP27 export and is required for efficient herpes simplex virus 1 replication. J. Virol. 83, 5309–5320 10.1128/JVI.00801-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souki S. K., Sandri-Goldin R. M. (2009). Arginine methylation of the ICP27 RGG box regulates the functional interactions of ICP27 with SRPK1 and Aly/REF during herpes simplex virus 1 infection. J. Virol. 83, 8970–8975 10.1128/JVI.00801-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., d’agay M.-F., Clauvel J. P., Raphael M., Degos L., Sigaux F. (1995). Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86, 1276–1280 [PubMed] [Google Scholar]
- Swaminathan S. (2005). Post-transcriptional gene regulation by gamma herpesviruses. J. Cell. Biochem. 95, 698–711 10.1002/jcb.20465 [DOI] [PubMed] [Google Scholar]
- Taylor A., Jackson B. R., Noerenberg M., Hughes D. J., Boyne J. R., Verow M., Harris M., Whitehouse A. (2011). Mutation of a C-terminal motif affects KSHV ORF57 RNA binding, nuclear trafficking and multimerisation. J. Virol. 85, 7881–7891 10.1128/JVI.00077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z., Stamminger T. (2008). The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13, 2939–2949 10.2741/2899 [DOI] [PubMed] [Google Scholar]
- Tunnicliffe R. B., Hautbergue G. M., Kalra P., Jackson B. R., Whitehouse A., Wilson S. A., Golovanov A. P. (2010). Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog. 7, e1001244. 10.1371/journal.ppat.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia P., Dias A. P., Reed R. (2008). Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 105, 3386–3391 10.1073/pnas.0800250105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D., Bais S., Gaillard M., Swaminathan S. (2010). Epstein-Barr virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J. Virol. 84, 11781–11789 10.1128/JVI.01359-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D., Swaminathan S. (2008). Epstein-Barr virus SM protein functions as an alternative splicing factor. J. Virol. 82, 7180–7188 10.1128/JVI.00344-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse A., Cooper M., Meredith D. M. (1998). The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J. Virol. 72, 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008). GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell Proteomics 7, 1598–1608 10.1074/mcp.M700574-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang S. E., Hayward G. S. (2005). The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22, 59–70 10.1016/j.immuni.2005.01.018 [DOI] [PubMed] [Google Scholar]