Abstract
(See the editorial commentary by Katz, on pages 867–9 and see the article by Stramer et al, on pages 886–94.)
Background. Genetic variations of human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV) can affect diagnostic assays and therapeutic interventions. Recent changes in prevalence of subtypes/genotypes and drug/immune-escape variants were characterized by comparing recently infected vs more remotely infected blood donors.
Methods. Infected donors were identified among approximately 34 million US blood donations, 2006–2009; incident infections were defined as having no or low antiviral antibody titers. Viral genomes were partially sequenced.
Results. Of 321 HIV strains (50% incident), 2.5% were non-B HIV subtypes. Protease and reverse transcriptase (RT) inhibitor resistance mutations were found in 2% and 11% of infected donors, respectively. Subtypes in 278 HCV strains (31% incident) yielded 1a>1b>3a>2b>2a>4a>6d, 6e: higher frequencies of 3a in incident cases vs higher frequencies of 1b in prevalent cases were found (P = .04). Twenty subgenotypes among 193 HBV strains (26% incident) yielded higher frequencies of A2 in incident cases and higher frequencies of A1, B2, and B4 in prevalent cases (P = .007). No HBV drug resistance mutations were detected. Six percent of incident vs 26% of prevalent HBV contained antibody neutralization escape mutations (P = .01).
Conclusions. Viral genetic variant distribution in blood donors was similar to that seen in high-risk US populations. Blood-borne viruses detected through large-scale routine screening of blood donors can complement molecular surveillance studies of highly exposed populations.
Volunteer (nonremunerated) blood donors provide whole blood and apheresis blood components used for transfusions as well as plasma for manufacture into therapeutic plasma derivatives. Past injection drug users (IDUs) and men who have had sex with men (MSM) since 1977 are currently excluded from donating. All donations in the United States are screened for antibodies to human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV), as well as for HIV and HCV RNA and HBV surface antigen (HBsAg); HBV DNA screening was widely implemented in 2009. Antibody and/or RNA/DNA/antigen-positive donors are notified, counseled, and excluded from further donation; all donations from viral marker-reactive donors are destroyed or used for research.
Blood donor screening can provide an efficient geographic and demographic sampling of individuals infected with these viruses that differ from the predominantly high-risk MSM and IDU populations that are the subjects of most viral molecular epidemiological studies. By routinely identifying recently acquired or incident infections, blood donation screening also provides an opportunity to analyze very recently transmitted viral strains at the forefront of currently active transmission chains. In this study, plasma samples from donors containing HIV, HBV, and HCV nucleic acids were classified as either recently acquired (incident) infections containing no or low concentrations of viral antibodies, or longer-term seropositive (prevalent) infections. Viral subtypes, drug resistance, and immune escape mutations were determined from representative incident and prevalent infections.
MATERIALS AND METHODS
Subjects, Specimens, and Case Definitions
This study included qualifying donations from 1 January 2006 through 31 December 31 2009 from 3 Retrovirus Epidemiology Donor Study-II (REDS-II) blood centers (Blood Centers of the Pacific, Blood Center of Wisconsin, and Hoxworth Blood Center/University of Cincinnati), all American Red Cross (ARC) Blood Services regions with data and samples provided through the Scientific Support Office in Gaithersburg, Maryland, and United Blood Services regions and the New York Blood Center (NYBC), with samples provided by Blood Systems Laboratory in Tempe, Arizona. Together, these centers account for approximately 70% of the US blood supply.
Data provided for all confirmed-positive donations included the date of donation, first-time/repeat donor status, date of birth, state of residency, race/ethnicity (if available), and sex. Screening and confirmatory test results for HIV and HCV nucleic acid testing (NAT), HIV and HCV antibody (Ab), HBsAg, and anti-HBV core antibody (anti-HBc) were also provided. Serologic and NAT screening and confirmatory testing were performed according to previously described algorithms using Food and Drug Administration (FDA)–licensed assays with documented performance characteristics including analytical sensitivities of NAT assays ([1–4]; see Appendix). Centers were requested to send residual test samples or samples from retrieved plasma units for all donors who qualified for study. Samples were stored at ≤−20°C prior to testing.
Informed Consent and IRB Approvals
Institutional review board (IRB)–approved information sheets were provided to donors explaining that (1) surplus samples of their donations may be used for research purposes, (2) future research may be performed with the donor’s blood without further consent if the IRB considers the research to be of negligible risk, (3) he or she will be contacted for additional consent depending on the nature of further research as determined by the IRB, and (4) the donor will be notified of any medically relevant information. IRBs representing each blood organization and the REDS-II Data Coordinating Center (Westat) approved the study protocol and determined that data related to HIV drug resistance mutations should be provided to donors as this may impact their treatment strategies; blood donors were not notified of other study results.
Sample Selection
The goal was to sequence samples from 150 incident and 150 prevalent cases for each virus over the 4-year study period. Samples with adequate volume from cases were consecutively selected from the contributing sites starting with donations made on January 1 2006 until the desired number of cases from a site was reached. Due to large numbers of HCV- and HBV-prevalent cases, site-specific sample numbers were based on proportions established by weighting the number of such donations detected by center in 2004 prior to study initiation. For donors selected for study with reported coinfections, each infecting virus was sequenced.
Viral Nucleic Acid Testing
Qualitative assays for HBV DNA (HBV Ampliscreen; Roche) were performed on HBsAg-positive/anti-HBc–negative donations to exclude possible incident cases that were likely due to false HBsAg positive results [1]. Viral load distributions were determined for all available samples that could not be amplified for sequence analysis, as well as a representative subset of successfully genotyped cases (Abbott RealTime HIV-1, HCV, and HBV Assays, Abbott Laboratories). All tests for this study, except for viral load determinations performed at Abbott Laboratories and routine donor screening and confirmation performed at blood center testing labs, were performed at the Blood Systems Research Institute.
Incidence Testing
Incident infections among HIV RNA-positive donors were defined as samples that were HIV-antibody negative or HIV-antibody positive with a less sensitive (LS) or “detuned” enzyme immunoassay (EIA: Vironostika HIV-1 MicroElisa; bioMérieux) with standardized-optical-density (SOD) ratio <1.0 using the serologic testing algorithm for recent HIV seroconversion, indicating that the infection was probably acquired <6 months prior to blood donation [5–7]. Conversely, HIV antibody–positive donations from first-time donors with LS-EIA SOD ratios ≥1.0 were defined as HIV-prevalent infections [8]. HCV RNA-positive and HBsAg-positive donations were classified as incident if anti-HCV and anti-HBc antibody tests, respectively, were nonreactive. HCV RNA and antibody positive donations were considered HCV-prevalent cases. HBsAg-positive/anti-HBc-reactive donations were considered HBV-prevalent infections. Starting with 2008 donations, HBV- and HCV-seropositive samples from repeat donors with a prior negative donation within the prior 2 years were also included as incident cases.
HIV, HCV, and HBV Sequencing
Total nucleic acid was extracted from 140 to 280 μL of plasma with QiaAMP Viral RNA Mini Kit or on a 96-well robotic platform (QIAxtractor with Reagent Pack VX, Qiagen). Complementary DNA for HIV and HCV was synthesized using M-MLV reverse transcriptase and random primer (0.5 μg/μL) according to the manufacturer’s instructions (Promega) and stored at −20°C.
Nested PCR was used to amplify an informative region of each virus. For HIV, a fragment of 1275 base pairs (bp) was amplified, including the protease and reverse transcriptase genes, using previously described PCR primers and conditions [9]. For HCV, a fragment of 363 bp in the core gene was amplified [10]. For HBV, a fragment of 2015 bp, including the envelope and polymerase genes, was amplified (Supplementary Methods in Appendix).
Sequence Analysis
Sequences were edited using Sequencher (version 4.9, Gene Codes Corporation). For HIV, the calibrated population resistance tool [11, 12] available through the Stanford University HIV Drug Resistance Database [13, 14] (http://cpr.stanford.edu/cpr.cgi) was used to determine subtype and identify transmitted drug resistance mutations in untreated persons. Mutations listed as causing or contributing to resistance are nonpolymorphic in untreated persons and apply to all HIV-1 subtypes in accordance with World Health Organization guidelines.
HCV sequences were subtyped using 2 online tools: the Oxford HCV subtyping tool (http://www.bioafrica.net/rega-genotype/html/subtypinghcv.html), a method based on phylogenetic analysis, and the NCBI viral genotyping tool, based on a sliding-window BLAST comparison (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi).
HBV genotypes were determined using 2 online tools: Oxford HBV subtyping tool (http://www.bioafrica.net/rega-genotype/html/citetoolhbv.html), a method based on phylogenetic analysis [15, 16], and the STAR genotyping tool available online at the University College London Center for Infection and Immunity (http://www.vgb.ucl.ac.uk/starn.shtml). The STAR tool uses distances to reference genomes and a statistical model to assign genotypes [17]. The polymerase sequence was checked for drug resistance mutations using the mutation annotator tool available online at the HepSEQ-Research Database System website (http://www.hepseq.org/Public/Web_Front/main.php). To identify potential antibody neutralization escape mutations, a list of mutations from the literature was compiled [18, 19], and sequences were manually aligned and compared with the reference list. GenBank accession numbers are JN214594-JN215208 and JN604118-JN604319.
Statistical Methods
Fisher exact test was used to compare the variant (subtype, genotype, and drug resistance mutation) distribution among incident cases to that among prevalent cases for each virus. Because molecular characterization could not be performed for all submitted cases, logistic regression was used to assess the ability to successfully characterize viral strains as a function of viral load and donor type (ie, incident vs prevalent cases). Analyses were conducted using SAS 9.2 software (SAS Institute).
RESULTS
HIV, HCV, and HBV Infection Rates and Demographic Characteristics
From 1 January 2006 through 31 December 2009, the participating blood organizations screened 33 947 146 allogeneic donations, including 5 968 986 (17.6%) from first-time and 27 950 520 (82.3%) from repeat donors; prior donation status information was not provided for 27 640 (0.1%) donations. For each virus, frequencies were generated by donor status (first-time vs repeat) and donor demographic characteristics (sex, race/ethnicity, geographic region, and age) (Table 1). A disproportionate risk of infection by all 3 viruses was noted for first-time, male, black, and Hispanic donors, except for HBV where infection rates were the highest among Asians, consistent with prior findings (Table 1) [1, 2, 20–22].
Table 1.
Numbers and Rates of Total Allogeneic Donations From Human Immunodeficiency Virus, Hepatitis C Virus, and Hepatitis B Virus Confirmed-Positive Donors by Donor Status (First-Time, Repeat) and Donor Demographic Characteristics Collected From 2 January 2006 Through 31 December 2009
| Characteristics | Total Allogeneic Donations | All HIV Positive | Rate per 100 000 Donations | All HCV Positive | Rate per 100 000 Donations | All HBV Positive | Rate per 100 000 Donations |
| Total | 33 947 146 | 1056 | 3.1 | 8015 | 23.6 | 3061 | 9.0 |
| Donor statusa | |||||||
| First-time donors | 5 968 986 | 633 | 10.6 | 6741 | 112.9 | 2561 | 42.9 |
| Repeat donors | 27 950 520 | 423 | 1.5 | 1274 | 4.6 | 500 | 1.8 |
| Sexa | |||||||
| Female | 15 850 421 | 251 | 1.6 | 3047 | 19.2 | 1063 | 6.7 |
| Male | 18 054 540 | 805 | 4.5 | 4968 | 27.5 | 1995 | 11.0 |
| Race or ethnicitya | |||||||
| White | 27 112 643 | 331 | 1.2 | 4399 | 16.2 | 651 | 2.4 |
| Asian | 510 633 | 24 | 4.7 | 81 | 15.9 | 848 | 166.1 |
| Black | 1 242 416 | 366 | 29.5 | 916 | 73.7 | 543 | 43.7 |
| Hispanic | 1 354 391 | 81 | 6.0 | 668 | 49.3 | 166 | 12.3 |
| Other | 628 302 | 37 | 5.9 | 211 | 33.6 | 189 | 30.1 |
| Not available | 3 071 121 | 217 | 7.1 | 1740 | 56.7 | 664 | 21.6 |
| CDC regionsa | |||||||
| Midwest | 10 523 749 | 172 | 1.6 | 900 | 8.6 | 292 | 2.8 |
| Northeast | 8 226 559 | 210 | 2.6 | 1957 | 23.8 | 913 | 11.1 |
| South | 8 911 822 | 474 | 5.3 | 3327 | 37.3 | 933 | 10.5 |
| West | 5 901 802 | 149 | 2.5 | 1760 | 29.8 | 900 | 15.2 |
| Other | 353 635 | 50 | 14.1 | 66 | 18.7 | 19 | 5.4 |
| Age categories, yearsa | |||||||
| <20 | 4 782 307 | 196 | 4.1 | 422 | 8.8 | 653 | 13.7 |
| 20–29 | 4 547 134 | 309 | 6.8 | 779 | 17.1 | 655 | 14.4 |
| 30–39 | 4 367 417 | 224 | 5.1 | 1023 | 23.4 | 576 | 13.2 |
| 40–49 | 7 080 519 | 201 | 2.8 | 2486 | 35.1 | 521 | 7.4 |
| 50–59 | 7 730 474 | 96 | 1.2 | 2696 | 34.9 | 428 | 5.5 |
| 60–69 | 3 927 177 | 26 | 0.7 | 478 | 12.2 | 163 | 4.2 |
| ≥70 | 1 482 979 | 4 | 0.3 | 131 | 8.8 | 65 | 4.4 |
Abbreviations: CDC, Centers for Disease Control and Prevention; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus.
Donations that did not have information for donor status, sex, race/ethnicity, CDC region, or age were excluded.
HIV Subtypes and Drug Resistance Profiles
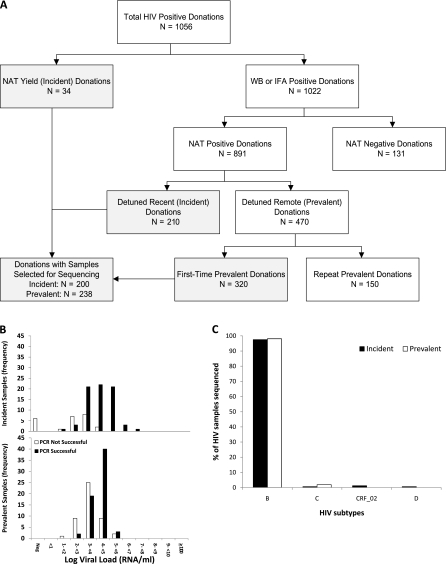
A total of 438 donation samples from HIV confirmed-positive donors were selected for sequencing from the 1056 submitted samples (Figure 1A). The 200 incident cases selected for viral sequencing included 34 RNA-positive, antibody-negative (NAT yield) donations and 166 of 210 donations classified as recent seroconvertors based on low antibody titers by LS-EIAtesting. A total of 238 of 320 prevalent HIV infections from first-time donors with high-titer antibody reactivity indicating long-standing infections were selected for further testing.
Figure 1.
A, Algorithm for classification of human immunodeficiency virus (HIV)–positive donations as incident or prevalent infections for selection of cases for sequencing. Boxes in gray indicate the donations from which samples were selected for sequencing for this study. B, Histograms showing success of polymerase chain reaction (PCR) relative to viral loads; frequencies of donations from incident and prevalent cases are shown separately. C, HIV subtypes of sequenced samples by incident and prevalent case status. Abbreviations: IFA, immunofluorescence assay; NAT, nucleic acid testing; WB, Western blot.
Of the 438 processed samples, 321 (73%) were successfully amplified and sequenced including 159 incident and 162 prevalent cases. Successful PCR amplification correlated with the viral load (P < .0001) Incident infections were 2.8-fold (95% confidence interval, 1.6–4.7) more likely to be genotyped than prevalent infections due to their higher viral loads (Figure 1B). Combining incident and prevalent cases, the success of obtaining sequences was approximately 10% if viral load was <1000 copies/mL, approximately 50% at 1000–10 000 copies/mL, and >90% if >10 000 copies/mL.
Of sequenced HIV strains, 97.5% (313) belonged to subtype B and 2.5% (n = 7) belonged to non-B subtypes: 4 subtype C (3 prevalent and 1 incident), 2 recombinant subtype CRF-02 (both incident) and 1 subtype D (incident) (Figure 1C). The number of non-B subtypes was too low to evaluate differences in frequency between incident and prevalent cases.
Four strains, all from incident cases, contained only protease inhibitor resistance mutations, and 33 strains (21 incident and 12 prevalent cases) contained only reverse transcriptase (RT) inhibitor resistance mutations (including 18 strains with only non-nucleoside reverse transcriptase inhibitor resistance mutation K103N). Two prevalent infections contained both protease and RT inhibitor resistance mutations (pro-M46I,I84V,L90M and RT-M41L,D67N,Y181C,M184V,G190A,L210W,T215Y; and pro-M46I,L90M and RT-M41L,Y181C,T215D). Overall, 39 of 321 (12%) sequenced HIV strains showed the presence of a drug resistance mutation, including 6 (2%) directed to protease inhibitors and 35 (11%) to RT inhibitors; these were from 25 of 159 (15.7%) incident and 14 of 162 (8.6%) prevalent cases (P = .06).
HCV Subtypes
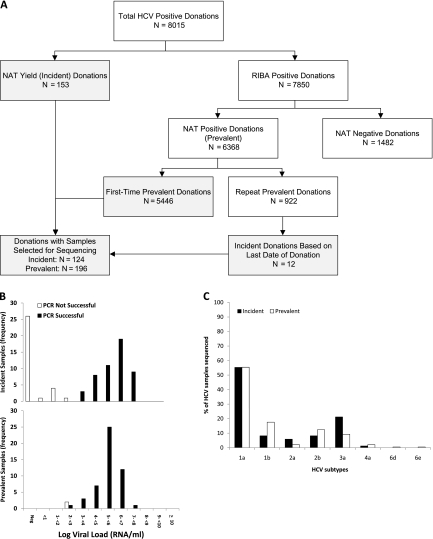
A total of 320 donation samples were selected for sequencing from 8015 HCV confirmed-positive donations (Figure 2A), including 112 of 153 HCV antibody-negative incident cases that had plasma aliquots available for testing. In addition, 12 incident cases were included based on antibody seroconversion within the previous 2 years, resulting in a total of 124 incident HCV cases. Of the 5446 HCV RNA and antibody-positive donations from first-time donors with prevalent infections, 196 representative samples were selected for molecular testing.
Figure 2.
A, Algorithm for classification of hepatitis C virus (HCV)–positive donations as incident or prevalent infections for selection of cases for sequencing. Boxes in gray indicate the donations from which samples were selected for sequencing for this study. B, Histograms showing success of polymerase chain reaction (PCR) relative to viral loads; frequencies of donations from incident and prevalent cases are shown separately. C, HCV subtypes of sequenced samples by incident and prevalent case status. Abbreviations: NAT, nucleic acid testing; RIBA, recombinant immunoblot assay.
Of the 320 samples processed for PCR amplification, 278 (87%) were successfully amplified and sequenced including 85 of 112 (68.5%) incident and 193 of 196 (99%) prevalent cases. Two of the successfully amplified HCV prevalent cases were coinfections; one donor also had an incident HIV infection and the second had a prevalent HBV infection. The probability of successful PCR amplification and sequencing was associated with viral load in the donors’ plasma for both incident and prevalent cases (P < .0001) but not with whether the donor was classified as a prevalent or incident case (Figure 2B). All samples with viral loads <100 copies/mL were negative for HCV core amplicons, approximately 50% of samples with viral loads of 100–1000 copies/mL were successfully amplified and sequenced, and all samples with viral loads >1000 copies/mL yielded sequence data.
Eight subtypes of HCV were present with 1a being the dominant type in both incident and prevalent cases. The subtype distribution was 55% (n = 154) 1a, 15% (n = 41) 1b, 13% (n = 36) 3a, 11% (n = 31) 2b, 3% (n = 9) 2a, 2% (n = 5) 4a, and <1% 6d or 6e (n = 1 each) (Figure 2C). Incident donors had significantly more subtype 3a strains (21% vs 9%) but significantly fewer subtype 1b strains (8% vs 18%) than did prevalent donors (P = .04).
HBV Genotype, Drug Resistance, and Immune Escape Profiles
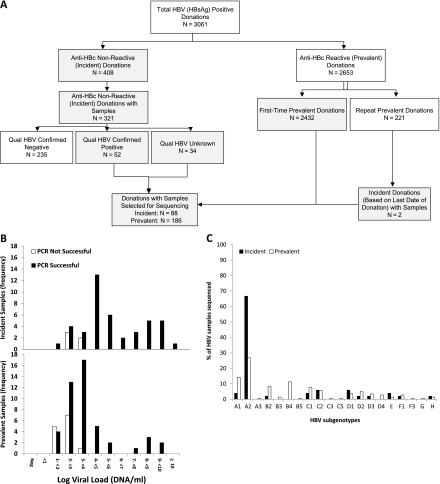
Of 3061 confirmed HBsAg-positive donations, 321 were anti-HBc-nonreactive and available (Figure 3A). However, only 52 of these possible incident infections were HBV DNA positive, whereas 235 tested HBV DNA negative, indicating either false-positive HBsAg neutralization results or recent receipt of the HBV vaccine with detection vaccine-derived HBsAg [1]. The 52 DNA-positive donations, plus the remaining 34 that were not tested for HBV DNA, along with 2 additional cases where HBV seroconversion occurred within 2 years of the index donation, comprised the 88 incident cases. Of the 2432 HBsAg confirmed-positive, anti-HBc-reactive donations from first-time donors, 186 representative samples were selected as prevalent infections for molecular testing.
Figure 3.
A, Algorithm for classification of hepatitis B virus (HBV)–positive donations as incident or prevalent infections for selection of cases for sequencing. Boxes in gray indicate the donations from which samples were selected for sequencing for this study. B, Histograms showing success of polymerase chain reaction (PCR) relative to viral loads; frequencies of donations from incident and prevalent cases are shown separately. C, HBV subgenotypes of sequenced samples by incident and prevalent case status. Abbreviations: HBc, anti-HBV core antibody; HBsAg, HBV surface antigen.
Of the 274 HBsAg-positive plasma samples selected, 193 (70%) had successful amplification and sequencing of the envelope and polymerase regions. As with HIV and HCV, the probability of successful PCR amplification and sequencing correlated with viral load (P < .0001) but not with incident/prevalent status. Approximately 50% of samples that had <100 copies/mL were negative for HBV PCR amplicons; in contrast, 70% of samples with 100–1000 copies/mL and 98% of those with >1000 copies/mL yielded informative sequence data.
The 193 donor strains successfully sequenced included 51 (26%) antibody-negative incident cases and 142 seropositive prevalent cases (Figure 3B). A total of 20 HBV subgenotypes were identified consisting of 72 A2, 22 A1, 16 B4, 13 B2, 13 C1, 11 C2, 8 D1, 8 D2, 6 D3, 5 F1, 4 D4, 4 F1, 3 H, 2 B3, and 1 each of the following subgenotypes: A3, B5, C3, C5, F3, and G (Figure 3C). Incident donors had significantly higher frequencies of subgenotype A2 (67%) vs those in prevalent donors (27%) who showed higher frequencies of subgenotypes A1, B2, C1, and D2 (8%–14% vs 0%–4%; P = .007).
Sequence analysis of the polymerase region of the 193 HBV strains did not detect any drug resistance–associated mutations. In the envelope sequences, 34 strains showed antibody neutralization escape–associated mutations, including 31 of 142 (22%) prevalent cases and a significantly lower proportion (3 of 52 [6%]) of incident cases (P = .01).
DISCUSSION
HIV subtype distribution varies in the United States depending on the population screened. Generally, the frequency of non-B subtypes has remained low in high-risk groups such as MSM and IDUs, as well as in non-IDU heterosexuals and blood donors [20–23]. The frequency of non-B subtype infections was greater in populations enriched for immigrants from nonclade B epidemic countries or military personnel who became infected overseas [24–28]. The frequency of non-B subtype in blood donors appears to be increasing at only a modest rate over the last 2 decades. Studies from the 1980s of seropositive donors and recipients of blood products found no nonclade B infections [23], whereas studies of seropositive donors from the 1990s identified approximately 1% nonclade B infections [7]; more recent studies of infected donors identified since 2000 reported rates of nonclade B infection in the 2%–5% range [3, 20, 22], similar to the 2.5% rate documented here.
Because the samples analyzed here were from asymptomatic blood donors who denied knowledge of their HIV infection, their drug resistance mutations are likely attributed to resistant virus acquired from their sources of infection who are presumed to have been on antiviral therapies. The frequency of HIV drug resistance mutations among blood donors trended but was not significantly higher in incident vs prevalent infections (P = .06). A stable frequency of drug resistance mutations also applied when resistance to the more recently introduced protease and the longer used RT inhibitors were analyzed separately. The frequency of transmitted drug resistance mutations appears to be stable among blood donors based on comparisons of rates among incident and prevalent infections in this study and in prior studies of HIV in US blood donors [3, 7, 20], an observation in keeping with reported rates of transmitted drug resistance mutations in high-risk untreated groups [29–32].
There are currently 7 HCV genotypes that are further subdivided into 83 subtypes (http://hcv.lanl.gov/content/sequence/HCV/classification/genotable.html) [33] that can vary widely in their geographic distribution (http://hcv.lanl.gov/components/sequence/HCV/geo/geo.comp). In high-risk groups in the United States, subtypes 1a and 1b predominate, whereas in most other countries the majority of HCV infections belong to other subtypes. Because HCV transmissions in the United States occur mainly among young IDUs [34] and reinfections can displace the original resident strain [35, 36], the distribution of HCV genotypes may rapidly change. Eight HCV subtypes were identified here with 1a (55%) and 1b (15%) predominating. The subtype distribution in prevalent cases was nearly identical to that reported for HCV-seropositive samples collected in 1988–1994 from a population reflecting that of the US [37], which supports the validity of our sampling strategy. In this study, we document a higher frequency of subtype 3a (21% vs 9%) and a lower frequency of 1b (8% vs 18%) in incident vs prevalent donors in keeping with a recent analysis showing decreasing genotype 1 frequencies in younger vs older IDUs [38].
HBV genotypes also vary greatly in their geographic distribution. Currently there are 8 genotypes that can be further subdivided into at least 24 subgenotypes defined as having >4% nucleotide difference [39]. Twenty of these 24 subgenotypes were identified among the 193 sequenced HBV strains. When the frequencies of the subgenotypes were compared, A2 occurred more frequently in incident cases (67% vs 27%) while A1, B2, and B4 frequencies were higher in prevalent cases (8%–14% vs 0%–4%).
No HBV antiviral drug resistance mutations were observed. Drug-resistant HBV variants may be inefficient at transmission and/or establishment of a chronic infection or may be underrepresented in the pool of HBV being actively transmitted by sexual or parenteral routes. Neutralization escape mutations in the HBV envelope protein were heavily overrepresented in prevalent vs incident HBV infections (22% vs 6%). This observation is consistent with these mutations having been more strongly selected for in long-term infected donors in whom a strong antibody response develops than in very recent, anti-HBc–negative incident cases [40–43].
This study of viral diversity has several limitations. First, the analysis was restricted to infections detected as NAT or HBsAg positive by current blood supply screening assays, most of which were also confirmed antibody positive. Consequently, infections by highly divergent variants that would not be detected by these assays would not be identified. Given efforts of test manufacturers and regulators to ensure that blood donor screening and confirmatory tests are sensitive to viral variants, we believe that this issue has limited impact on our findings. Second, a moderate proportion of donations selected for molecular analysis were not able to be characterized due to failure of long-amplicon PCR. These results were largely explained by absence of detectable nucleic acid or lower viral load in the PCR-refractory samples. It is also well recognized that all donor screening assays have low but significant rates of false positivity, especially if the classification is made only upon the routine testing results and does not include further testing of an independent sample such as the retrieved frozen plasma unit or follow-up donor sample. This is a particular problem with possible NAT yield samples (ie, seronegative and reactive by a single NAT assay), as evidenced by the high rate of incident cases with negative PCR results in this study (Figure 2B), many of which are likely due to false- NAT results. Third, we performed bulk sequencing of PCR products and therefore may not have detected cases of dual infection or minor populations of drug resistance or immune escape variants represented in viral quasi-species.
Overall, our analysis indicates that the HIV epidemic is relatively stable in terms of subtypes and transmitted drug resistance mutations. The HCV data provide evidence of differences in subtypes between incident and prevalent cases that may be stochastically driven by random founder effects and/or result from immigration of infected individuals to the United States. The HBV subgenotypes also showed evidence of change, possibly driven by similar epidemiological factors. The relative frequencies of different viral genetic clades and resistance patterns observed in our study population showed general concordance with those in populations with admitted high-risk behavior [44, 45]. Molecular characterization of recently transmitted blood-borne viruses detected through the large-scale routine NAT and antibody screening of blood donors is therefore a good complement to studies in highly exposed populations. As predominant viral strains change over time, sequence data generated by such blood donor molecular surveillance studies may be of use to adjust primers used in nucleic acid detection methods [46, 47], as well as the specificities of antibodies and antigens used in serologic assays [48–50] in order to maintain the high sensitivity of blood donation screening assays.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
We thank the staff at the following blood centers: ARC, Blood Systems Inc., NYBC, Blood Systems Laboratories, Blood Systems Research Institute (BSRI), and the ARC Scientific Support Office. We also thank Shihai Huang at Abbott Molecular Inc for performing viral load testing. Without their help, this study would not have been possible.
At the time of this writing, the Retrovirus Epidemiology Donor Study–II (REDS-II) was the responsibility of the following persons and organizations:
R. Cable, J. Rios, and R. Benjamin at ARC Blood Services, New England Region, Farmington, Connecticut; J. D. Roback at ARC Blood Services, Southern Region/Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia; R. A. Sacher, S. L. Wilkinson, and P. M. Carey at the Hoxworth Blood Center, University of Cincinnati Academic Health Center, Cincinnati, Ohio; E. L. Murphy, B. Custer, and N. Hirschler at the University of California, San Francisco, and BSRI and Blood Centers of the Pacific, San Francisco, California; D. Triulzi, R. Kakaiya, and J. Kiss at The Institute for Transfusion Medicine, Pittsburgh, Pennsylvania; J. Gottschall and A. Mast at the Blood Center of Wisconsin, Milwaukee; J. Schulman and M. King at Westat, Inc (Coordinating Center), Rockville, Maryland; G. Nemo and S. A. Glynn at the National Heart, Lung, and Blood Institute, National Institutes of Health, Rockville, Maryland; M. P. Busch and P. J. Norris at the Central Laboratory, BSRI.
Financial support.
This work was supported by National Institutes of Health, Heart, Lung, and Blood Institute (contracts N01-HB-47168, -47169, -47170, -47171, -47172, -47174, -47175, and -57181).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Zou S, Stramer SL, Notari EP, et al. Current incidence and residual risk of hepatitis B infection among blood donors in the United States. Transfusion. 2009;49:1609–20. doi: 10.1111/j.1537-2995.2009.02195.x. [DOI] [PubMed] [Google Scholar]
- 2.Zou S, Dorsey KA, Notari EP, et al. Prevalence, incidence, and residual risk of human immunodeficiency virus and hepatitis C virus infections among United States blood donors since the introduction of nucleic acid testing. Transfusion. 2010;50:1495–504. doi: 10.1111/j.1537-2995.2010.02622.x. [DOI] [PubMed] [Google Scholar]
- 3.Brennan CA, Stramer SL, Holzmayer V, et al. Identification of human immunodeficiency virus type 1 non-B subtypes and antiretroviral drug-resistant strains in United States blood donors. Transfusion. 2009;49:125–33. doi: 10.1111/j.1537-2995.2008.01935.x. [DOI] [PubMed] [Google Scholar]
- 4.Stramer SL, Wend U, Candotti D, et al. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236–47. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- 5.Busch MP, Pilcher CD, Mastro TD, et al. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS. 2010;24:2763–71. doi: 10.1097/QAD.0b013e32833f1142. [DOI] [PubMed] [Google Scholar]
- 6.Janssen RS, Satten GA, Stramer SL, et al. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA. 1998;280:42–8. doi: 10.1001/jama.280.1.42. [DOI] [PubMed] [Google Scholar]
- 7.Machado DM, Delwart EL, Diaz RS, et al. Use of the sensitive/less-sensitive (detuned) EIA strategy for targeting genetic analysis of HIV-1 to recently infected blood donors. AIDS. 2002;16:113–19. doi: 10.1097/00002030-200201040-00014. [DOI] [PubMed] [Google Scholar]
- 8.Rawal BD, Degula A, Lebedeva L, et al. Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J Acquir Immune Defic Syndr. 2003;33:349–55. doi: 10.1097/00126334-200307010-00009. [DOI] [PubMed] [Google Scholar]
- 9.Shafer RW, Hertogs K, Zolopa AR, et al. High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J Clin Microbiol. 2001;39:1522–9. doi: 10.1128/JCM.39.4.1522-1529.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardin F, Tobler L, Walsh I, Williams JD, Busch M, Delwart E. Clearance of hepatitis C virus RNA from the peripheral blood mononuclear cells of blood donors who spontaneously or therapeutically control their plasma viremia. Hepatology. 2008;47:1446–52. doi: 10.1002/hep.22184. [DOI] [PubMed] [Google Scholar]
- 11.Shafer RW, Rhee SY, Pillay D, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–23. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gifford RJ, Liu TF, Rhee SY, et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics. 2009;25:1197–8. doi: 10.1093/bioinformatics/btp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shafer RW, Jung DR, Betts BJ. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat Med. 2000;6:1290–2. doi: 10.1038/81407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Oliveira T, Deforche K, Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 16.Alcantara LC, Cassol S, Libin P, et al. A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res. 2009;37:W634–42. doi: 10.1093/nar/gkp455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers R, Clark C, Khan A, Kellam P, Tedder R. Genotyping hepatitis B virus from whole- and sub-genomic fragments using position-specific scoring matrices in HBV STAR. J Gen Virol. 2006;87:1459–64. doi: 10.1099/vir.0.81734-0. [DOI] [PubMed] [Google Scholar]
- 18.Gauthier M, Bonnaud B, Arsac M, et al. Microarray for hepatitis B virus genotyping and detection of 994 mutations along the genome. J Clin Microbiol. 2010;48:4207–15. doi: 10.1128/JCM.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran N, Berne R, Chann R, et al. European multicenter evaluation of high-density DNA probe arrays for detection of hepatitis B virus resistance mutations and identification of genotypes. J Clin Microbiol. 2006;44:2792–800. doi: 10.1128/JCM.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan CA, Yamaguchi J, Devare SG, Foster GA, Stramer SL. Expanded evaluation of blood donors in the United States for human immunodeficiency virus type 1 non-B subtypes and antiretroviral drug-resistant strains: 2005 through 2007. Transfusion. 2010;50:2707–12. doi: 10.1111/j.1537-2995.2010.02767.x. [DOI] [PubMed] [Google Scholar]
- 21.Delwart E, Kuhns MC, Busch MP. Surveillance of the genetic variation in incident HIV, HCV, and HBV infections in blood and plasma donors: implications for blood safety, diagnostics, treatment, and molecular epidemiology. J Med Virol. 2006;78(Suppl 1):S30–5. doi: 10.1002/jmv.20604. [DOI] [PubMed] [Google Scholar]
- 22.Delwart EL, Orton S, Parekh B, Dobbs T, Clark K, Busch MP. Two percent of HIV-positive U.S. blood donors are infected with non-subtype B strains. AIDS Res Hum Retroviruses. 2003;19:1065–70. doi: 10.1089/088922203771881149. [DOI] [PubMed] [Google Scholar]
- 23.de Oliveira CF, Diaz RS, Machado DM, et al. Surveillance of HIV-1 genetic subtypes and diversity in the US blood supply. Transfusion. 2000;40:1399–406. doi: 10.1046/j.1537-2995.2000.40111399.x. [DOI] [PubMed] [Google Scholar]
- 24.Thomson MM, Najera R. Travel and the introduction of human immunodeficiency virus type 1 non-B subtype genetic forms into Western countries. Clin Infect Dis. 2001;32:1732–7. doi: 10.1086/320764. [DOI] [PubMed] [Google Scholar]
- 25.Achkar JM, Burda ST, Konings FA, et al. Infection with HIV type 1 group M non-B subtypes in individuals living in New York City. J Acquir Immune Defic Syndr. 2004;36:835–44. doi: 10.1097/00126334-200407010-00011. [DOI] [PubMed] [Google Scholar]
- 26.Lin HH, Gaschen BK, Collie M, et al. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquir Immune Defic Syndr. 2006;41:399–404. doi: 10.1097/01.qai.0000200663.47838.f1. [DOI] [PubMed] [Google Scholar]
- 27.Singer DE, Bautista CT, O'Connell RJ, et al. HIV infection among U.S. Army and Air Force military personnel: sociodemographic and genotyping analysis. AIDS Res Hum Retroviruses. 2010;26:889–94. doi: 10.1089/aid.2009.0289. [DOI] [PubMed] [Google Scholar]
- 28.Brodine SK, Starkey MJ, Shaffer RA, et al. Diverse HIV-1 subtypes and clinical, laboratory and behavioral factors in a recently infected US military cohort. AIDS. 2003;17:2521–7. doi: 10.1097/00002030-200311210-00016. [DOI] [PubMed] [Google Scholar]
- 29.Turner D, Wainberg MA. HIV transmission and primary drug resistance. AIDS Rev. 2006;8:17–23. [PubMed] [Google Scholar]
- 30.Pillay D. Current patterns in the epidemiology of primary HIV drug resistance in North America and Europe. Antivir Ther. 2004;9:695–702. [PubMed] [Google Scholar]
- 31.Girardi E. Epidemiological aspects of transmitted HIV drug resistance. Scand J Infect Dis Suppl. 2003;106:17–20. doi: 10.1080/03008870310009597. [DOI] [PubMed] [Google Scholar]
- 32.Wensing AM, Boucher CA. Worldwide transmission of drug-resistant HIV. AIDS Rev. 2003;5:140–55. [PubMed] [Google Scholar]
- 33.Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–73. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 34.Hahn JA, Page-Shafer K, Lum PJ, et al. Hepatitis C virus seroconversion among young injection drug users: relationships and risks. J Infect Dis. 2002;186:1558–64. doi: 10.1086/345554. [DOI] [PubMed] [Google Scholar]
- 35.Herring BL, Page-Shafer K, Tobler LH, Delwart EL. Frequent hepatitis C virus superinfection in injection drug users. J Infect Dis. 2004;190:1396–403. doi: 10.1086/424491. [DOI] [PubMed] [Google Scholar]
- 36.Page K, Hahn JA, Evans J, et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis. 2009;200:1216–26. doi: 10.1086/605947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alter MJ, Kruszon-Moran D, Nainan OV, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–62. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 38.Telles Dias P, Hahn JA, Delwart E, et al. Temporal changes in HCV genotype distribution in three different high risk populations in San Francisco, California. BMC Infect Dis. 2011;11:208. doi: 10.1186/1471-2334-11-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267–80. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 40.Protzer-Knolle U, Naumann U, Bartenschlager R, et al. Hepatitis B virus with antigenically altered hepatitis B surface antigen is selected by high-dose hepatitis B immune globulin after liver transplantation. Hepatology. 1998;27:254–63. doi: 10.1002/hep.510270138. [DOI] [PubMed] [Google Scholar]
- 41.Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237–47. doi: 10.1007/BF02256597. [DOI] [PubMed] [Google Scholar]
- 42.Song BC, Kim SH, Kim H, et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76:194–202. doi: 10.1002/jmv.20354. [DOI] [PubMed] [Google Scholar]
- 43.Echevarria JM, Avellon A. Hepatitis B virus genetic diversity. J Med Virol. 2006;78(Suppl 1):S36–42. doi: 10.1002/jmv.20605. [DOI] [PubMed] [Google Scholar]
- 44.Grant RM, Hecht FM, Warmerdam M, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–8. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 45.Weinstock HS, Zaidi I, Heneine W, et al. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1-infected persons in 10 US cities. J Infect Dis. 2004;189:2174–80. doi: 10.1086/420789. [DOI] [PubMed] [Google Scholar]
- 46.Brennan CA, Bodelle P, Coffey R, et al. HIV global surveillance: foundation for retroviral discovery and assay development. J Med Virol. 2006;78(Suppl 1):S24–9. doi: 10.1002/jmv.20603. [DOI] [PubMed] [Google Scholar]
- 47.Peeters M, Aghokeng AF, Delaporte E. Genetic diversity among human immunodeficiency virus-1 non-B subtypes in viral load and drug resistance assays. Clin Microbiol Infect. 2010;16:1525–31. doi: 10.1111/j.1469-0691.2010.03300.x. [DOI] [PubMed] [Google Scholar]
- 48.Jongerius JM, Wester M, Cuypers HT, et al. New hepatitis B virus mutant form in a blood donor that is undetectable in several hepatitis B surface antigen screening assays. Transfusion. 1998;38:56–9. doi: 10.1046/j.1537-2995.1998.38198141499.x. [DOI] [PubMed] [Google Scholar]
- 49.Avellon A, Echevarria JM, Weber B, et al. European collaborative evaluation of the Enzygnost HBsAg 6.0 assay: performance on hepatitis B virus surface antigen variants. J Med Virol. 2011;83:95–100. doi: 10.1002/jmv.21943. [DOI] [PubMed] [Google Scholar]
- 50.Scheiblauer H, El-Nageh M, Diaz S, et al. Performance evaluation of 70 hepatitis B virus (HBV) surface antigen (HBsAg) assays from around the world by a geographically diverse panel with an array of HBV genotypes and HBsAg subtypes. Vox Sang. 2010;98:403–14. doi: 10.1111/j.1423-0410.2009.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.