Abstract
Due to its highly reproducible and quantitative nature, and minimal requirements for sample preparation or separation, 1H nuclear magnetic resonance (NMR) spectroscopy is widely used for profiling small-molecule metabolites in biofluids. However 1H NMR spectra contain many overlapped peaks. In particular, blood serum/plasma and diabetic urine samples contain high concentrations of glucose, which produce strong peaks between 3.2 ppm – 4.0 ppm. Signals from most metabolites in this region are overwhelmed by the glucose background signals and become invisible. We propose a simple “Add to Subtract” background subtraction method, and show that it can reduce the glucose signals by 98% to allow retrieval of the hidden information. This procedure includes adding a small drop of concentrated glucose solution to the sample in the NMR tube, mixing, waiting for an equilibration time, and acquisition of a second spectrum. The glucose-free spectra are then generated by spectral subtraction using Bruker Topspin software. Subsequent multivariate statistical analysis can then be used to identify biomarker candidate signals for distinguishing different types of biological samples. The principle of this approach is generally applicable for all quantitative spectral data and should find utility in a variety of NMR-based mixture analyses as well as in metabolite profiling.
Keywords: 1H NMR, metabolomics, metabolite profiling, glucose, signal suppression, mixture analysis, blood, urine
Introduction
Metabolomics, also referred to as metabonomics1 or metabolic profiling, is the study of the concentrations and fluxes of small molecules in biological systems in different states. Metabolomics provides an indispensable complement to the fields of genomics and proteomics for understanding complex biochemical networks and for providing insight to many important problems, such as human disease diagnosis, prognosis and therapeutic development.2–13
Nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) are the two major analytical techniques used in the field of metabolomics to characterize many metabolites in parallel and deliver a wealth of information with high throughput.6, 14–20 NMR provides highly reproducible and relatively easily quantified data,21, 22 and usually requires minimal or no sample preparation or separation. NMR analysis is also non-destructive to the sample. Extensive application of 1H NMR has been focused on the study of small molecules in biofluids to solve various problems in metabolomics. Urine and blood serum/plasma are the most studied, because they both contain hundreds to thousands of detectable metabolites and can be easily obtained.5, 15, 23 Typically, a urine or blood 1H NMR spectrum can be acquired within a few minutes, and several tens of metabolites can be identified and quantified. The application of multivariate statistical pattern recognition methods to 1H NMR spectra of biofluids has greatly improved the ability to extract useful information such as potential biomarkers of disease.
The 1H NMR-based metabolomics is however seriously limited by the spectral complexity and overlap. In a typical 1H NMR spectrum, signals are observed between 0 ppm and 9 ppm and most of them are crowded into two spectral regions that roughly span 5 ppm (0.8 – 4.4 ppm, 6.8 – 8.0 ppm). Due to serious signal overlap, the identification and quantification of certain metabolites of interest often becomes impossible even with the help of chemometric methods.24–28 2D NMR experiments29–39 provide extra resolution, reduce the overlap and partly circumvent this bottleneck, however they are typically lower in either sensitivity or throughput. An alternative is to use selective TOCSY experiments;27, 28, 39 however, these experiments usually detect a few targeted metabolites.
D-Glucose, a six-carbon monosaccharide, is probably the most important and most studied carbohydrate in biology. Produced through photosynthesis by living plants, D-glucose serves as the energy source for living cells of both prokaryotes and eukaryotes through oxidative metabolism. Since L-glucose does not occur naturally and has no biological activity, D-glucose is very often referred to as glucose. Upon dissolution in water, glucose starts to form hemiacetal ring structures; equilibrium is then established between α-D-glucopyranose and β-D-glucopyranose. As the result, 36% of the glucose is in α-form, 64% is in β-form and 0.02% is open-chain. Multiple J-couplings exist among the 1H nuclei in all forms of glucose and produce very complicated 1H NMR signals.40 Since the concentrations of glucose in human blood (3.9 - 6.1 mM)41 and diabetic urine samples (~ 10 mM)42 are much higher than the concentrations of most other metabolites, the spectral region of 3.2 - 4.0 ppm is usually dominated by glucose signals. The signals of other metabolites in this region therefore cannot be well studied, unless the glucose signals can be removed or sufficiently suppressed. Similar situations are seen in the 1H NMR spectra of fruit juice, wines, milk and tissues.
Solvent suppression has been an important topic in 1H NMR sequence development. Most solvent suppression methods can successfully reduce the signal of water by more than 1000-fold and allow the detection of analytes at µM levels.43–46 These methods are based on the difference between the resonance frequencies of solvent and solute, although differences in relaxation times have been used as well. The application of solvent-suppression type pulse sequences that could remove the glucose background would also remove the signals of interest.
Options to achieve the goal of removing glucose signals without affecting other signals include the physical removal of glucose from the sample and background subtraction. Modern separation techniques such as ion chromatography and capillary electrophoresis can separate glucose from the rest of the biofluid.47 However, this approach would involve various sample treatments to meet the compatibility requirements of the separation method and likely a solvent change following the separation to ensure good quality NMR spectra. Along with the separation, these steps add to the complexity and labor involved in the analysis, greatly reduce the throughput, and introduce more errors into the spectral data. Using certain compounds or microorganisms to consume glucose through chemical or biological processes is likely accompanied with the production or consumption of other small molecules and can impact the results unpredictably.
A background subtraction that has been widely used for acquiring UV-Vis, IR and Raman spectra would seem to provide an easier and faster solution. The removal of complex signals from the NMR spectrum is however quite different from its conventional counterparts in optical spectroscopy. Because of the slow relaxation times in NMR (measured in seconds instead of picoseconds), 1H NMR signals have very narrow line widths in contrast to UV-Vis and IR signals, usually a few Hz that correspond to less than 0.01 ppm for a typical modern NMR spectrometer. This characteristic makes NMR spectral subtraction very sensitive to small variations in frequencies and line shapes. These small variations, mostly related to matrix effects, compromise the conventional background subtraction approach that uses the spectrum of a standard glucose sample, and produce artifacts of considerable levels. In contrast, we report an improved background subtraction method to effectively reduce the glucose signals by approximately 98%, and thereby reveal the signals of other metabolites in the spectral region of 3.2 – 4.0 ppm. This approach can be applied broadly and should aid in a number of spectral analysis problems in NMR.
Theoretical Background
The proposed approach “Add to Subtract” is based on the insight that the background peaks from an analyte such as glucose can be distinguished from other signals by (1) adding the analyte to the sample, and (2) computationally interpreting the difference between the NMR spectra with and without the added analyte. The approach assumes that the external introduction of the background analyte does not change the sample matrix, and therefore does not change the line shapes and the frequencies of the signals. This can be achieved experimentally by adding highly concentrated analyte, preserving constant buffer concentrations, and using the same NMR tubes for both samples.
The proposed mathematical interpretation of the acquired spectra is as follows. We denote Ii as the intensity of the spectrum of the original sample at a frequency i. In regions of the spectra where the signals from glucose and from other metabolites overlap, the intensity can be decomposed into the contributions of these sources as in Eqn. 1, where Gi are the contributions of glucose, and Mi are the contributions of other metabolites and the main quantities of interest. εi are the independent non-systematic stochastic deviations from the signals, i.e., noise.
| (1) |
We then denote Ii' as the intensity at frequency i of the spectrum of the sample with added glucose. The intensity is now decomposed as in Eqn. 2, and is affected by a factor a, which represents the added background analyte. It is also affected by a factor b, which represents a possible slight change in the sensitivity between the two 1H NMR spectra due to the tiny changes in sample volume and instrument variation. ε'i are the independent non-systematic stochastic deviations in the second spectrum.
| (2) |
The factors a and b are assumed constant across all i. If they were known, the signals of interest could be determined by combining the systematic parts of Eqn. 1 and 2, eliminating Gi and solving for Mi to yield
| (3) |
where ‘^’ indicates that the quantity is estimated from the data. In practice a and b are unknown, and also need to be estimated from the spectra. This can be done using regions of the spectra with no glucose, and with the signal from glucose alone, as follows.
To estimate b, consider a set Met of frequencies i with metabolite signal only. In these regions, Eqn. 1 and 2 simplify to
| (4) |
The factor b can be estimated using a standard least squares procedure, which minimizes the sum of the squared distances between the systematic parts of the two spectra over all the frequencies.
| (5) |
To estimate a, consider a set Glu of frequencies i with glucose signal only. In these regions, Eqn. 1 and 2 simplify to
| (6) |
The factor a can now also be estimated by the least squares procedure, while plugging in the estimator b̂ from Eqn. (5)
| (7) |
As the result, the final estimate of the metabolite signal Mi is obtained as
| (8) |
and the final estimate of the glucose signal Gi is obtained as
| (9) |
In practice, manual iterative fitting was used rather than least squares regression because of the extraordinary linear detection of 1H NMR across the entire spectral width. Manual fitting does not estimate the individual values of a and b, but the values of ab and b(a − 1) to yield the “glucose-free” spectrum. The spectrum of the original sample was scaled by a factor z interactively to minimize glucose signals zIi − Ii' (i ∈ Glu) in the difference spectrum. The minimum was reached when z = âb̂,
| (10) |
and the spectral intensities at all frequencies with signals of other metabolites present then became b̂(â − 1)M̂.
| (11) |
The scaling effect of b̂(â − 1) was easily removed by a normalization step with respect to a clean signal not overlapping with glucose signals, such as the chemical shift reference. This iterative fitting procedure was done using Bruker spectrometer software Topspin without the need of additional software. More details are provided in the experimental section.
Experimental Section
Chemical and Biological Samples
Potassium phosphate monobasic, potassium phosphate dibasic, D-(+)-glucose were purchased from Mallinckrodt Baker Inc. (Phillipsburg, NJ); deuterium oxide was purchased from Cambridge Isotope Laboratories (Andover, MA). Other chemicals were obtained from Sigma-Aldrich Co. (St. Louis, MO). All chemicals were used without further purification. Sample set 1: A human urine sample from a healthy volunteer was obtained in accordance with the Institutional Review Board protocol at Purdue University, and was diluted five-fold; glucose was added to reach 10 mM in order to mimic a typical sample of a subject with diabetes. Sample set 2: A human serum sample was purchased from Innovative Research (Novi, MI) and aliquoted. One aliquot was used without further processing for CPMG experiments. For comparison, proteins in a second aliquot were removed by adding methanol to the serum in a 2:1 (v/v) ratio, vortexing and incubation at −20 °C for 20 min. After centrifugation at 13200 g for 10 min, the clear supernatant was dried in vacuum and then re-dispersed in water. Sample set 3: A human urine sample from a healthy volunteer was obtained in accordance with the Institutional Review Board at Purdue University. Sodium azide was added to every sample to result in a 0.1% (wt. /vol.) concentration in order to inhibit bacterial growth. All samples were stored frozen at −80 °C until analyzed.
NMR Spectroscopy
All NMR experiments were carried out on a Bruker DRX-500 spectrometer equipped with an inverse room temperature triple axis gradient probe operating at 298 K. Urine samples were analyzed using water pre-saturation (Pre-Sat). Serum samples were analyzed using the Carr-Purcell-Meiboom-Gill (CPMG) sequence. The inter-scan delay/Pre-Sat time was 3 s, the spectral width was 5 kHz; and the size of the FID was set to 64 k points unless otherwise noted. The number of scans (64), receiver gain, and excitation pulse angle (45° for Pre-Sat experiments, 90° for the CPMG experiment) were kept the same for all samples. The CPMG pulse sequence consisted of 100 spin echoes of 500 µs each. To ensure good quality spectra, every sample was manually tuned and matched to minimize the RF reflection, and then automatically shimmed three times using gradient 1H shimming after the predefined shim file was loaded.
Experimental Procedure for “Add to Subtract”
Phosphate buffer (1 M, pH = 7.4) was added to all biological samples to reach a final concentration of 100 mM, and D2O containing 0.5 mM 3-(trimethylsilyl)-1-propanesulfonic acid-d6 sodium salt (DSS) was added to reach a final concentration of 10% (v/v). Glucose stock solution (2.5 M) was made with the same concentrations of phosphate buffer and D2O. The 1H NMR spectrum of a glucose-containing sample (serum or urine) was first acquired to find the glucose level. An appropriate volume of 2.5 M glucose stock solution was then added to the sample in the 5 mm standard NMR tube to roughly double the glucose level. The NMR tube was then capped, inverted and shaken a few times and set at room temperature for at least 2 hrs to equilibrate different forms of glucose (which interconvert according to the glucose mutarotation kinetics). A second 1H NMR spectrum was then acquired with the same settings. Both spectra were phase- and baseline-corrected and then superimposed under the “multiple display” mode of Bruker Topspin 3.0 program with their difference spectrum displayed. The spectra were aligned with respect to the glucose signal at 3.83 ppm; a scout scaling (increase the intensity of the first spectrum until the difference spectrum showed no negative signals) then removed the majority of glucose signals and spectral regions with glucose components only were visually determined. After magnifying these regions, the residual glucose signals were further reduced by fine shifting and scaling. The resulted difference spectrum was saved as the “glucose-free” spectrum, and thus provided the value for “z” as described in the Theoretical Background section. This subtraction procedure is shown in Scheme 1. Similarly, the spectrum of “glucose in the biological sample” can be obtained by minimizing the signals of other metabolites in the difference spectrum, and thereby provides the scaling factor “b”. For quantitative analysis of the Sample Set 3, the difference spectra were normalized with respect to the internal standard using MestReNova 5.3.1 (Mestrelab Research SL, Santiago de Compostela, Spain) to account for the scaling during the generation of the difference spectra; and saved in the format of ASCII text for statistical analysis. NMR signals were tentatively assigned based on chemical shifts and confirmed using spiked synthetic standards.
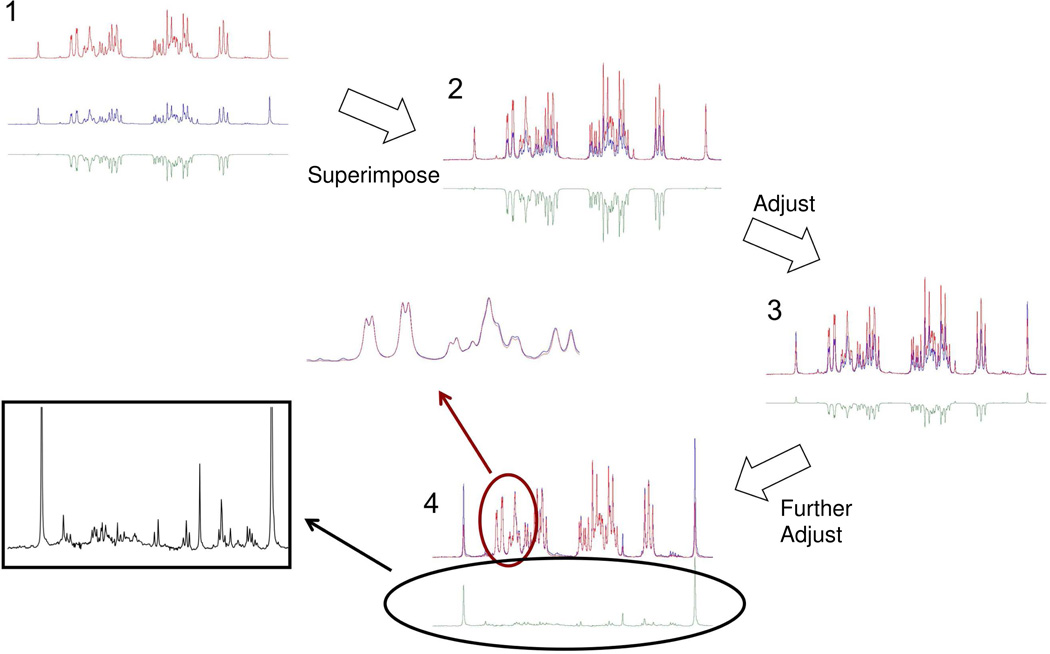
Scheme 1.
“Add to Subtract” interactive spectral subtraction. 1: The two spectra acquired before (blue, middle) and after (red, upper) adding external glucose to the biological sample were processed using the Bruker Topspin program multiple display mode with their difference spectrum (green, lower) displayed. 2: The two spectra were superimposed. 3: By interactively scaling and shifting the spectrum with no external glucose (blue), the glucose background in the difference spectrum (green) is minimized. 4: The residual glucose signal has been minimized in the final spectrum. Inset shows the “glucose free” spectrum (boxed) and a partial spectrum of the glucose region showing the matched intensity between the original spectrum and the glucose added spectrum.
Subtraction of Glucose Background Through Interactive Curve-fitting
The phased and baseline corrected spectrum of a glucose-containing sample was processed using Chenomx NMR Suite Processer to calibrate its shim and chemical shape profile based on the signal of DSS. The file was saved and opened in Chenomx NMR Suite Profiler for removal of glucose signals. The concentration and cluster frequencies were first automatically fitted and then fine-tuned manually for the best subtraction result.
Subtraction of Glucose Background Using a Pure Glucose Spectrum
The 1H NMR spectrum of a 5 mM glucose solution with the same added contents (0.1% sodium azide, 100 mM phosphate buffer, 10% D2O, 0.05 mM DSS) as the glucose-containing biological sample was acquired. After phasing and baseline correction, the two spectra were superimposed along with their difference spectrum displayed. Similar to the “Add to Subtract” procedure, the glucose signals in the difference spectrum were minimized by interactively shifting and scaling the pure glucose spectrum; the resulting difference spectrum was then saved as the “glucose-free” spectrum.
Principal Component Analysis
The 1H NMR spectra of each data set were aligned with respect to the internal reference DSS signal. The full resolution spectral data points between 3.15 ppm and 4.0 ppm, where most glucose signals appear, were mean-centered and subject to principal component analysis (PCA) using EigenVector PLS_ToolBox software package (Version 4.1.1, Eigenvector Research, Inc., Wenatchee, WA).
Results
We evaluated the performance of the proposed approach “Add to Subtract” on three sets of ad-hoc biological samples with known components.
Sample set 1, human urine
An ad hoc sample was created artificially from a urine sample of a healthy individual. It was diluted, and mixed with a high concentration of glucose, to mimic a typical urine sample of a diabetic patient. The 1H NMR spectrum of this sample is shown in Figure 1(a). A well-performing method will correctly remove the background glucose and correctly uncover the spectrum of the diluted urine sample free of glucose shown in Figure 1(b), which is therefore viewed as the control.
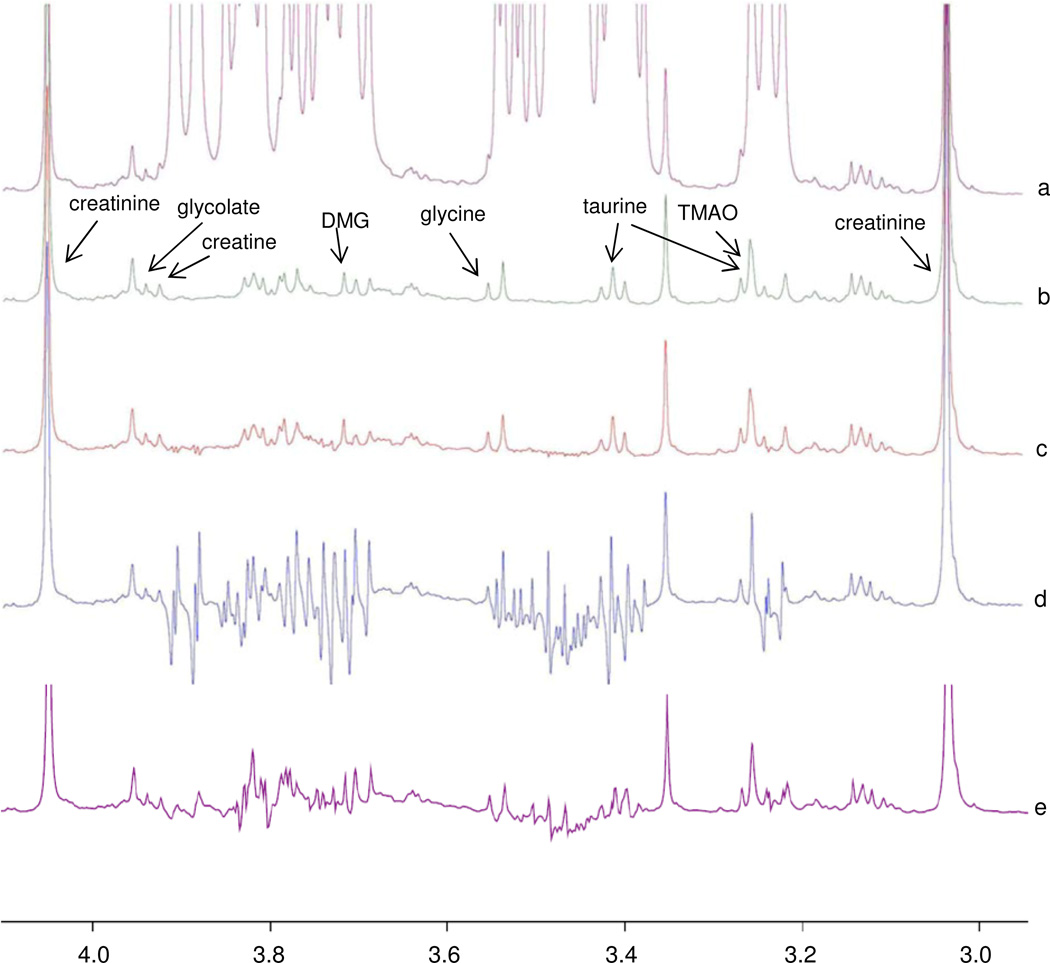
Figure 1.
Evaluation of the proposed NMR “Add to Subtract” approach on sample set 1. (a) Spectrum of a diluted urine sample with a high concentration of glucose, that mimics a typical sample from a patient with diabetes; (b) Spectrum of the original healthy sample (control); (c) Spectrum in (a), after removing the glucose background by “Add to Subtract”; (d) Spectrum in (a), after removing the glucose background by subtraction of a pure glucose solution with the same buffer and D2O concentrations; (e) Spectrum in (a), after removing the glucose background by semi-automatic curve-fitting using Chenomx NMR Suite 7.1.
Figures 1(c)–(e) compare the performance of “Add to Subtract” to the performance of direct background subtraction using a pure glucose spectrum, or using a curve-fitted glucose spectrum by Chenomx. As can be seen, approximately a 98% reduction of the glucose signals (estimated from the residues at 3.9 and 3.5 ppm) was achieved, which produced a spectrum highly similar to the control spectrum. This was more than ten times better than the results from direct subtractions using a pure glucose spectrum, or a curve-fitted glucose spectrum generated using Chenomx, in which the residual glucose signals were 10% to 50% of the original levels, and still overshadowed all other signals in the same region.
The key to the successful background subtraction observed in Figure 1(c) was the almost identical chemical shifts and line shapes of glucose signals in the two spectra used by “Add to Subtract.” In order to achieve optimal results, special care was taken to produce highly similar sample matrices and minimize possible variation. First, a concentrated glucose solution (2.5 M) was used to spike the samples, rather than a diluted glucose solution or glucose crystals, such that the sample volume and concentrations of other metabolites did not vary by more than 0.5%. Second, the concentrations of buffer and D2O were the same for all samples and the glucose solution. Third, the same NMR sample tube was used for acquiring the two spectra for each biological sample. And fourth, the magnetic field was automatically shimmed using 1H gradient shimming three times to achieve nearly identical line shapes for the two spectra.
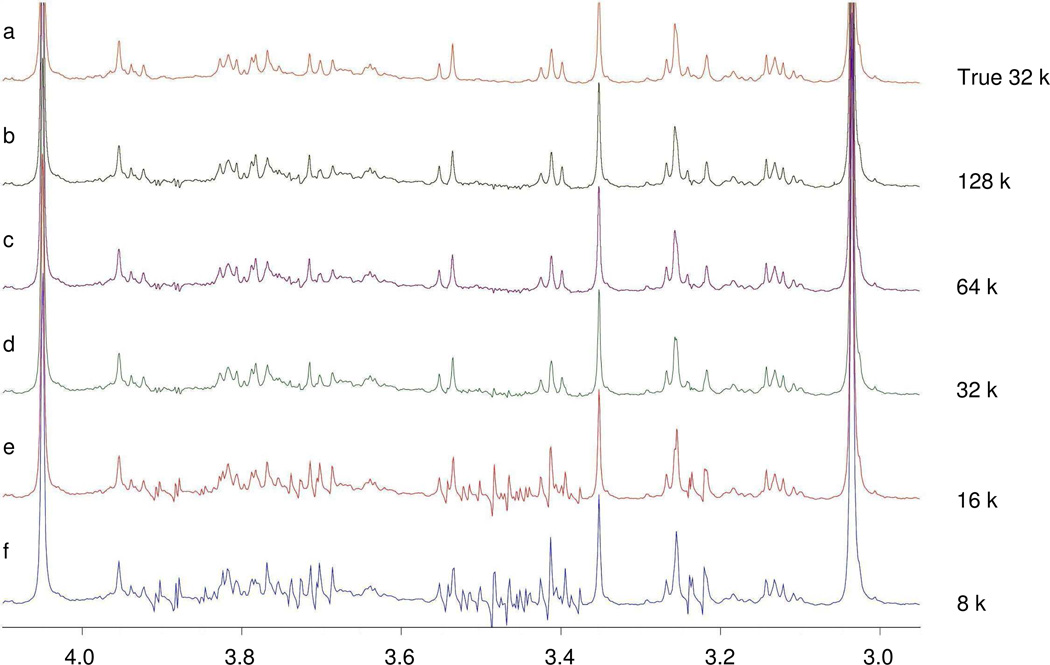
High spectral resolution was also found to be a critical factor in obtaining good results. To gain insight into the impact of spectral resolution, we re-applied the procedure to the same two FIDs utilized by “Add to Subtract” above, but processed at different digital resolutions. As shown in Figure 2, the 16 k and 8 k data sizes of the 5 kHz spectra did not provide enough resolution (3.2 and 1.6 spectral data points per Hz, respectively) for optimal glucose background removal. At the low resolution, there were too few data points to allow accurate alignment of signals; therefore subtractions were performed between data points at different frequencies. When the digital resolution was raised to 32 k or above, the intensities of residual glucose signals were reduced by 60–90%. The higher resolution provides a more accurate and precise signal depiction and results in better alignment for spectral subtraction.
Figure 2.
Impact of spectral resolution on the performance of “Add to Subtract” on sample set 1. (a) Spectrum of the original healthy sample (control); (b)–(f) Metabolite signal uncovered by “Add to Subtract” for the original FIDs (64 k data size, 5 k Hz spectral width) with different digital resolution. Spectra with the two lowest resolutions were enhanced by zero filling the FID before Fourier transformation. The 16 k and 8 k spectra did not provide enough resolution for correct glucose background removal.
Sample set 2, human serum
The second set of ad hoc biological samples were used to evaluate the ability of the method to subtract the background in serum. A major difference between blood and urine samples for NMR analysis is that blood contains large concentrations of macromolecules such as proteins and lipids, and the macromolecules produce broad lines in the 1H NMR spectrum. The signals of small-molecule metabolites bound to macromolecules are also subject to broadening. Two experimental approaches allowed us to address these artifacts. First, the proteins can be effectively removed from serum samples using various precipitation or filtration methods. Second, the CPMG pulse sequence suppresses the broad lines, and is the more convenient choice for acquiring the spectra of small-molecule metabolites in serum samples.48 Spectra acquired using these two approaches are generally quite similar;16 however some differences exist. For example, small molecules can be released during the protein removal step and become visible by NMR.
Here we evaluate the performance of “Add to Subtract” using both the Pre-sat spectra of a protein-precipitated sample and the CPMG spectra of the intact sample. Figures 3 (a) and (e) show that similar spectra were observed by protein precipitation and by protein signal suppression. Since glucose is present in every serum sample, it is impossible to find a natural glucose-free blood that can be viewed as a control. However, a successful background removal should produce similar results for both Pre-sat and CPMG spectra, and should have a low level of residual glucose signals and spectral artifacts. Figures 3 (b)–(d) and (f)–(h) compare the performance of “Add to Subtract” to the performances of direct subtraction and curve-fitted background subtraction for these spectra. In Figures 3 (b) and 3 (f), it can be observed that “Add to Subtract” left much less residual glucose signal than either the direct subtraction approach in Figures 3 (c) and 3 (g), or the curve-fitted subtraction approach in Figures 3 (d) and 3 (h). Therefore, it is compatible with the CPMG sequence and, although protein precipitation also provides excellent results, the protein precipitation step is not required.
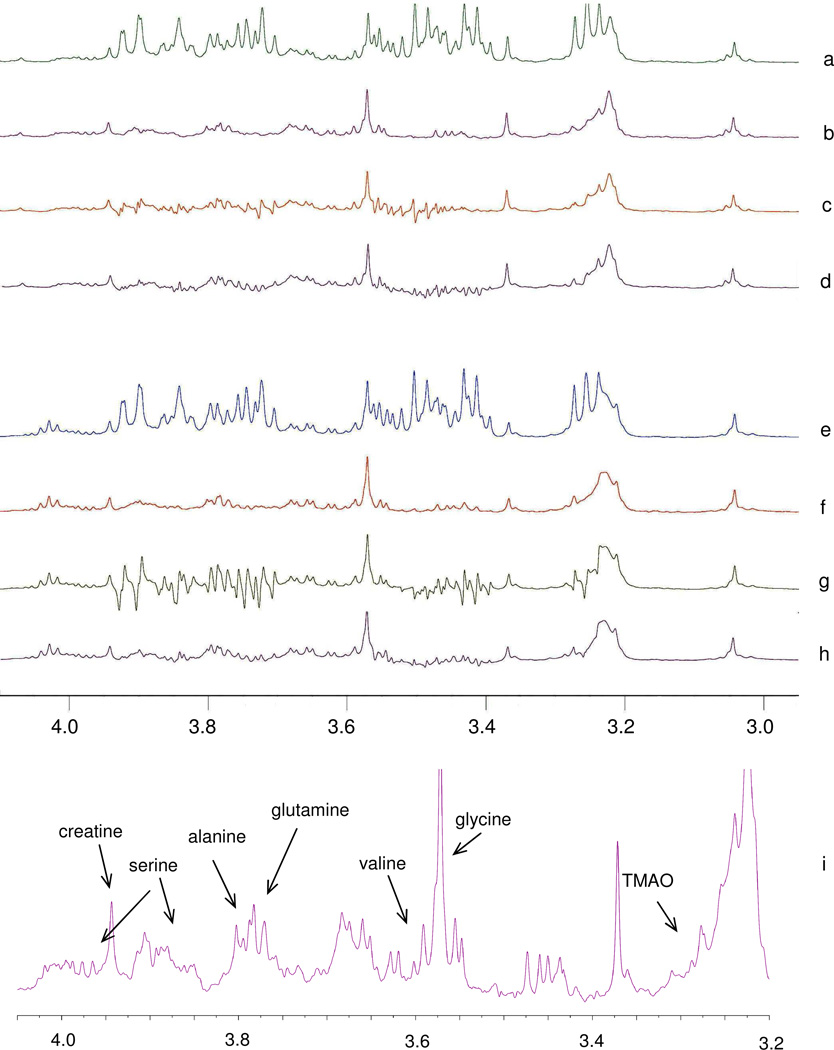
Figure 3.
Evaluation of the proposed “Add to Subtract” approach on sample set 2. (a) Pre-sat spectrum of the protein-precipitated human blood serum; (b) Spectrum after removing the glucose background by “Add to Subtract”; (c) Spectrum after removing the glucose background by subtraction of the spectrum of a pure glucose solution; (d) Spectrum after removing the glucose background by semi-automatic curve-fitting using Chenomx NMR 5.1 professional; (e) CPMG spectrum of an intact serum sample without protein preciptation; (f)–(h): results of background subtraction, as in (b)–(d), but applied to the spectrum in (e); (i): a magnified view of (b) with peak assignments. Subtraction using Chenomx NMR Suite 7.1 evaluation resulted in spectra similar to (d) and (h), but with higher residual glucose signals.
Sample set 3, multivariate analysis
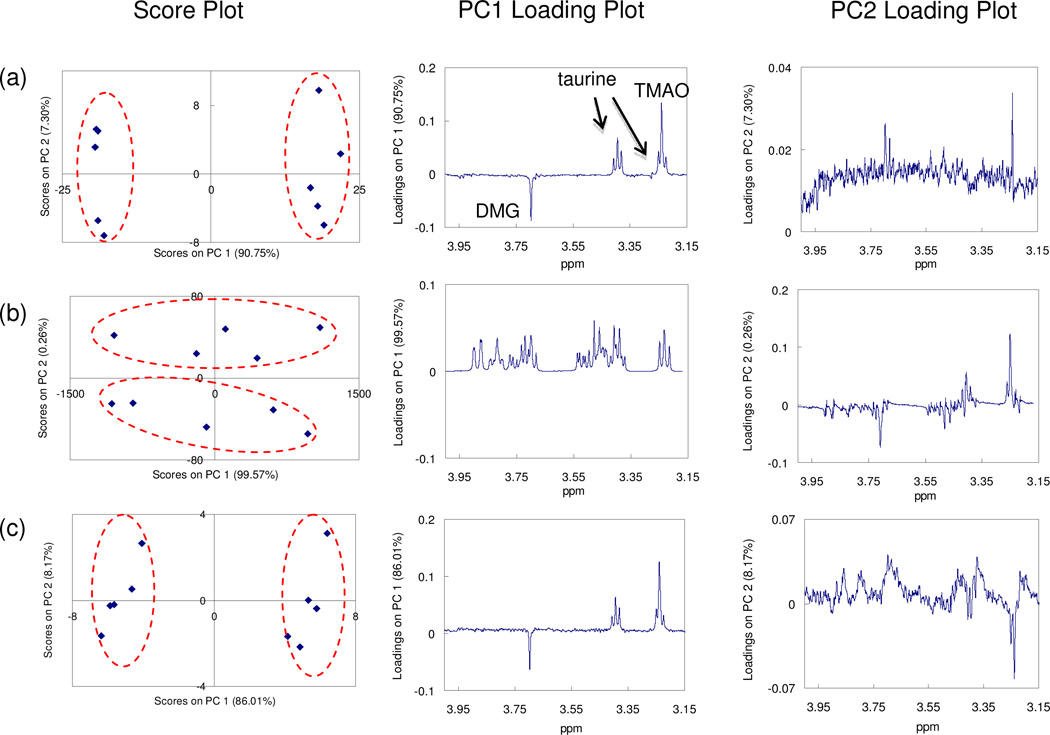
The third set of ad hoc biological samples evaluates the impact of removing glucose signals on the downstream multivariate statistical analysis. The set contains two groups of 5 samples. All the samples share a complex background of a urine sample from a healthy subject. The five samples in Group 1 were spiked with N-dimethylglycine (DMG) to a final concentration of 3.4 µM ± 4.8%, and the five samples in Group 2 were spiked with taurine (135 µM ± 9.2%) and trimethylamine-N-oxide (TMAO) (20 µM ± 8.8%). All ten samples were spiked with glucose (15 mM ± 20%). 1H NMR spectra from the samples were processed with “Add to Subtract,” and then subjected to principal component analysis (PCA). As a control, a duplicate series of ten samples were prepared in the same manner, but free of any additional glucose.
Figure 4 compares the results of PCA for spectra of the control samples, and the glucose-rich samples before and after background subtraction. As can be seen, the control and the background-corrected set of samples are clearly separated along the first principal component (PC1). The loadings of PC1 indicated that the signals of the three spiked compounds (DMG, taurine and TMAO) drove the separation, and were accompanied with very low levels of noise. In contrast, the glucose-rich samples without background subtraction were not separated along PC1; the PC1 loading indicated a large variation in the glucose signals, and signals of the spiked compounds were found along PC2 as secondary variations with high noise levels.
Figure 4.
Evaluation of the proposed “Add to Subtract” approach on the urine spike-in (with added TMAO, DMG and taurine) sample set 3. Results of PCA for (a) spike-in control samples; (b) glucose-rich spike-in samples of interest before background subtraction; and (c) glucose-rich spike-in samples of interest after “Add to Subtract.”
Discussion
Spectral subtraction has been a widely used approach to remove interfering background signals in spectroscopic analyses. The challenges in achieving a successful background subtraction of glucose from NMR spectra are the slight changes in chemical shift and line shape/width observed in a complex biological sample versus a buffered solution, which are mostly due to a variety of matrix effects (pH, ionic strength, complex formation, etc.). Standard addition is a common practice in many chemical analyses, including atomic absorption spectroscopy (AAS) and gas chromatography (GC) analysis to eliminate matrix effects and achieve accurate quantitation. The same concept was applied here to eliminate the matrix effect present in the 1H NMR signals of glucose. When more glucose is added to the glucose-containing biological sample, glucose signals in the 1H NMR spectrum will show higher intensities but the same chemical shifts and line shapes. By careful subtraction as described above, a difference signal can be obtained that is free of any glucose signal. Because of the highly quantitative nature and very broad dynamic range inherent in 1H NMR spectroscopy, only a single addition is sufficient, instead of a series of additions of glucose. The difference between the two spectra containing different amounts of glucose is therefore the spectrum of “glucose in the biological sample.”
Two alternative approaches can also provide the spectrum of “glucose in the biological sample.” Selective total correlation spectroscopy (TOCSY) is a simple NMR experiment that shows only the signals within a coupled 1H spin system.27, 28 It provides spectra that are quantitative measure of glucose as well as on other selected metabolites. However the line shapes and widths of a selective TOCSY spectrum are quite different from those of a simple 1H NMR spectrum. This makes the selective TOCSY spectrum of “glucose in the biological sample” incompatible to the desired background subtraction. The second approach is to simulate the glucose signals based on the chemical shift and line shape of a clean signal that belongs to a reference compound such as TSP (3-(trimethylsilyl)propionic acid-d4 sodium salt), DSS (3-(trimethylsilyl)-1-propanesulfonic acid-d6 sodium salt) or formate in the same sample, along with the knowledge regarding signals of both glucose and the reference compound in different sample matrices. This approach had been very successful in identifying and quantifying metabolites.19, 49 However even after thorough optimization of parameters, it is currently impossible to achieve a perfect match among all the simulated and real signals as shown in Figures 1(e), 3(d) and 3(h). Nevertheless, we expect that future algorithms and models will improve the performance.
For metabolite profiling applications, the effect of efficient background (glucose) subtraction has been illustrated in Figure 4 for the case of PCA. A key issue for PCA and other multivariate dimension reduction methods is that they tend to focus on large and variable signals rather than weak signals. As biomarkers or metabolites of important biological meanings are not always the large and variable compounds in biofluids, they are very likely ignored. The high glucose background in the 1H NMR spectra of blood serum, plasma and diabetic urine therefore not only hides signals of many metabolites in the spectra but also dominates the statistical analysis by PCA. The removal of this background will not only benefit the identification and quantification of low level metabolites but may also lead to the discovery of additional biomarker candidates through multivariate statistical methods.
One limitation is the time it takes for glucose to reach its equilibrium among its different isomeric forms (mutarotation), which is generally on the order of hours. This time can likely be minimized by matching the temperatures of the sample and added glucose solution, although we did not explore this approach in detail. For applications involving other high concentration species this equilibration time would be different and typically not as long as for glucose.
In conclusion, a simple method has been developed to remove the complex spectral background from high levels of glucose in 1H NMR spectra of biofluids with high efficiency. The reduction of the glucose background by 98% allows retrieval of many weak signals in the region between 3.2 and 4.0 ppm. This method has been studied and evaluated using human urine and blood serum samples, and its impact on multivariate statistical analysis (PCA) has been demonstrated. Upon removal of the glucose background, metabolite signals at µM levels can be identified in the corresponding spectral region either in individual spectra or by using statistical methods. In this work, the subtraction has been manually done in an interactive way, but can be fully automated by implementation of existing algorithms.50, 51 Though only the removal of the glucose signals from biofluid 1H NMR spectra has been demonstrated here, the “Add to Subtract” methodology can be applied to the removal of other background signals from the 1H NMR spectra of various complex mixtures. In fact, the concept is not limited to NMR spectroscopy but can be more generally applied to many types of analytical methods providing quantitative spectral data. We expect this concept of “Add to Subtract” to find numerous applications in the analysis of mixtures.
Acknowledgements
This work was supported by the National Institute of Health Grant (R01GM085291) to DR and NSF CAREER grant (DBI-1054826) to OV. Daniel Raftery is a member of the Purdue Center for Cancer Research and the Oncological Sciences Center. The authors thank Dr. Dor Ben-Amotz and Dr. Huaping Mo for advice and discussion.
References
- 1.Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Fiehn O. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 3.Fell DA. J. Exp. Bot. 2005;56:267–272. doi: 10.1093/jxb/eri011. [DOI] [PubMed] [Google Scholar]
- 4.Saghatelian A, Cravatt BF. Curr. Opin. Chem. Biol. 2005;9:62–68. doi: 10.1016/j.cbpa.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Gowda GAN, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Expert Rev. Mol. Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin JL. Curr. Opin. Chem. Biol. 2003;7:648–654. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Holmes E, Wilson ID, Nicholson JK. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 8.Assfalg M, Bertini I, Colangiuli D, Luchinat C, Schafer H, Schutz B, Spraul M. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1420–1424. doi: 10.1073/pnas.0705685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Nat. Chem. Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Werf MJ, Overkamp KM, Muilwijk B, Koek MM, van der Werff-van der Vat BJC, Jellema RH, Coulier L, Hankemeier T. Mol. Biosyst. 2008;4:315–327. doi: 10.1039/b717340g. [DOI] [PubMed] [Google Scholar]
- 11.Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Greef J, Hankemeier T, McBurney RN. Pharmacogenomics. 2006;7:1087–1094. doi: 10.2217/14622416.7.7.1087. [DOI] [PubMed] [Google Scholar]
- 13.Kell DB. Curr. Opin. Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Pan Z, Raftery D. Anal. Bioanal. Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 15.Ye T, Zhang S, Gowda GAN, Raftery D. Encyclopedia of Analytical Chemistry. John Wiley and Sons, Ltd.; 2010. [Google Scholar]
- 16.Beckonert O, Keun HC, Ebbels TMD, Bundy JG, Holmes E, Lindon JC, Nicholson JK. Nat. Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 17.Kim HK, Choi YH, Verpoorte R. Nat. Protoc. 2010;5:536–549. doi: 10.1038/nprot.2009.237. [DOI] [PubMed] [Google Scholar]
- 18.Serkova NJ, Niemann CU. Expert Rev. Mol. Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 19.Wishart DS. Trends Anal. Chem. 2008;27:228–237. [Google Scholar]
- 20.Goodpaster AM, Romick-Rosendale LE, Kennedy MA. Anal. Biochem. 2010;401:134–143. doi: 10.1016/j.ab.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Mo H, Raftery D. Anal. Chem. 2008;80:9835–9839. doi: 10.1021/ac801938j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mo H, Harwood J, Zhang S, Xue Y, Santini R, Raftery D. J. Magn. Reson. 2009;200:239–244. doi: 10.1016/j.jmr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Gowda GAN, Ye T, Raftery D. Analyst. 2010:135. doi: 10.1039/c000091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Anal. Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 25.Zheng C, Zhang S, Ragg S, Raftery D, Vitek O. Bioinformatics. 2011;27:1637–1644. doi: 10.1093/bioinformatics/btr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Graaf RA, Chowdhury GMI, Behar KL. Anal. Chem. 2011;83:216–224. doi: 10.1021/ac102285c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandusky P, Raftery D. Anal. Chem. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- 28.Sandusky P, Raftery D. Anal. Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 29.Chikayama E, Suto M, Nishihara T, Shinozaki K, Kikuchi J. PLoS One. 2008;3:e3805. doi: 10.1371/journal.pone.0003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, Westler WM, Sussman MR, Markley JL. Anal. Chem. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shanaiah N, Desilva MA, Gowda GAN, Raftery MA, Hainline BE, Raftery D. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye T, Mo H, Shanaiah N, Gowda GAN, Zhang S, Raftery D. Anal Chem. 2009;81:4882–4888. doi: 10.1021/ac900539y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye T, Zhang S, Mo H, Tayyari F, Gowda GAN, Raftery D. Anal. Chem. 2010;82:2303–2309. doi: 10.1021/ac9024818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan TWM, Bandura LL, Higashi RM, Lane AN. Metabolomics. 2005;1:325–339. [Google Scholar]
- 35.Hyberts SG, Heffron GJ, Tarragona NG, Solanky K, Edmonds KA, Luithardt H, Fejzo J, Chorev M, Aktas H, Colson K, Falchuk KH, Halperin JA, Wagner G. J. Am. Chem. Soc. 2007;129:5108–5116. doi: 10.1021/ja068541x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chylla RA, Hu K, Ellinger JJ, Markley JL. Anal. Chem. 2011;83:4871–4880. doi: 10.1021/ac200536b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaise BJ, Navratil V, Domange C, Shintu L, Dumas M-E, Elena-Herrmann B, Emsley L, Toulhoat P. J. Proteome Res. 2010;9:4513–4520. doi: 10.1021/pr1002615. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig C, Viant MR. Phytochem. Anal. 2010;21:22–32. doi: 10.1002/pca.1186. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig C, Ward DG, Martin A, Viant MR, Ismail T, Johnson PJ, Wakelam MJO, Günther UL. Magn. Reson. Chem. 2009;47:S68–S73. doi: 10.1002/mrc.2519. [DOI] [PubMed] [Google Scholar]
- 40.Gurst JE. J. Chem. Educ. 1991;68:1003–1004. [Google Scholar]
- 41.Suckale J, Solimena M. Front. Biosci. 2008;13:7156–7171. doi: 10.2741/3218. [DOI] [PubMed] [Google Scholar]
- 42.Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ. Clin. Chem. 1984;30:426–432. [PubMed] [Google Scholar]
- 43.Mo H, Raftery D. J. Biomol. NMR. 2008;41:105–111. doi: 10.1007/s10858-008-9246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mo H, Raftery D. J. Magn. Reson. 2008;190:1–6. doi: 10.1016/j.jmr.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balayssac S, Delsuc M-A, Gilard V, Prigent Y, Malet-Martino M. J. Magn. Reson. 2009;196:78–83. doi: 10.1016/j.jmr.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 46.Simpson AJ, Brown SA. J. Magn. Reson. 2005;175:340–346. doi: 10.1016/j.jmr.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Owens JA, Robinson JS. J. Chromatogr. 1985;338:303–314. doi: 10.1016/0378-4347(85)80101-8. [DOI] [PubMed] [Google Scholar]
- 48.Rabenstein DL, Millis KK, Strauss EJ. Anal Chem. 1988;60:1380A–1391A. doi: 10.1021/ac00175a001. [DOI] [PubMed] [Google Scholar]
- 49.Crockford DJ, Keun HC, Smith LM, Holmes E, Nicholson JK. Anal. Chem. 2005;77:4556–4562. doi: 10.1021/ac0503456. [DOI] [PubMed] [Google Scholar]
- 50.Loethen YL, Zhang D, Favors RN, Basiaga SBG, Ben-Amotz D. Appl Spectrosc. 2004;58:272–278. doi: 10.1366/000370204322886609. [DOI] [PubMed] [Google Scholar]
- 51.Perera PN, Fega KR, Lawrence C, Sundstrom EJ, Tomlinson-Phillips J, Ben-Amotz D. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12230–12234. doi: 10.1073/pnas.0903675106. [DOI] [PMC free article] [PubMed] [Google Scholar]