Abstract
On a global scale, crustal fluids fuel a large part of the deep-subseafloor biosphere by providing electron acceptors for microbial respiration. In this study, we examined bacterial cultures from sediments of the Juan de Fuca Ridge, Northeast Pacific (IODP Site U1301). The sediments comprise three distinctive compartments: an upper sulfate-containing zone, formed by bottom-seawater diffusion, a sulfate-depleted zone, and a second (∼140 m thick) sulfate-containing zone influenced by fluid diffusion from the basaltic aquifer. In order to identify and characterize sulfate-reducing bacteria, enrichment cultures from different sediment layers were set up, analyzed by molecular screening, and used for isolating pure cultures. The initial enrichments harbored specific communities of heterotrophic microorganisms. Strains affiliated to Desulfosporosinus lacus, Desulfotomaculum sp., and Desulfovibrio aespoeensis were isolated only from the top layers (1.3–9.1 meters below seafloor, mbsf), while several strains of Desulfovibrio indonesiensis and a relative of Desulfotignum balticum were obtained from near-basement sediments (240–262 mbsf). Physiological tests on three selected strains affiliated to Dv. aespoeensis, Dv. indonesiensis, and Desulfotignum balticum indicated that all reduce sulfate with a limited number of short-chain n-alcohols or fatty acids and were able to ferment either ethanol, pyruvate, or betaine. All three isolates shared the capacity of growing chemolithotrophically with H2 as sole electron donor. Strain P23, affiliating with Dv. indonesiensis, even grew autotrophically in the absence of any organic compounds. Thus, H2 might be an essential electron donor in the deep-subseafloor where the availability of organic substrates is limited. The isolation of non-sporeforming sulfate reducers from fluid-influenced layers indicates that they have survived the long-term burial as active populations even after the separation from the seafloor hundreds of meters above.
Keywords: Desulfovibrio, Desulfotignum, diversity, deep biosphere, Juan de Fuca Ridge, hydrogen, chemolithoautotrophy, IODP
Introduction
The subseafloor biosphere is probably the largest reservoir for prokaryotic life on Earth (Whitman et al., 1998; Heberling et al., 2010). It extends several hundred meters into deeply buried sediments (Parkes et al., 1994; Roussel et al., 2008) and even further down into the upper layers of the oceanic crust (Thorseth et al., 1995; Furnes and Staudigel, 1999; Ehrhardt et al., 2007). Recently, it was estimated that the ocean crust contains a similar amount of microorganisms as the entire volume of the world’s oceans (Heberling et al., 2010). The continuous circulation of seawater within the upper crust turns these voluminous, porous, and permeable basalts into the largest globally connected aquifer (Johnson and Pruis, 2003; Johnson et al., 2006).
Intense fluid circulation is a consequence of specific geological settings evolved during crust formation at ocean-spreading centers. It is especially pronounced at ocean ridges such as the Juan de Fuca Ridge in the Northeast Pacific (Johnson et al., 2006). This area is one of the most intensively studied locations in terms of heat-driven fluid flow (Fisher et al., 2003; Hutnak et al., 2006). While cold bottom-seawater is recharged at seamounts, it warms up within the oceanic crust beneath the sediments before being discharged again at other rocky outcrops exposed at the seafloor. The chemical composition of these low-temperature hydrothermal fluids [<150°C (Cowen, 2004)] is altered during long-term circulation through the basalt due to continuous abiotic water–rock interaction (Edwards et al., 2003) especially with increasing basement temperature (Wheat and Mottl, 1994; Wheat et al., 2000), or as a response to volcanic eruption (Butterfield et al., 1997). Additionally, microbial activity of crust-hosted communities contributes to changes in fluid composition by removing seawater constituents such as sulfate as indicated by sulfur-isotope measurements (Rouxel et al., 2008). However, due to a limitation in electron donors, crustal fluids are not fully reduced and still contain suitable electron acceptors, such as sulfate, for anaerobic respiration (Wheat and Mottl, 1994; Wheat et al., 2000; Cowen et al., 2003; Edwards et al., 2005).
It was postulated that basement fluids not only supply electron donors and acceptors to microbial life within the crust, but also to the microbial communities in the overlying sediments by diffusion from below (Cowen et al., 2003; DeLong, 2004; D’Hondt et al., 2004). We tested this hypothesis during an expedition to the eastern flank of the Juan de Fuca Ridge (IODP Exp. 301) by analyzing a 265-m-long sediment column of IODP site U1301. Sampling included material taken only two meters above the sediment–basement interface (Expedition 301 Scientists, 2005). At this site, sulfate diffuses into the sediments from both the seafloor (∼27 mM) and the underlying basement (∼16 mM). As a precondition for a sound microbiological and geochemical analysis, contamination controls were performed directly onboard the drillship JOIDES Resolution and proved the pristine character of the sediment samples (Lever et al., 2006).
Our previous work has shown that fluids from the oceanic crust do support microbial life in the overlying sediments (Engelen et al., 2008). Exoenzyme activities and sulfate reduction rates were not only elevated near the seafloor but also at the bottom of the sediment column which correlated well with the overall geochemical settings. We detected enhanced microbial abundance in sediment layers above the basement by direct counting and the cultivation-based most probable number (MPN) technique. Microbial growth in anoxic MPN dilution series from sediment layers near the oceanic crust indicated considerable amounts of viable microbial populations. Thus, the detection of a deep sulfate reduction zone and the successful enrichment of anaerobic microorganisms was the motivation for isolating sulfate-reducing bacteria (SRB) especially from fluid-influenced sediment layers. Identifying defined physiological adaptations of indigenous microorganisms to environmental conditions can be achieved best when pure cultures are available.
Even though sulfate reduction is supposed to be an important process in deeply buried sediments, only few isolates are available in strain collections. The type strain of Desulfovibrio profundus was isolated from 500 m depth in sediments of the Japan Sea (Parkes et al., 1995; Bale et al., 1997). Other piezophilic isolates closely related to Dv. profundus were cultivated from 222 m deep sediments of the Cascadia margin of the Pacific Ocean (Barnes et al., 1998). However, cultivation-based studies on the marine deep biosphere are still limited to a few sampling sites representing pinpricks in the ocean floor. So far, isolates from the marine subsurface were obtained from sediment samples retrieved from Mediterranean sediments (Süss et al., 2004) and from various sites in the Pacific Ocean: The Sea of Okhotsk, north of Japan (Inagaki et al., 2003), the Nankai Trough south–east of Japan (Mikucki et al., 2003; Toffin et al., 2004a,b, 2005; Kendall et al., 2006), the Equatorial Pacific, and the Peru Margin (D’Hondt et al., 2004; Biddle et al., 2005; Lee et al., 2005; Batzke et al., 2007). Recently, several heterotrophic bacteria and methanogenic Archaea were isolated from up to 106 mbsf deep sediments off Shimokita Peninsula, Japan using a continuous-flow bioreactor (Imachi et al., 2011).
In this study, we extended our previous investigations on IODP Site U1301 to determine the microbial diversity within different sediment layers of the deep subsurface. We hypothesize, that zones with different sulfate concentrations harbor different populations of SRB due to varying substrate availabilities. A cultivation-based approach in combination with molecular screening tools was chosen to isolate and compare SRB from fluid-influenced sediments and near-surface layers. The metabolic properties of the isolates might provide new insights on the impact of crustal fluids on microbial metabolism in the deep-subseafloor biosphere where substrates are recalcitrant but electron acceptors are still available.
Materials and Methods
Sample material
Sediment samples were recovered from the eastern flank of the Juan de Fuca Ridge by the drill ship “JOIDES Resolution” during IODP Expedition 301 in 2004. Characteristics of IODP Site U1301 were described in the expedition report (Expedition 301 Scientists, 2005). Sediment sampling, contamination tests, and subsampling for further analyses were described in detail by Engelen et al. (2008). All samples proved to be free of contamination as previously described by Lever et al. (2006).
Initial enrichments of deep-biosphere bacteria
To elucidate the diversity of cultured bacteria, a total of 736 initial enrichment cultures were set up directly onboard. Sediment slurries from 17 representative depth intervals (Engelen et al., 2008) were prepared immediately after sample recovery with anoxic artificial seawater medium (Süss et al., 2004). MPN series for anoxic and oxic microorganisms from these slurries were performed in 10-fold steps within 96-deep-well microtiter plates as previously described (Engelen et al., 2008). In addition, liquid dilution series in 20 ml-glass tubes were inoculated, flushed with N2 and sealed with butyl rubber stoppers. Anoxic substrate gradient tubes were prepared with undisturbed 1-cm3-sediment subcores from hole U1301C, only by embedding them within agar-solidified artificial seawater media (Köpke et al., 2005). In general, a mixture of the following substrates were supplied to stimulate microbial growth: glycerol, glucose, lactate, fumarate, malate, succinate, methanol, ethanol, 1-propanol, 1-butanol, formate, acetate, propionate, butyrate, valerate, caproate, and all the 20 l-amino acids (final concentration of each compound: 0.1 mM). For a better comparison of all enrichments, incubation was performed at 20°C.
Anoxic and oxic MPN viable counts were determined after 14 weeks of incubation to quantify the cultured part of the microbial communities within the sampled sediment layers. Procedure and results have already been published by Engelen et al. (2008). For the present cultivation study, all dilution cultures showing growth were transferred into 20 ml-glass tubes containing freshly prepared media and further incubated for at least five months at 20°C. Since cell densities were generally low, growth was determined several times during incubation by epifluorescence microscopy using Sybr®GreenI as a fluorescent dye. Growth of sulfate reducers was monitored by measuring the formation of sulfide (Cord-Ruwisch, 1985). Gradient cultures were incubated for approximately one year without interruption. Stimulation of growth within the sediment subcore was analyzed by microscopy and molecular methods. Finally, a total of 116 positive cultures were analyzed by means of molecular biological methods as described below to identify the cultivated microorganisms and to select enrichments for further isolation processes.
Isolation of pure cultures
Pure cultures from SRB and other anaerobes were isolated and maintained in a slightly different artificial seawater media. One liter of this basal medium contained 24.32 g NaCl, 10.0 g MgCl2·6H2O, 1.5 g CaCl2·2H2O, 4.0 g Na2SO4, 0.66 g KCl, and 0.09 g KBr. Resazurin (1 mg/l) was added as redox-indicator. The media was autoclaved, cooled under a nitrogen atmosphere, and supplemented with the following sterile solutions: NH4Cl (2 mM), KH2PO4 (1 mM), CO2-saturated sodium bicarbonate (30 mM), and from sterile stocks: 1 ml/l of trace element solution SL10 (Widdel and Bak, 1992) 0.2 ml/l of selenite-tungsten solution (Widdel and Bak, 1992) and 2 ml/l of a solution of 10 vitamins (Balch et al., 1979). The anoxic medium was reduced by addition of Na2S (final concentration: ∼1 mM) and few crystals of sodium dithionite. The pH was adjusted to 7.2–7.5 with 4 M NaOH. To increase cell density of all subcultures, a 10-fold higher concentrated substrate mix was provided (i.e., final concentration of each compound: 1 mM).
Repeated application of the deep-agar dilution method (Widdel and Bak, 1992) or dilution-to-extinction was performed to isolate deep-biosphere bacteria from liquid enrichments. Sediment subcores from gradient cultures were homogenized and slurried with 4 ml anoxic artificial seawater to further establish subcultures as gradient dilution series (up to 10−6). Aerobic microorganisms were subcultured for isolation by the liquid dilution-to-extinction method with subsequent purification on agar plates using a HEPES/bicarbonate-buffered oxic seawater medium. The purity of all isolates was checked by microscopy and molecular analysis as described below. Furthermore, the cultures were transferred to a complex HEPES-buffered oxic seawater medium containing yeast extract (0.03 g/l), glucose (1 mM), lactate (5 mM), and peptone (0.06 g/l) as substrates to check for contamination.
Molecular screening of enrichment cultures
The above described enrichment and isolation procedure was monitored and directed by molecular screening to identify unique phylotypes. Positive dilutions or growing colonies were analyzed by using polymerase chain reaction (PCR) of 16S rRNA gene-fragments, denaturing gradient gel electrophoresis (DGGE), and subsequent sequencing of re-amplified DGGE bands. DNA from liquid cultures was extracted using a protocol combining bead-beating with phenol/chloroform/isoamyl alcohol treatment and isopropanol/sodium acetate precipitation (Stevens et al., 2005). Nucleic acid extraction from substrate gradient cultures was performed by using the UltraClean™ Soil DNA Isolation Kit (MoBio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturers’ instructions.
Polymerase chain reaction-amplification of bacterial 16S rRNA genes was conducted in 50-μl volumes containing the following components: 1–2 μl of DNA-template, 10 pmol of each primer, 0.2 mM of each dNTP, 0.5–2 μl of bovine serum albumin (BSA, 10 mg/ml), 5 μl of 10×-ThermoPol reaction buffer and 1 U/μl of Taq Polymerase (New England Biolabs, Inc., Ipswich, MA, USA) and nuclease-free water. For DGGE analysis, the almost complete 16S rRNA genes were amplified with the primer set 8f/1492r (Overmann and Tuschak, 1997). The samples were incubated in a thermal cycler (Mastercycler, Eppendorf, Hamburg, Germany) under the following conditions: initial denaturation at 95°C for 5 min, 28 cycles of amplification by denaturation at 95°C for 30 s, annealing at 40°C for 60 s, and elongation at 72°C for 3 min. Terminal elongation was performed at 72°C for 10 min. The resulting amplicons were used as templates for a nested PCR. Shorter 16S rRNA gene-fragments were amplified (Wilms et al., 2006a) using the universal bacterial primer set GC-341f and 907r (Overmann and Tuschak, 1997). All PCR products were always visualized by agarose-gel electrophoresis (Wilms et al., 2006a). DGGE was performed with a gradient from 40 to 70% (Süss et al., 2004). PCR products were mixed with loading buffer before loading onto the gel (Wilms et al., 2006a).
Sequencing of DGGE bands and pure cultures
For sequence analysis of DGGE bands, distinctive bands were excised, eluted in 50 μl nuclease-free water, re-amplified in a 25-μl-PCR (primers 341f/907r, Wilms et al., 2006b), and purified (Wilms et al., 2006a) using the QIAquick PCR purification Kit (Qiagen GmbH, Hilden, Germany) or the PCR-Purifying-Kit (SeqLab, Göttingen, Germany) and sequenced with a IRDyeTM800 labeled 907r-primer (Süss et al., 2004). For phylogenetic identification of pure cultures, genomic DNA was extracted from the isolated strains using a freeze and thaw procedure. From picked colonies or 2 ml of liquid cultures, 1 μl of a cell pellet was resuspended with 100 μl of filter-sterilized Tris-buffer (50 mM, pH 7.4). The suspension was frozen at −80°C for 3 min and heated at 85°C for 3 min. This procedure was repeated five times, and 2 μl of the final extract were added to 48 μl of PCR mixture. Partial or nearly full-length bacterial 16S rRNA gene sequences were amplified using the bacteria-specific primer set 341f/907r and 8f/1492r, respectively, and sequenced as described above. In case of sulfate-reducing strains, DNA was sequenced in both directions using the respective PCR primers and the service of GATC Biotech AG (Konstanz, Germany). Consensus sequences were constructed after alignment by using the BioEdit software tool version 7.0.91. All 16S rRNA gene sequences obtained in this study were compared for their affiliation to the closest relatives using the BLASTN program 2.2.26+ (Altschul et al., 1990; Morgulis et al., 2008)2. The partial 16S rRNA gene sequences of all 40 isolates are deposited in GenBank database under the accession numbers JQ411257–JQ411296.
Physiological characterization of sulfate-reducing isolates
Physiological tests were generally performed in sealed glass tubes containing 10 ml of artificial seawater medium. Sulfidogenic growth was tested with 18 different substrates at final concentrations between 1 and 5 mM in the presence of sulfate. Fermentative growth with betaine, ethanol, malate, or pyruvate (2–5 mM, each) was tested in medium without additional electron acceptors. The cultures were incubated for at least 4 weeks at 20°C in the dark. Chemolithotrophic growth with H2 as electron donor was tested with a headspace (2/3 of the culture volume) filled with a mixture of H2/CO2 (80/20 v/v, 1 kPa). Those cultures were incubated horizontally at 20°C. Growth was checked by visual inspection of turbidity, by phase contrast microscopy, and by sulfide formation (Cord-Ruwisch, 1985). Substrate utilization was defined to be positive after the third successful transfer into fresh media.
The capability of anaerobic respiration was tested in sulfate-free medium with ethanol or lactate (5 mM, each) as electron donor in combination with six different electron acceptors. Reduction of Fe(III) was indicated by the formation of black precipitates under the expense of the reddish ferric hydroxide. Mn(IV) utilization was shown by the disappearance of brown manganese carbonates and the occurrence of white precipitates. The production of sulfide as a result of the reduction of thiosulfate or sulfite was measured at 480 nm using a Shimadzu UV-1202 photometer (Cord-Ruwisch, 1985). In addition, cultures were checked microscopically for the presence of bacterial cells.
Growth experiments for autotrophic growth were performed at 35°C, the optimum temperature for growth of our test strain P23. Growth rates were calculated from linear regression of produced sulfide (Cord-Ruwisch, 1985) and formed cell protein (Bradford, 1976) as function of time.
The temperature range for growth of SRB was tested from 4 to 55°C with lactate (10 mM) as electron donor. Growth was followed at OD436 via sulfide production and by photometrical determination of protein concentrations (Bradford, 1976).
For phase contrast microscopy, agarose-coated slides were used. To prepare those, slides were thoroughly cleaned and preheated by infrared light in order to get a smooth agarose film. Then, 1 ml of the hot agarose solution (2% w/v) was dispensed on the warm slides. Before usage, the agarose slides were air dried. Upon placing a drop of a bacterial culture to a coated slide, the liquid diffuses into the dry agarose, while the cells are gently squeezed under the cover slip and get fixed in the same plane. Cell dimensions were determined using a Leitz DMRB microscope (Wetzlar, Germany).
Transmission-electron microscopy was performed as follows: a 400-mesh Formvar copper grid (Plano) was placed on a drop of cell suspension for 10 min. Cells adsorbed to the grid were stained with 0.5% aqueous uranyl acetate for 1 min, washed twice in a drop of water for a few seconds and examined with a transmission-electron microscope (EM 902A, Zeiss). A Proscan High Speed SSCCD camera system with iTEMfive software was used for images acquisition.
Results
Geochemical profiles divide the sediment column into three distinctive zones
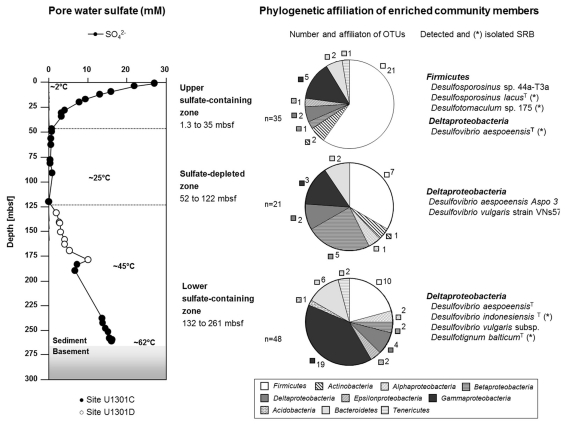
The geomorphological structure of the eastern flank of the Juan de Fuca Ridge leads to a hydrological situation where sulfate-containing fluids from the oceanic crust diffuse ∼140 m into overlying sediment layers. The effect of this heat-driven fluid circulation was reflected by the temperature gradient within the sediments of 2°C at the seafloor to approximately 62°C above the basement (Expedition 301 Scientists, 2005). Using the porewater profile of sulfate, the sediment column can be separated into three zones (Figure 1). The upper sulfate-containing zone was formed by bottom-seawater diffusion showing decreasing concentrations from 27 mM at the top to 3 mM in 35 mbsf. Below, a sulfate-depleted zone was located between 47 and 121 mbsf (<1 mM). The lower sulfate-containing zone was characterized by increasing sulfate concentrations from 2 to 16 mM toward the basement at ∼265 mbsf due to sulfate diffusion from crustal fluid flow into the overlying sediments.
Figure 1.
Zonation of the 265-m-long sediment column of the eastern flank of the Juan de Fuca Ridge, Northeast Pacific (IODP Site U1301), and phylogenetic affiliation of enriched and isolated marine subsurface bacteria with special emphasis on sulfate-reducing bacteria. Operational taxonomic units (OTUs) detected via PCR–DGGE are defined at 97% sequence similarity.
Shifts in microbial diversity between the initial enrichment cultures from the different zones
Anoxic and oxic MPN series, liquid dilution series in tubes and the substrate gradient technique were used to enrich and further isolate deep-biosphere bacteria. The cultivation progress was monitored by microscopy and PCR–DGGE. Unique DGGE bands were subsequently sequenced to identify the community composition within the enrichments. A total of 135 partial 16S rRNA gene sequences were obtained after DGGE analysis of growing cultures. The technique was not only chosen to prevent multiple isolation of one strain and to check the purity of cultures, but also to identify community members that could not be isolated. This molecular-directed cultivation indicated the presence of diverse viable microbial populations within the different zones of the investigated sediment column.
The phylogenetic screening of the initial enrichments identified different bacterial populations among the growing cultures obtained from the three sediment zones (Figure 1). A typical decrease in cultivation success with respect to the conditions set in our growth media was observed for the two upper zones, which correlates with the general depletion of electron donors and acceptors. Within the top 30 m of the sediment column, 35 different operational taxonomic units (OTUs, defined at 97% sequence similarity) were detected via PCR–DGGE in enrichments from the respective sediment layers. From the sulfate-depleted zone, 21 OTUs were retrieved. For samples from the deep, fluid-influenced sediment zone, the cultivation success increased again with 48 identified OTUs.
In general, the number of OTUs belonging to the Firmicutes decreased with sediment depth from 60% in enrichments from the upper sulfate-containing zone to 21% in the lower sulfate zone. In addition, Gammaproteobacteria accounted for 40% of all OTUs retrieved from enrichments of the lower sulfate zone. Bacteria belonging to the phylum Acidobacteria, Bacteroidetes, and the classes Beta- and Epsilonproteobacteria were enriched as identified by molecular methods but could not be isolated or were lost during purification procedures. The majority of the enriched but not isolated organisms were phylogenetically affiliated to uncultured bacteria from different terrestrial and marine environments (data not shown in detail).
Diversity of isolated pure cultures
From the 116 initial enrichments that were tested positively for growth, 40 strains could be isolated (14 from the upper sulfate-containing zone, 8 from the sulfate-depleted zone, and 18 from the lower sulfate-containing zone). Based on 16S rRNA gene sequences, the 40 pure cultures could be affiliated to the phyla Actinobacteria, Firmicutes, and Tenericutes or the classes Alphaproteobacteria, Gammaproteobacteria, Deltaproteobacteria (Table 1). The majority of isolates (32 of 40) were obtained from liquid dilution series that were initially inoculated with hundred to million fold diluted sediment (10−2 to 10−6), indicating a significant number of cells in situ. Nearly all isolates were closely related to cultivated species from sediments or soils, fluids, or other aquatic environments. Among them, 13 were strict anaerobes. With exception of the sporeforming Firmicutes all other pure cultures including those obtained from oxic media were considered to be facultatively anaerobic, since they originated from anoxic sediment horizons. While some isolates seem to be ubiquitous within the sediment column [e.g., Shewanella frigidimarina, 98–99% sequence similarity or Bacillus spp. (96–100%)], others were retrieved from single sediment layers, only (e.g., Anaerovirgula multivorans or Marinobacter flavimaris, both 99% sequence similarity).
Table 1.
Origin and phylogenetic affiliation of isolated strains from IODP Site U1301, a 265-m-long sediment column of the eastern flank of the Juan de Fuca Ridge, Northeast Pacific.
| Phylogenetic group, closest relative* in GenBank (accession no.) | Similarity (%) | Sediment depth (mbsf) | No. of isolates | Habitat of closest relatives*** |
|---|---|---|---|---|
| ACTINOBACTERIA | ||||
| Bacterium Ellin5115 (AY234532) | 99 | 112 | 1 | Soil, Australia |
| [Actinomycetospora chibensisT (AB514517)] | 99 | Paddy soil, Japan | ||
| Iron-reducing enrichment clone Cl-A3 (DQ676995) | 99 | 31 | 1** | Estuary sediment, Europe |
| [Propionicimonas paludicolaT (FR733712)] | 99 | Rice-field soil, Japan | ||
| FIRMICUTES | ||||
| Anaerovirgula multivoransT (NR_041291) | 99 | 1.3 | 1** | Owens Lake, USA |
| Bacillus siralisT (NR_028709) | 98, 96 | 112, 132 | 2 | General study |
| Bacillus sp. AS7 HS-2008 (AM950301) | 98, 99 | 99, 112 | 2 | Brine Lake Sediment, Mediterranean |
| Bacillus sp. Hs56 (JF803865) | 99 | 1.3, 169 | 2 | Marine sponge, Bay in Ireland |
| Desulfosporosinus lacusT (NR_042202) | 97 | 1.3 | 1** | Sediments of Lake Stechlin, Germany |
| Desulfotomaculum sp. 175 (AF295656) | 98, 99 | 1.3, 9.1 | 2** | Aquifer/lignite seam, Germany |
| Marinilactibacillus sp. A5 (DQ344853) | 98, 99 | 75, 99 | 2** | Deep-sea sediment, Pacific |
| Paenibacillus sp. UXO5-11 (DQ522106) | 99 | 31 | 1 | Marine sediments, Hawaii |
| Bacillus circulans USC24 (HQ441221) | 99 | 9.1 | 1 | Fish tank sediment, Spain |
| Uncult. bacterium clone LCKS880B24 (EF201766) | 98 | 9.1 | 1** | Lake Chaka, China |
| [Desulfonispora thiosulfatigenesT (NR_026497)] | 90 | Sewage plant, Germany | ||
| ALPHAPROTEOBACTERIA | ||||
| Pelagibacterium halotoleransT (EU709017) | 99 | 132 | 1 | Ocean water, China |
| GAMMAPROTEOBACTERIA | ||||
| Alteromonas sp. USC168 (HQ441215) | 99 | 31, 52 | 2 | Mediterranean surface water, Spain |
| Halomonas sp. 1B.4 (HQ427421) | 99 | 141 | 1 | Ocean crust, JdF Ridge. Pacific |
| Halomonas axialensisT (NR_027219) | 100 | 163 | 1 | Hydrothermal fluid, JdF Ridge, Pacific |
| Halomonas sp. PEB09 (GU213166) | 99 | 163 | 1 | Estuarine microbial mat, Spain |
| Marinobacter flavimarisT (NR_025799) | 99 | 141 | 1 | Sea water, Yellow Sea in Korea |
| Pseudoalteromonas sp. D20 (AY582936) | 99 | 150 | 1 | Deep-sea sediment, Pacific |
| Pseudomonas sp. G12a-1 (FN397994) | 99 | 141, 150 | 2 | Deep-sea sediment, Indian Ocean |
| Shewanella frigidimarina ACAM 584 (U85902) | 98–99 | 1.3, 31, 52, 260 | 4 | Southern Ocean waters |
| Vibrio diazotrophicusT (NR_026123) | 98 | 150 | 1 | General study |
| Vibrio pelagiusT (X74722) | 98 | 31, 150 | 2 | General study |
| DELTAPROTEOBACTERIA | ||||
| Desulfotignum balticumT (NR_041852) | 99 | 260 | 1** | Marine mud, Denmark |
| Desulfovibrio aespoeensisT (NR_029307) | 98 | 1.3 | 1** | Aespoe hard rock borehole, Sweden |
| Desulfovibrio indonesiensisT (NR_044916) | 99 | 240, 252, 260 | 3** | Corroding ship, Indonesia |
| TENERICUTES | ||||
| Anaerobic bacterium MO-XQ (AB598274) | 99 | 260 | 1 | Subseafloor sediments, Japan |
| [Acholeplasma palmaeT (NR_029152)] | 93 | Plant surface | ||
*In case of environmental clones the next cultivated organism is indicated in square brackets.
**Strictly anaerobic isolates.
***Based upon the results of the megaBLAST search (NCBI).
Sulfate-reducing bacteria were isolated from both sulfate-containing zones
The sulfate reducers isolated from the upper 10 m predominantly belonged to the Firmicutes (Figures 1 and 2; Table 1). Three strains were identified as members of the genera Desulfotomaculum and Desulfosporosinus. The latter shared 97% sequence similarity with its closest described relative Desulfosporosinus lacus, firstly isolated from freshwater lake sediments (Ramamoorthy et al., 2006). The Desulfotomaculum strains were phylogenetically related to isolates originally obtained from a terrestric aquifer system (Detmers et al., 2001). Another isolate from a near-surface layer (strain P20) was closely affiliated to Desulfovibrio aespoeensis. This Deltaproteobacterium was also enriched in co-culture with strains related to Desulfovibrio indonesiensis (culture P34 and P19) from 240 and 260 mbsf, respectively, as identified by DGGE and subsequent sequencing of the bands. Two sequences affiliated to sulfate reducers were also detected in enrichment cultures from sediments of the sulfate-depleted zone (Figure 1). However, no isolates could be retrieved. SRB isolated from the deepest sediments above the basement solely belonged to the Deltaproteobacteria, namely Desulfotignum balticum (strain P18; 260 mbsf) and Dv. indonesiensis (strains P12, P19-1, P23, P33, and P34). The latter phylotype was highly abundant in the lower sulfate-containing zone as it was frequently retrieved from different fluid-influenced layers (240, 252, 260 mbsf). Furthermore, strains P18, P23, and P34 were isolated from million fold diluted MPN-cultures, allowing the assumption, that they must be present in higher numbers within the respective sediment layer, where they probably play an active role.
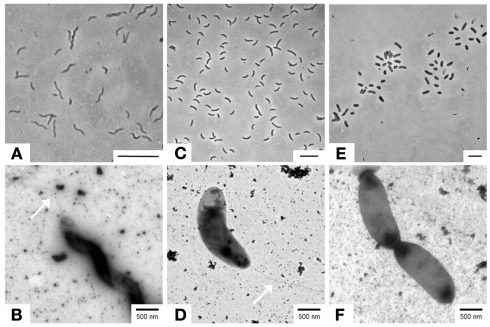
Figure 2.
Microscopic images from three sulfate-reducing isolates obtained from sediments of IODP Site U1301. (A,B) Desulfovibrio aespoeensis strain P20 (1.3 mbsf); (C,D) Desulfovibrio indonesiensis strain P23 (260.4 mbsf); (E,F) Desulfotignum balticum strain P18 (260.4 mbsf). Upper images: phase contrast (bar = 5 μm); lower images: transmission-electron microscopy, TEM (bar = 500 nm). Arrows in (B,D) indicate flagella.
Morphological and physiological characteristics of three representative sulfate-reducing isolates
Strains affiliated to Dv. aespoeensis (strain P20), Dv. indonesiensis (strain P23), and Desulfotignum balticum (strain P18) were morphologically and physiologically investigated in more detail (Figure 2; Table 2). Strain P20 was used for further analysis since it was the only available pure culture related to Dv. aespoeensis that was obtained in this study. Other relatives of this species were enriched from near-basement layers, but only in co-culture with strains affiliated to Dv. indonesiensis. Various efforts to separate the two species failed. Strain P23, obtained from the deepest sediment layer (260 mbsf), was chosen as a representative for strains related to Dv. indonesiensis, since the other closely related isolates showed nearly identical characteristics under the growth conditions tested.
Table 2.
Comparison of characteristics of sulfate-reducing isolates from IODP Site U1301: temperature range of growth, morphology, substrate utilization, and alternative electron acceptors.
| Isolated strain | P20 | P23 | P18 |
|---|---|---|---|
| Closest relative in GenBank | Desulfovibrio aespoeensisT | Desulfovibrio indonesiensisT | Desulfotignum balticumT |
| Sediment depth (mbsf) | 1.30 | 260.43 | 260.43 |
| Trange | 20–35°C | 10–48°C | 4–48°C |
| Topt | 25°C | 25–35°C | 25–35°C |
| Morphology | Highly motile, thin, vibrio like, spirilloid cells, 3.8 μm (±0.9 μm) long, 0.4 μm (±0.1 μm) thick | Motile, vibrio shaped cells, 2.7 μm (±0.4 μm) long, 0.7 μm (±0.1 μm) thick | Non-motile, short thick rods with rounded ends, 2.3 μm (±0.4 μm) long, 1.0 μm (±0.1 μm) thick |
| ELECTRON DONORS AND SUBSTRATES IN THE PRESENCE OF SULFATE | |||
| H2, CO2 + acetate (1 mM) | + | + | +** |
| H2, CO2 (excess) | − | +* | +** |
| Acetate (5 mM) | − | − | (+) |
| Benzoate (2.5 mM) | − | − | + |
| Betaine (2 mM) | n.t. | − | + |
| Butanol (5 mM) | − | + | + (No H2S) |
| Butyrate (5 mM) | − | − | (+) |
| Ethanol (5 mM) | − | + | − |
| Formate (5 mM) | + | + | (+) Slow |
| Fumarate (5 mM) | − | + | + |
| Lactate (5 mM) | + | + | + |
| Propanol (5 mM) | − | (+) | − |
| Pyruvate (5 mM) | − | + | + |
| Succinate (5 mM) | − | − | (+) Slow |
| FERMENTATION | |||
| Ethanol (5 mM) | − | (+) Slow | (+) Slow |
| Betaine (2 mM) | n.t. | n.t. | + |
| Pyruvate (5 mM) | (+) | + | − |
| ELECTRON ACCEPTORS*** | |||
| Sulfate (28 mM) | + | + | + |
| Sulfite (10 mM) | + | + | + |
| Thiosulfate (10 mM) | + | + | + |
| Fe(III) hydroxide (∼40 mM) | − | (+) | (+) |
| Mn(IV) (20 mM) | − | (+) | (+) |
+, Substrate used for growth as indicated by turbidity increase and production of H2S in the presence of sulfate, sulfite, or thiosulfate; (+), poor growth, no turbidity increase, but significant production of H2S, in the presence of sulfate; −, no growth; n.t., not tested.
*Even in absence of vitamins and resazurin as redox-indicator; **In presence of vitamins only.
***In presence of N2/CO2 or H2/CO2 and lactate or ethanol.
The culture medium contained 28 mM sulfate as electron acceptor. For fermentation tests and utilization of alternative electron acceptors, a sulfate-free culture medium was used. No strain grew on amino acid mix (1 mM), glucose (5 mM), malate (5 mM), methanol (5 mM), propionate (2 mM), or yeast extract (0.005% v/v). None of the strains fermented malate (5 mM) or used nitrate (10 mM) as alternative electron acceptor.
For all investigated strains, colonies formed in deep-agar dilution series exhibited yellowish to brownish colors. The Desulfovibrio affiliated strains showed curved, motile cells (Figures 2A,C) with single polar flagella as identified by electron microscopy of negatively stained cells (Figures 2B,D). The relative of the non-motile Desulfotignum balticum formed ∼2–3 μm short thick rods with rounded ends (Figures 2E,F).
Desulfovibrio aespoeensis strain P20 grew within a temperature range of 20–35°C with an optimum at 25°C. Desulfotignum strain P18 and Dv. indonesiensis strain P23 instead exhibited growth within a broad temperature range from 4 to 48°C and 10 to 48°C, respectively, with the optimal growth temperature lying between 25 and 35°C.
All strains were capable of using sulfite or thiosulfate as alternative electron acceptor other than sulfate. Slow growth by iron or manganese reduction on lactate was observed for Desulfotignum balticum strain P18 and Dv. indonesiensis strain P23. Growth was not as fast as with sulfate as electron acceptor and high cell densities were not achieved. However, growth on metal oxides occurred even after the third transfer. None of the strains used nitrate as electron acceptor for anaerobic respiration.
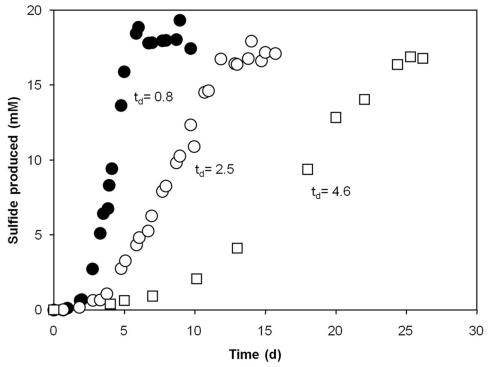
Of all substrates provided, Dv. aespoeensis strain P20 only utilized lactate and formate for growth in the presence of sulfate. In contrast, the type strain of Dv. aespoeensis only grew on lactate as sole substrate (Motamedi and Pedersen, 1998). The two other strains tested (P18 and P23) showed a slightly broader substrate spectrum. Strain P23, for instance, grew on different n-alcohols (C2–C4), formate, fumarate, lactate, and pyruvate. Only Desulfotignum strain P18 grew on acetate, benzoate, betaine, butyrate, and succinate, whereas fast growth and high cell densities were achieved with betaine, which was also fermented. Fermentative growth with pyruvate occurred in Desulfovibrio strains, only. All strains used hydrogen as electron donor. Desulfotignum balticum strain P18 grew autotrophically but only in the presence of vitamins. This was already known for the type strain of Desulfotignum balticum, which was described to grow on H2 and CO2 (Kuever et al., 2001). Surprisingly, autotrophic growth for Dv. indonesiensis strain P23 was observed in media that did not contain any organic additives such as vitamins, resazurin, or yeast extract and after at least 10 transfers to eliminate carbon sources from initial cultures (Figure 3). Growth rates (based on protein production) for strain P23 were 0.12 d−1 under autotrophic conditions, and approximately three times higher (0.30 d−1) when 1 mM of acetate was added.
Figure 3.
Comparison of heterotrophic and autotrophic growth of Desulfovibrio indonesiensis strain P23 at 35°C (◯) under autotrophic conditions using hydrogen, CO2 and sulfate (28 mM); (●) after addition of 1 mM acetate, both in the presence of resazurin and vitamins; (□) under autotrophic conditions in media without vitamins and resazurin. Doubling times (td) in days are indicated.
Discussion
Organic matter and sulfate availability generate the three different zones of the sediment column
The stratification of the different sediment compartments has an imprint on the life conditions. In both, the seawater- and fluid-influenced layers, the availability of electron acceptors stimulates microbial growth and activity of indigenous microorganisms (Engelen et al., 2008). In terms of electron donors, bacteria that thrive in the upper 30 m of the sediments are supported by burial of relatively young organic carbon (Fisher et al., 2003; Johnson et al., 2006). Therefore, they are used to a higher supply of electron donors and adapt much better to the given cultivation conditions. In deeper sediment horizons, indigenous bacteria have to survive long-term burial by adapting to a minimum supply of substrates and electron acceptors. Their limited availability strongly influences the metabolic activities in the deep marine subsurface. Indeed, based on geochemical porewater profiles, it has been concluded that the metabolic activities of subseafloor prokaryotes are very low (D’Hondt et al., 2002, 2004). They probably have developed different life strategies such as slow growth or survival as spores. The latter were presumably stimulated to germinate during our cultivation experiments since a major part of 16S rRNA gene sequences detected in all enrichment cultures affiliated to sporeforming Firmicutes (Figure 1). However, the decreasing number of Firmicutes with depth indicates that not all of them survive the long-term burial as spores as they might have germinated stochastically over geological time scales (Epstein, 2009).
Other subsurface organisms that are adapted to low organic carbon concentrations might not be able to grow under the given laboratory conditions. Even though the composition of our culture media was designed to provide organic substrates in sub-millimolar concentrations, a substrate shock (Straskrabová, 1983) might not have been circumvented. For instance, we were not able to grow any Archaea (data not shown) even though they are proposed to represent a substantial part of the deep biosphere as indicated by intact-lipid analysis (Lipp et al., 2008).
The supply of electron acceptors into the sediment column by crustal fluid diffusion dramatically changes the situation for microbial life within these deeply buried layers. The large numbers of non-sporeforming Gammaproteobacteria that were enriched from near-basement layers indicate the presence of viable cells. Many Gammaproteobacteria are adapted to elevated substrate concentrations (Lauro et al., 2009) and are therefore readily cultivated using our media. Some of them might even be typical for oceanic ridge systems. Halomonas and Marinobacter species were found to be present in hydrothermal fluids collected at the Juan de Fuca Ridge (Kaye et al., 2011). They were enriched during in situ colonization experiments on basaltic crust (Smith et al., 2011) and have also been detected in basaltic seafloor lavas and overlying seawater at the East Pacific Rise (Santelli et al., 2008).
The upper and lower sulfate-containing zones harbor different sulfate-reducing bacteria
The majority of sequences obtained from upper sediment horizons that were affiliated to SRB have Desulfosporosinus and Desulfotomaculum species as closest relatives, both sporeforming Firmicutes. However, it is unclear if they contribute to the high sulfate reduction rates of up to 8 nmol cm-3 d-1 determined for the upper sulfate-containing zone of IODP Site U1301 (Engelen et al., 2008). This would only be the case if these SRB are present as viable cells. It cannot be specified if they are metabolically active or if they only survive as spores within these layers.
In contrast, fluid-influenced sediments exclusively harbor sulfate reducers that are members of the Deltaproteobacteria, which are not known to form any resting stages. These viable populations contribute to sulfate reduction rates of up to 3 pmol cm-3 d-1 within the lower sulfate reduction zone (Engelen et al., 2008). Due to their high abundance, this activity might derive from sulfate reducers affiliated to Dv. indonesiensis. This is quite surprising since the in situ temperature is around 60°C and most Desulfovibrio species are not active above 40°C (Widdel and Bak, 1992). However, a broad temperature range of growth was not only found for our isolates, but also for the Japan Sea isolates of Dv. profundus (Bale et al., 1997) and might represent an adaptation to the conditions in the deep biosphere.
Thus, one reason for the divergence in the SRB communities detected in both sulfate-containing zones might be the different temperature and pressure regimes present at the top and bottom of the sediment column. Surprisingly, the isolates from the deepest fluid-influenced layers did not grow at in situ temperatures of approximately 60°C. This might be due to the chosen initial incubation conditions at 20°C and ambient hydrostatic pressure instead of the in situ pressure of ∼30 MPa. As temperature and pressure counteract on the cell membrane composition (Mangelsdorf et al., 2005), an insufficient combination of both parameters might result in membrane disintegration. This assumption is supported by the fact that no isolates were obtained from enrichment cultures that were incubated under in situ temperatures (data not shown). In future experiments, pressure incubations might help to overcome such problems in cultivation efficiencies.
Sulfate-reducing bacteria from the lower zone have relatives in deep terrestrial aquifers
Previous microbiological investigations on crustal fluids from the Juan de Fuca Ridge have identified several isolates (Nakagawa et al., 2006) and 16S rRNA clones (Cowen et al., 2003; Huber et al., 2006) that were affiliated to SRB. In general, the overlap between these studies compared with our culture collection from fluid-influenced sediments is quite low. Only relatives of Desulfotomaculum and Desulfonatronovibrio species were detected in two studies on the adjacent ODP Site 1026. One 16S rRNA gene sequence that is affiliated to Desulfobacterium species was found in fluids that discharge at “Baby bare seamount.” A possible explanation for this discrepancy might be that most of our isolates represent typical sediment inhabitants, which do not necessarily occur in the upper oceanic crust. However, our Deltaproteobacteria that were isolated from the lower sulfate-containing zone are facing similar physico-chemical conditions in the highly compacted sediments above the basement as in the crustal aquifer.
A close relation of deep marine with terrestrial aquifers is indicated by the cultivation of Dv. aespoeensis strains from the fluid-influenced layers. Dv. aespoeensis is the most abundant sulfate reducer within formation waters of deep terrestrial boreholes at the Aespoe hard rock laboratory in Sweden (Motamedi and Pedersen, 1998). Those aquifers are also inhabited by complex microbial communities that are comparable to those thriving within the ocean crust (Pedersen, 2000). The energetical constrains are similar and select for, e.g., iron-reducing bacteria, acetogens, methanogens, and sulfate reducers (Pedersen, 1997).
Our most frequently isolated strains from up to 260 m deep fluid-influenced sediments that are affiliated to Dv. indonesiensis also have close relatives within the deep terrestrial biosphere. Even though the type strain was originally isolated from a biofilm on a corroded ship off the Indonesian coast (Feio et al., 1998, 2000), relatives were obtained from porewater brines of a deep terrestrial gas-reservoir (Sass and Cypionka, 2004). Furthermore, these organisms are supposedly involved in iron corrosion as determined during a study on hydrogen-consuming microorganisms in oil facilities from Japan (Mori et al., 2010). Biocorrosive capabilities (Feio et al., 1998) of Dv. indonesiensis might be an indication for a crustal origin of this species as this process plays an important role in the weathering of basalts (Edwards et al., 2005). Under anoxic conditions, SRB, and especially Desulfovibrio species are responsible for the corrosion of metal surfaces in consuming cathodic hydrogen (Pankhania, 1988; Dinh et al., 2004). This process might occur in the habitat as well as in our metabolic tests. As all isolates deriving from the fluid-influenced zone were capable of using hydrogen as electron donor, they might even exhibit a chemolithoautotrophic life-mode in situ.
Chemolithoautotrophy within the deep biosphere
Autotrophic, hydrogen-consuming microorganisms were repeatedly detected in deep continental aquifers and can even outnumber heterotrophs (Stevens and McKinley, 1995). The assumption that autotrophy is also a common metabolic attribute within the crust at IODP Site U1301, is supported by the isolation of a novel member of the genus Archaeoglobus from a fluid-influenced sample of ODP Site 1226 (Steinsbu et al., 2010). Archaeoglobus sulfaticallidus sp. nov., is a thermophilic and facultatively lithoautotrophic sulfate reducer and was isolated from black rust formations on top of a leaking borehole seal.
Although there is no clear evidence available for lithoautotrophy within the subseafloor (Stevens, 1997), there are numerous studies that deal with hydrogen as suitable source for deep subsurface life. In these habitats, hydrogen can originate from many sources (Nealson et al., 2005) such as the fermentation of organic matter or mechanochemical processes due to the tectonic action of the Earth (Parkes et al., 2011), degassing from the Earth’s mantle during serpentinization of ultramafic rocks (McCollom and Bach, 2009), or even by radiolysis of water (Blair et al., 2007; D’Hondt et al., 2009). Furthermore, in the presence of sulfate, the oxidation of hydrogen is thermodynamically favored at high temperatures (Orcutt et al., 2010).
Thus, in many deep subsurface habitats, hydrogen might become apparently the biochemically most important electron donor and carbon dioxide is a ubiquitous carbon source. For example, both gases were found in micro-molar concentrations in deep igneous-rock aquifers (Pedersen, 1997) and deep aquifers of the Columbia river basalt which is located close to our investigated site (Stevens and McKinley, 1995). For both sites, the authors have proposed a model for a hydrogen-driven biosphere. They assume autotrophic acetogens to form acetate from hydrogen and carbon dioxide. Acetoclastic methanogens can utilize acetate to produce methane or hydrogenotrophic methanogens might directly use hydrogen and CO2. At relatively young ridge-flank systems, hydrogen-utilizing sulfate reducers will outcompete methanogens as sulfate is still available within the fluids.
Conclusion
Even though cultivation might not cover the whole microbial diversity of a given habitat, we were able to isolate and physiologically characterize indigenous microorganisms that are numerically and metabolically important for the marine deep subsurface. Thus, cultivation-based studies offer the opportunity to complement molecular techniques. In our study, the isolation of SRB from deep sediment layers was the precondition to answer questions concerning specific metabolic adaptations to the conditions at the sediment–basement interface.
The isolation of facultatively autotrophic sulfate reducers from near-basement layers strongly suggests that these organisms survive due to their capability of consuming hydrogen after organic compounds have been depleted or become too recalcitrant for microbial degradation. The continuous supply of sulfate from the aquifer below supports their viability within their respective sediment layers even after the separation from organic matter input at the seafloor due to sediment accumulation. When organic substrate availability from the ocean becomes a limiting factor, hydrogen becomes the most important electron donor.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Katharina Schmidt, Heiner Hartwich, Dewi Nasima, and Martin Mierzejewski assisted in molecular analyses and cultivation studies. Andrea Schlingloff is thanked for her help in DNA sequencing and Erhard Rhiel is acknowledged for preparing TEM photos. This study used samples and data provided by the Integrated Ocean Drilling Program from IODP Exp. 301. The work was financially supported by the German Research Foundation (DFG).
Footnotes
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. (1979). Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43, 260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S. J., Goodman K., Rochelle P. A., Marchesi J. R., Fry J. C., Weightman A. J., Parkes R. J. (1997). Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int. J. Syst. Bacteriol. 47, 515–521 [DOI] [PubMed] [Google Scholar]
- Barnes S. P., Bradbrook S. D., Cragg B. A., Marchesi J. R., Weightman A. J., Fry J. C., Parkes R. J. (1998). Isolation of sulfate-reducing bacteria from deep sediment layers of the Pacific Ocean. Geomicrobiol. J. 15, 67–83 10.1080/01490459809378066 [DOI] [Google Scholar]
- Batzke A., Engelen B., Sass H., Cypionka H. (2007). Phylogenetic and physiological diversity of cultured deep-biosphere bacteria from equatorial Pacific Ocean and Peru Margin sediments. Geomicrobiol. J. 24, 261–273 10.1080/01490450701456453 [DOI] [Google Scholar]
- Biddle J. F., House C. H., Brenchley J. E. (2005). “Enrichment and cultivation of microorganisms from sediment from the slope of the Peru Trench (ODP Site 1230),” in Proceedings of Ocean Drilling Program, Scientific Results, 201, eds Jørgensen B. B., D’Hondt S. L., Miller D. J. (College Station, TX: Ocean Drilling Program; ), Available at: http://www-odp.tamu.edu/publications/201_SR/107/107.htm [Google Scholar]
- Blair C., D’Hondt S., Spivack A., Kingsley R. (2007). Radiolytic hydrogen and microbial respiration in subsurface sediments. Astrobiology 7, 951–970 10.1089/ast.2007.0150 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Butterfield D. A., Jonasson I. R., Massoth G. J., Feely R. A., Roe K. K., Embley R. E., Holden J. F., McDuff R. E., Lilley M. D., Delaney J. R. (1997). Seafloor eruptions and evolution of hydrothermal fluid chemistry. Philos. Trans. R. Soc. Lond. A 355, 369–386 10.1098/rsta.1997.0013 [DOI] [Google Scholar]
- Cord-Ruwisch R. (1985). A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4, 33–36 10.1016/0167-7012(85)90005-3 [DOI] [Google Scholar]
- Cowen J. P. (2004). The microbial biosphere of sediment-buried oceanic basement. Res. Microbiol. 155, 497–506 10.1016/j.resmic.2004.03.008 [DOI] [PubMed] [Google Scholar]
- Cowen J. P., Giovannoni S. J., Kenig F., Johnson H. P., Butterfield D., Rappe M. S., Hutnak M., Lam P. (2003). Fluids from aging ocean crust that support microbial life. Science 299, 120–123 10.1126/science.1075653 [DOI] [PubMed] [Google Scholar]
- DeLong E. F. (2004). Microbial life breathes deep. Science 306, 2198–2200 10.1126/science.1107241 [DOI] [PubMed] [Google Scholar]
- Detmers J., Schulte U., Strauss H., Kuever J. (2001). Sulfate reduction at a lignite seam: microbial abundance and activity. Microb. Ecol. 42, 238–247 10.1007/s00248-001-1014-8 [DOI] [PubMed] [Google Scholar]
- D’Hondt S., Jørgensen B. B., Miller D. J., Batzke A., Blake R., Cragg B. A., Cypionka H., Dickens G. R., Ferdelman T., Hinrichs K. U., Holm N. G., Mitterer R., Spivack A., Wang G. Z., Bekins B., Engelen B., Ford K., Gettemy G., Rutherford S. D., Sass H., Skilbeck C. G., Aiello I. W., Guèrin G., House C. H., Inagaki F., Meister P., Naehr T., Niitsuma S., Parkes R. J., Schippers A., Smith D. C., Teske A., Wiegel J., Padilla C. N., Acosta J. L. S. (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2221 10.1126/science.1101155 [DOI] [PubMed] [Google Scholar]
- D’Hondt S., Rutherford S., Spivack A. J. (2002). Metabolic activity of subsurface life in deep-sea sediments. Science 295, 2067–2070 10.1126/science.1064878 [DOI] [PubMed] [Google Scholar]
- D’Hondt S., Spivack A., Pockalny R., Ferdelman T., Fischer J., Kallmeyer J., Abrams L., Smith D., Graham D., Hasiuk F., Schrum H., Stancin A. (2009). Subseafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. U.S.A. 106, 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh H. T., Kuever J., Mussmann M., Hassel A. W., Stratmann M., Widdel F. (2004). Iron corrosion by novel anaerobic microorganisms. Nature 427, 829–832 10.1038/nature02321 [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Bach W., McCollom T. M. (2005). Geomicrobiology in oceanography: mineral-microbe interactions at and below the seafloor. Trends Microbiol. 13, 449–459 10.1016/j.tim.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Bach W., Rogers D. R. (2003). Geomicrobiology of the ocean crust: a role for chemoautotrophic Fe-bacteria. Biol. Bull. 204, 180–185 10.2307/1543555 [DOI] [PubMed] [Google Scholar]
- Ehrhardt C. J., Haymon R. M., Lamontagne M. G., Holden P. A. (2007). Evidence for hydrothermal Archaea within the basaltic flanks of the East Pacific Rise. Environ. Microbiol. 9, 900–912 10.1111/j.1462-2920.2006.01211.x [DOI] [PubMed] [Google Scholar]
- Engelen B., Ziegelmüller K., Wolf L., Köpke B., Gittel A., Cypionka H., Treude T., Nakagawa S., Inagaki F., Lever M. A., Steinsbu B. O. (2008). Fluids from the oceanic crust support microbial activities within the deep biosphere. Geomicrobiol. J. 25, 56–66 10.1080/01490450701829006 [DOI] [Google Scholar]
- Epstein S. S. (2009). Microbial awakenings. Nature 457, 1083. 10.1038/4571083a [DOI] [PubMed] [Google Scholar]
- Expedition 301 Scientists. (2005). “Site U1301,” in Proceedings of Integrated Ocean Drilling Program, 301, eds Fisher A. T., Urabe T., Klaus A. and Expedition 301 Scientists (College Station, TX: Integrated Ocean Drilling Program Management International, Inc.) [Google Scholar]
- Feio M. J., Beech I. B., Carepo M., Lopes J. M., Cheung C. W. S., Franco R., Guezennec J., Smith J. R., Mitchell J. I., Moura J. J. G., Lino A. R. (1998). Isolation and characterisation of a novel sulphate-reducing bacterium of the Desulfovibriogenus. Anaerobe 4, 117–130 10.1006/anae.1997.0142 [DOI] [PubMed] [Google Scholar]
- Feio M. J., Beech I. B., Carepo M., Lopes J. M., Cheung C. W. S., Franco R., Guezennec J., Smith J. R., Mitchell J. I., Moura J. J. G., Lino A. R. (2000). “ Desulfovibrio indonesiensis corrig. sp. nov.,” in validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 75. Int. J. Syst. Evol. Microbiol. 50, 1415–1417 10.1099/00207713-50-4-1415 [DOI] [PubMed] [Google Scholar]
- Fisher A. T., Davis E. E., Hutnak M., Spiess V., Zühlsdorff L., Cherkaoui A., Christiansen L., Edwards K., Macdonald R., Villinger H., Mottl M. J., Wheat C. G., Becker K. (2003). Hydrothermal recharge and discharge across 50 km guided by seamounts on a young ridge flank. Nature 421, 618–621 10.1038/nature01352 [DOI] [PubMed] [Google Scholar]
- Furnes H., Staudigel H. (1999). Biological mediation in ocean crust alteration: how deep is the deep biosphere? Earth Planet. Sci. Lett. 166, 97–103 10.1016/S0012-821X(99)00005-9 [DOI] [Google Scholar]
- Heberling C., Lowell R. P., Liu L., Fisk M. R. (2010). Extent of the microbial biosphere in the oceanic crust. Geochem. Geophys. Geosyst. 11, 1–15 10.1029/2009GC002968 [DOI] [Google Scholar]
- Huber J. A., Johnson H. P., Butterfield D. A., Baross J. A. (2006). Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8, 88–99 10.1111/j.1462-2920.2005.00872.x [DOI] [PubMed] [Google Scholar]
- Hutnak M., Fisher A. T., Zühlsdorff L., Spiess V., Stauffer P. H., Gable C. W. (2006). Hydrothermal recharge and discharge guided by basement outcrops on 0.7–3.6 Ma seafloor east of the Juan de Fuca Ridge: observations and numerical models. Geochem. Geophys. Geosyst. 7, 1–36 10.1029/2006GC001242 [DOI] [Google Scholar]
- Imachi H., Aoi K., Tasumi E., Saito Y., Yamanaka Y., Saito Y., Yamaguchi T., Tomaru H., Takeuchi R., Morono Y., Inagaki F., Takai K. (2011). Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J. 5, 1913–1925 10.1038/ismej.2011.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F., Suzuki M., Takai K., Oida H., Sakamoto T., Aoki K., Nealson K. H., Horikoshi K. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of Okhotsk. Appl. Environ. Microbiol. 69, 7224–7235 10.1128/AEM.69.12.7224-7235.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. P., Baross J. A., Bjorklund T. A. (2006). On sampling the upper crustal reservoir of the NE Pacific Ocean. Geofluids 6, 251–271 [Google Scholar]
- Johnson H. P., Pruis M. J. (2003). Fluxes of fluid and heat from the oceanic crustal reservoir. Earth Planet. Sci. Lett. 216, 565. 10.1016/S0012-821X(03)00545-4 [DOI] [Google Scholar]
- Kaye J. Z., Sylvan J. B., Edwards K. J., Baross J. A. (2011). Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol. Ecol. 75, 123–133 10.1111/j.1574-6941.2010.00984.x [DOI] [PubMed] [Google Scholar]
- Kendall M. M., Liu Y., Sieprawska-Lupa M., Stetter K. O., Whitman W. B., Boone D. R. (2006). Methanococcus aeolicus sp. nov., a mesophilic, methanogenic archaeon from shallow and deep marine sediments. Int. J. Syst. Evol. Microbiol. 56, 1525–1529 10.1099/ijs.0.64216-0 [DOI] [PubMed] [Google Scholar]
- Köpke B., Wilms R., Engelen B., Cypionka H., Sass H. (2005). Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl. Environ. Microbiol. 71, 7819–7830 10.1128/AEM.71.12.7819-7830.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuever J., Könneke M., Galushko A., Drzyzga O. (2001). Reclassification of Desulfobacterium phenolicum as Desulfobacula phenolica comb. nov. and description of strain SaxT as Desulfotignum balticum gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 51, 171–177 [DOI] [PubMed] [Google Scholar]
- Lauro F. M., McDougald D., Thomas T., Williams T. J., Egan S., Rice S., DeMaere M. Z., Ting L., Ertan H., Johnson J., Ferriera S., Lapidus A., Anderson I., Kyrpides N., Munk A. C., Detter C., Han C. S., Brown M. V., Robb F. T., Kjelleberg S., Cavicchioli R. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533 10.1073/pnas.0903507106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-J., Wagner I., Brice M., Kevbrin V., Mills G., Romanek C., Wiegel J. (2005). Thermosediminibacter oceani gen. nov., sp. nov. and Thermosediminibacter litoriperuensis sp. nov., new anaerobic thermophilic bacteria isolated from Peru Margin. Extremophiles 9, 375–383 10.1007/s00792-005-0453-4 [DOI] [PubMed] [Google Scholar]
- Lever M. A., Alperin M., Engelen B., Inagaki F., Nakagawa S., Steinsbu B. O., Teske A., IODP Expedition 301 Scientists (2006). Trends in basalt and sediment core contamination during IODP Expedition 301. Geomicrobiol. J. 23, 517–530 10.1080/01490450600897245 [DOI] [Google Scholar]
- Lipp J. S., Morono Y., Inagaki F., Hinrichs K. U. (2008). Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994 10.1038/nature07174 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf K., Zink K. G., Birrien J. L., Toffin L. (2005). A quantitative assessment of pressure dependent adaptive changes in the membrane lipids of piezosensitive deep sub-seafloor bacterium. Org. Geochem. 36, 1459–1479 10.1016/j.orggeochem.2005.08.002 [DOI] [Google Scholar]
- McCollom T. M., Bach W. (2009). Thermodynamic constraints on hydrogen generation during serpentinization of ultramafic rocks. Geochim. Cosmochim. Acta 73, 856–875 10.1016/j.gca.2008.10.032 [DOI] [Google Scholar]
- Mikucki J. A., Liu Y., Delwiche M., Colwell F. S., Boone D. R. (2003). Isolation of a methanogen from deep marine sediments that contain methane hydrates, and description of Methanoculleus submarinus sp. nov. Appl. Environ. Microbiol. 69, 3311–3316 10.1128/AEM.69.6.3311-3316.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A., Coulouris G., Raytselis Y., Madden T. L., Agarwala R., Schäffer A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 15, 1757–1764 10.1093/bioinformatics/btn322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Tsurumaru H., Harayama S. (2010). Iron corrosion activity of anaerobic hydrogen-consuming microorganisms isolated from oil facilities. J. Biosci. Bioeng. 110, 426–430 10.1016/j.jbiosc.2010.04.012 [DOI] [PubMed] [Google Scholar]
- Motamedi M., Pedersen K. (1998). Desulfovibrio aespoeensis sp. nov., a mesophilic sulfate-reducing bacterium from deep groundwater at Äspö hard rock laboratory, Sweden. Int. J. Syst. Bacteriol. 48, 311–315 10.1099/00207713-48-1-311 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Inagaki F., Suzuki Y., Steinsbu B. O., Lever M. A., Takai K., Engelen B., Sako Y., Wheat C. G., Horikoshi K., IODP Expedition(301)Scientists (2006). Microbial community in black rust exposed to hot ridge flank crustal fluids. Appl. Environ. Microbiol. 72, 6789–6799 10.1128/AEM.01238-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H., Inagaki F., Takai K. (2005). Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): do they exist and why should we care? Trends Microbiol. 13, 405–410 10.1016/j.tim.2005.07.010 [DOI] [PubMed] [Google Scholar]
- Orcutt B. N., Bach W., Becker K., Fisher A. T., Hentscher M., Toner B. M., Wheat C. G., Edwards K. J. (2010). Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J. 5, 692–703 10.1038/ismej.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmann J., Tuschak C. (1997). Phylogeny and molecular fingerprinting of green sulfur bacteria. Arch. Microbiol. 167, 302–309 10.1007/s002030050448 [DOI] [PubMed] [Google Scholar]
- Pankhania I. P. (1988). Hydrogen metabolism in sulphate-reducing bacteria and its role in anaerobic corrosion. Biofouling 1, 27–47 10.1080/08927018809378094 [DOI] [Google Scholar]
- Parkes R. J., Cragg B. A., Bale S. J., Getliff J. M., Goodman K., Rochelle P. A., Fry J. C., Weightman A. J., Harvey S. M. (1994). Deep bacterial biosphere in Pacific Ocean sediments. Nature 371, 410–413 10.1038/371410a0 [DOI] [Google Scholar]
- Parkes R. J., Cragg B. A., Bale S. J., Goodman K., Fry J. C. (1995). A combined ecological and physiological approach to studying sulfate reduction within deep marine sediment layers. J. Microbiol. Methods 23, 235–249 10.1016/0167-7012(95)00036-K [DOI] [Google Scholar]
- Parkes R. J., Linnane C. D., Webster G., Sass H., Weightman A. J., Hornibrook E. R. C., Horsfield B. (2011). Prokaryotes stimulate mineral H2 formation for the deep biosphere and subsequent thermogenic activity. Geology 39, 219–222 10.1130/G31598.1 [DOI] [Google Scholar]
- Pedersen K. (1997). Microbial life in deep granitic rock. FEMS Microbiol. Rev. 20, 399–414 10.1111/j.1574-6976.1997.tb00325.x [DOI] [Google Scholar]
- Pedersen K. (2000). Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol. Lett. 185, 9–16 10.1111/j.1574-6968.2000.tb09033.x [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S., Sass H., Langner H., Schumann P., Kroppenstedt R. M., Spring S., Overmann J., Rosenzweig R. F. (2006). Desulfosporosinus lacus sp nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. Int. J. Syst. Evol. Microbiol. 56, 2729–2736 10.1099/ijs.0.63610-0 [DOI] [PubMed] [Google Scholar]
- Roussel E. G., Bonavita M. A. C., Querellou J., Cragg B. A., Webster G., Prieur D., Parkes R. J. (2008). Extending the sub-sea-floor biosphere. Science 320, 1046–1046 10.1126/science.1154545 [DOI] [PubMed] [Google Scholar]
- Rouxel O., Ono S., Alt J., Rumble D., Ludden J. (2008). Sulfur isotope evidence for microbial sulfate reduction in altered oceanic basalts at ODP Site 801. Earth Planet. Sci. Lett. 268, 110–123 10.1016/j.epsl.2008.01.010 [DOI] [Google Scholar]
- Santelli C. M., Orcutt B. N., Banning E., Bach W., Moyer C. L., Sogin M. L., Staudigel H., Edwards K. J. (2008). Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656 10.1038/nature06899 [DOI] [PubMed] [Google Scholar]
- Sass H., Cypionka H. (2004). Isolation of sulfate-reducing bacteria from the terrestrial deep subsurface and description of Desulfovibrio cavernae sp. nov. Syst. Appl. Microbiol. 27, 541–548 10.1078/0723202041748181 [DOI] [PubMed] [Google Scholar]
- Smith A., Popa R., Fisk M., Nielsen M., Wheat C. G., Jannasch H. W., Fisher A. T., Becker K., Sievert S. M., Flores G. (2011). In situ enrichment of ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem. Geophys. Geosyst. 12, 1–19 10.1029/2011GC003689 [DOI] [Google Scholar]
- Steinsbu B. O., Thorseth I. H., Nakagawa S., Inagaki F., Lever M. A., Engelen B., Øvreås L., Pedersen R. B. (2010). Archaeoglobus sulfaticallidus sp. nov., a novel thermophilic and facultatively lithoautotrophic sulfate-reducer isolated from black rust exposed to hot ridge flank crustal fluids. Int. J. Syst. Evol. Microbiol. 60, 2745–2752 10.1099/ijs.0.016105-0 [DOI] [PubMed] [Google Scholar]
- Stevens H., Brinkhoff T., Simon M. (2005). Composition of free-living, aggregate-associated and sediment surface-associated bacterial communities in the German Wadden Sea. Aquat. Microb. Ecol. 38, 15–30 10.3354/ame038015 [DOI] [Google Scholar]
- Stevens T. (1997). Lithoautotrophy in the subsurface. FEMS Microbiol. Rev. 20, 327–337 10.1111/j.1574-6976.1997.tb00318.x [DOI] [Google Scholar]
- Stevens T. O., McKinley J. P. (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270, 450–455 10.1126/science.270.5235.450 [DOI] [Google Scholar]
- Straskrabová V. (1983). The effect of substrate shock on populations of starving aquatic bacteria. J. Appl. Bacteriol. 54, 217–224 10.1111/j.1365-2672.1983.tb02610.x [DOI] [Google Scholar]
- Süss J., Engelen B., Cypionka H., Sass H. (2004). Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol. Ecol. 51, 109–121 10.1016/j.femsec.2004.07.010 [DOI] [PubMed] [Google Scholar]
- Thorseth I. H., Torsvik T., Furnes H., Muehlenbachs K. (1995). Microbes play an important role in the alteration of oceanic crust. Chem. Geol. 126, 137–146 10.1016/0009-2541(95)00114-8 [DOI] [Google Scholar]
- Toffin L., Bidault A., Pignet P., Tindall B. J., Slobodkin A., Kato C., Prieur D. (2004a). Shewanella profunda sp. nov., isolated from deep marine sediment of the Nankai trough. Int. J. Syst. Evol. Microbiol. 54, 1943–1949 10.1099/ijs.0.03007-0 [DOI] [PubMed] [Google Scholar]
- Toffin L., Webster G., Weightman A. J., Fry J. C., Prieur D. (2004b). Molecular monitoring of culturable bacteria from deep-sea sediment of the Nankai trough, Leg 190 Ocean Drilling Program. FEMS Microbiol. Ecol. 48, 357–367 10.1016/j.femsec.2004.02.009 [DOI] [PubMed] [Google Scholar]
- Toffin L., Zink K., Kato C., Pignet P., Bidault A., Bienvenu N., Birrien J. L., Prieur D. (2005). Marinilactibacillus piezotolerans sp. nov., a novel marine lactic acid bacterium isolated from deep sub-seafloor sediment of the Nankai trough. Int. J. Syst. Evol. Microbiol. 55, 345–351 10.1099/ijs.0.63236-0 [DOI] [PubMed] [Google Scholar]
- Wheat C. G., Elderfield H., Mottl M. J., Monnin C. (2000). Chemical composition of basement fluids within an oceanic ridge flank: implications for along-strike and across-strike hydrothermal circulation. J. Geophys. Res. 105, 13437–13447 10.1029/2000JB900070 [DOI] [Google Scholar]
- Wheat C. G., Mottl M. J. (1994). Hydrothermal circulation, Juan de Fuca Ridge eastern flank: factors controlling basement water composition. J. Geophys. Res. 99, 3067–3080 10.1029/93JB01612 [DOI] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F., Bak F. (1992). “Gram-negative mesophilic sulfate-reducing bacteria,” in The Prokaryotes, eds Balows A., Trüper H. G., Dworkin M., Harder W., Schleifer K. H. (New York: Springer-Verlag; ), 3352–3378 [Google Scholar]
- Wilms R., Köpke B., Sass H., Chang T. S., Cypionka H., Engelen B. (2006a). Deep biosphere-related bacteria within the subsurface of tidal flat sediments. Environ. Microbiol. 8, 709–719 10.1111/j.1462-2920.2005.00949.x [DOI] [PubMed] [Google Scholar]
- Wilms R., Sass H., Köpke B., Köster J., Cypionka H., Engelen B. (2006b). Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters. Appl. Environ. Microbiol. 72, 2756–2764 10.1128/AEM.72.4.2756-2764.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]