Abstract
(See the editorial commentary by Bagni and Whitby, on pages 873–4.)
Background. Candidemia is a severe invasive fungal infection with high mortality. Recognition of Candida species is mediated through pattern recognition receptors such as Toll-like receptors (TLRs). This study assessed whether genetic variation in TLR signaling influences susceptibility to candidemia.
Methods. Thirteen mostly nonsynonymous single nucleotide polymorphisms (SNPs) in genes encoding TLRs and signaling adaptors MyD88 and Mal/TIRAP were genotyped in 338 patients (237 white, 93 African American, 8 other race) with candidemia and 351 noninfected controls (263 white, 88 African American). The SNPs significant in univariate analysis were further analyzed with multivariable logistic regression to determine association with clinical outcomes. Functional consequences of these polymorphisms were assessed via in vitro stimulation assays.
Results. Analyses of TLR SNPs revealed that 3 TLR1 SNPs (R80T, S248N, I602S) were significantly associated with candidemia susceptibility in whites. This association was not found in African Americans, likely due to lower power in this smaller study population. Furthermore, these TLR1 polymorphisms displayed impaired cytokine release by primary monocytes. No associations with susceptibility to candidemia were observed for SNPs in TLR2, TLR4, TLR6, TLR9, MyD88, or TIRAP.
Conclusions. Nonsynonymous SNPs in TLR1 are associated with impaired TLR1 function, decreased cytokine responses, and predisposition to candidemia in whites.
Candidemia (ie, bloodstream infection with Candida species) is the fourth most common nosocomial infection in immunocompromised patients and the systemic infections with the highest mortality rate, reaching 30%–40% in intensive care unit (ICU) patients [1, 2]. It is well established that the main risk factors for candidemia are neutropenia, mucosal barrier injury, solid organ or stem cell transplantation, treatment with immunosuppressive drugs, use of broad-spectrum antibiotics, prolonged ICU stay, and central venous catheterization [3–5]. However, among patients with similar predisposing factors, the risk of systemic Candida infection and its clinical outcome vary significantly.
Several Candida species, of which C. albicans is the most prevalent, have been isolated from the bloodstream of patients with candidemia. However, in recent years there has been a trend toward increased prevalence of non-albicans species such as C. glabrata and C. krusei, with increasing reports of resistance to antifungal azoles [6, 7], which may be in part promoted by the frequent use of antifungal prophylaxis, preemptive therapeutic strategies in high-risk patients, or alteration of host status [8, 9]. Antifungal prophylaxis remains the most effective way to prevent systemic Candida infection because of the lack of specific clinical and laboratory parameters to diagnose candidemia at an early stage [10, 11]. Due to all these facts, there is urgent need to identify host factors that predispose to candidemia in order to establish a well-defined predictive profile for patients at risk for developing infection.
Toll-like receptors (TLRs) are transmembrane proteins on immune cells that recognize conserved microbial molecular motifs designated as pathogen-associated molecular patterns. Toll-like receptors interact with several adaptor proteins, including myeloid differentiation primary response gene 88 (MyD88) and MyD88 adaptor–like/Toll–interleukin 1 receptor domain–containing adaptor protein (Mal/TIRAP), to activate transcription factors, leading to production of proinflammatory cytokines and subsequent induction of adaptive immunity. Recognition of Candida species by innate immune cells has been demonstrated to be mediated through several TLRs, which in turn leads to a potent host defense activation against the yeast [12]. Genetic variants of TLRs and their adaptors MyD88 and TIRAP have been associated with increased susceptibility to bacterial infections [13–17]. A number of studies have proposed associations of TLR2 and TLR4 polymorphisms with increased risk of aspergillosis and candidemia [18–21]. However, most of these studies have been performed in very small cohorts of patients, and a systematic genetic study investigating variation in pattern recognition receptors in a large cohort of patients at risk of candidemia, comprising infected patients and noninfected control patients, has not been performed to robustly interrogate the importance of this innate pathway for protection against Candida species.
In the present study, we hypothesized that certain polymorphisms in TLR genes or in genes encoding their signaling adaptors may influence host susceptibility to the most common invasive fungal disease, candidemia. To test this hypothesis, we analyzed the prevalence of mostly nonsynonymous single nucleotide polymorphisms (SNPs) in 5 candidate TLR genes and genes encoding their adaptors MyD88 and TIRAP in a cohort of 338 candidemia patients and 351 noninfected controls that had similar underlying predisposing factors. The genetic study was further complemented and supported with functional assays that assessed the consequences of these specific polymorphisms on the cytokine production capacity of primary human blood mononuclear cells in order to validate the mechanistic association of the susceptibility trait.
METHODS
Ethics Statement and Subjects
Adult subjects were enrolled after informed consent (or waiver as approved by the institutional review board) at Duke University Hospital (DUMC, Durham, North Carolina) and Radboud University Nijmegen Medical Centre (RUNMC, Nijmegen, the Netherlands). Race of the patients was self-reported; all Dutch RUNMC patients were white. The study was approved by the institutional review boards at each study center, and enrollment occurred between January 2003 and January 2009. The study was performed in accordance with the Declaration of Helsinki.
Susceptibility to Infection
Infected subjects were enrolled consecutively and must have had ≥1 positive blood culture for a Candida species while hospitalized at the participating center. Noninfected controls must have been hospitalized with no history or evidence of candidemia or any invasive fungal infection. None of the control patients were on antifungal therapy/prophylaxis. Although it was impracticable to match controls on specific risk factors, noninfected controls were recruited consecutively from the same hospital wards/services as infected patients during the study period, with a similar balance of medical, surgical, and oncology patients in case and control groups. Immunocompromised status was a combined variable based on a broad definition including neutropenia, CD4+ lymphopenia (human immunodeficiency virus [HIV] associated or otherwise), chronic steroid use or receipt of immunosuppressive chemotherapy/immunosuppressant medications within 30 days prior to baseline, diabetes mellitus, and/or end-stage renal disease requiring hemodialysis. Acute renal failure was defined based on clinical diagnosis, including doubling of serum creatinine and/or requirement for acute dialysis (intermittent hemodialysis or continuous veno-venous hemofiltration dialysis). Intergroup comparisons between the 2 groups of noninfected subjects and between the 2 groups of infected subjects (at DUMC and RUNMC) were performed regarding similarity in genetic distribution of the studied SNPs prior to further statistical analysis of infected versus noninfected subjects.
Genetic Association With Clinical Outcomes
Due to the lack of detailed clinical data on the Dutch candidemia patients (n = 40) recruited at RUNMC, the correlations of the genetic data with clinical outcome was performed only for the patients (n = 298) from DUMC. Infected subjects at DUMC were followed prospectively for up to 12 weeks following diagnosis of candidemia to determine clinical outcome. Mortality data, including date of death, was ascertained from hospital records and/or the National Social Security Death Index. Disseminated infection was defined as the presence of Candida species at normally sterile body sites outside the bloodstream (excluding the urine). Persistent fungemia was defined as ≥5 days of persistently positive blood cultures.
Genetic Analysis
Genomic DNA was isolated from whole blood using standard procedures [22]. Coding nonsynonymous SNPs and a few SNPs in untranslated regions of the analyzed genes were selected based on previously published associations with human diseases and/or known functional effects on protein function or gene expression. A total of 13 SNPs in TLR1, TLR2, TLR4, TLR6, TLR9, TIRAP, and MyD88 were genotyped (Table 1) with the use of a mass spectrometry genotyping platform. Quality control was performed by duplicating samples within and across plates and by the incorporation of positive and negative control samples.
Table 1.
Genotyped Single Nucleotide Polymorphisms in Genes Encoding Toll-like Receptors and Associated Adaptor Molecules
| Gene | SNP ID | Gene Region | Amino Acid Change |
| MYD88 | rs4988453 | Promoter | … |
| rs6853 | 3′ UTR | … | |
| TIRAP | rs595022 | Intron 1 | … |
| rs8177374 | Exon 5 | Nonsynonymous S180L | |
| TLR1 | rs4833095 | Exon 4 | Nonsynonymous S248N |
| rs5743611 | Exon 4 | Nonsynonymous R80T | |
| rs5743618 | Exon 4 | Nonsynonymous I602S | |
| TLR2 | rs5743704 | Exon 3 | Nonsynonymous P631H |
| rs5743708 | Exon 3 | Nonsynonymous R753Q | |
| TLR4 | rs4986790 | Exon 3 | Nonsynonymous D299G |
| rs4986791 | Exon 3 | Nonsynonymous T399I | |
| TLR6 | rs5743810 | Exon 1 | Nonsynonymous S249P |
| TLR9 | rs5743836 | Promoter | … |
Abbreviations: SNP, single nucleotide polymorphism; TLR, Toll-like receptor; UTR, untranslated region.
In Vitro Peripheral Blood Mononuclear Cell Stimulation Assays
Isolation and stimulation of peripheral blood mononuclear cells (PBMCs) was performed as described previously [23]. In brief, PBMCs obtained from 49 healthy white volunteers were incubated at 37°C for 24 hours with culture medium, the TLR1/2 agonist Pam3Cys (10 μg/mL; EMC Microcollections) or the TLR4 agonist lipopolysaccharide (Escherichia coli O55:B5 LPS, 10 ng/mL; Sigma-Aldrich). Cytokine production of interleukin 1 β (IL-1β), interleukin 6 (IL-6), and interleukin 8 (IL-8) was measured in the PBMC supernatants by enzyme-linked immunosorbent assay (R&D Systems).
Statistical Analysis
Chi-square tests of deviation from Hardy-Weinberg equilibrium for all 13 SNPs were calculated in cases and controls separately using PLINK statistical software [24]. Single locus tests of association were performed with logistic regression (or exact logistic regression, in the case of African Americans) using a dominant model (eg, minor allele homozygote combined with the heterozygote as risk genotypes) using SAS software version 9.2 (SAS Corporation). Model selection was based upon genotype frequencies. Standard summary statistics (odds ratio [OR] and 95% confidence interval [CI]) were reported for these tests of association. Baseline demographic and clinical categorical variables in the case and control groups were compared by χ2 analysis or Fisher exact test (as appropriate), and variables with P < .05 were analyzed as confounders of any SNP significantly associated with susceptibility to infection using logistic regression (or exact logistic regression as appropriate). Standard summary statistics for pairwise linkage disequilibrium (LD), D’, and r2 were calculated using Haploview. Haplotype blocks were assigned using the D’ confidence interval algorithm created by Gabriel et al [25].
Data on susceptibility to infection were analyzed for whites separately from African Americans because allelic frequencies were expected to differ among these 2 populations and controls, and examination of race-stratified genotype frequencies for the tested polymorphisms confirms this expectation [26, 27]. Statistical analysis of the cytokine data was performed using the Mann–Whitney U test. A P value < .05 was considered statistically significant.
Within the infected DUMC cohort, allelic frequencies were further assessed in association with 3 prespecified clinical outcomes: (1) disseminated disease, (2) persistent fungemia, and (3) all-cause mortality at 30 days. This analysis was performed in the entire infected cohort of DUMC whites and African Americans because the progression of the disease once occurring is not expected to differ between white and African American patients and race was considered as covariate in the analysis. Variables with P < .2 were further assessed in a multivariable logistic regression model using backward elimination. Variables with P < .05 were retained in the final predictive model. Odds ratios and 95% CIs were reported for variables that remained significant in the final multivariable model.
RESULTS
Demographics
Three hundred thirty-eight adult patients (298 North American, 40 Dutch) and 351 adult controls (300 North American, 51 Dutch) had genetic and clinical data available for this analysis. Demographics for study subjects are presented in Table 2 (including Dutch and North American subjects), and major clinical features among the infected and noninfected cohort are presented in Table 3 (including the North American subjects only). As expected, the groups were similar in regard to primary admitting service/primary presenting problem.
Table 2.
Demographics for Duke University Hospital and Radboud University Nijmegen Medical Centre Adult Study Subjects
| Infected Cohort (n = 338), No. (%) | Noninfected Controls (n = 351), No. (%) | |
| Race | ||
| White | 237 (70%) | 263 (75%) |
| African American | 93 (28%) | 88 (25%) |
| Other | 8 (2%) | 0 (0%) |
| Sex | ||
| Male | 199 (59%) | 184 (52%) |
| Female | 139 (41%) | 167 (48%) |
Table 3.
Baseline Patient Characteristics for Duke University Hospital Infected Cohort and Control Cohort, Including White and African American Adult Patients
| Variable | Infected Cohort (n = 298) | Control Cohort (n = 300) | P Value |
| Median age, y (IQR) | 58.7 (43.0–68.1) | 58.7 (45.0–69.6) | .41 |
| Primary admitting service/primary problem | |||
| Medical | 50% | 54% | .31 |
| Surgical | 33% | 31% | .64 |
| Oncology | 17% | 15% | .36 |
| Median length of hospitalization, d (IQR)a | 9 (2–18) | 6 (4–12) | .80 |
| Immunocompromised state | 59% | 42% | <.0001 |
| Hematopoietic stem cell transplantation | 1% | 0% | .12 |
| Solid organ transplant | 12% | 2% | <.0001 |
| Active malignancyb | 32% | 20% | .002 |
| Solid tumor | 23% | 12% | .27 |
| Leukemia | 7% | 5% | .29 |
| Lymphoma | 4% | 4% | 1 |
| Chemotherapy within past 3 months | 16% | 11% | .03 |
| Neutropenia (ANC <500 cells/mm3) | 10% | 4% | .001 |
| HIV infected | 1% | 0.1% | .06 |
| Surgery within past 30 days | 43% | 48% | .25 |
| Receipt of total parenteral nutrition | 19% | 4% | <.0001 |
| Dialysis dependent | 12% | 7% | .02 |
| Acute renal failure | 34% | 22% | .001 |
| Liver disease | 25% | 4% | <.0001 |
| Intensive care unit admission within past 14 days | 49% | 33% | <.0001 |
| Central intravascular catheter | 82% | 41% | <.001 |
| Peripheral intravenous catheter | 100% | 97% | .004 |
| Baseline serum creatinine, mg/dL, median (IQR) | 1.3 (0.8–2.3) | 1.0 (0.8–1.5) | .001 |
| Median baseline WBC count, cells/mm3 (IQR) | 10.5 (6.0–17.1) | 8.6 (6.4–12.4) | .04 |
| Candida speciesc | |||
| albicans | 42% | … | |
| glabrata | 29% | … | |
| parapsilosis | 16% | … | |
| tropicalis | 13% | … | |
| krusei | 4% | … | |
| Other Candida species | 3% | … |
Abbreviations: ANC, absolute neutrophil count; HIV, human immunodeficiency virus; IQR, interquartile range; WBC, white blood cell.
Until index blood culture for cases.
Subjects could have >1.
16 subjects had >1 species isolated.
Genetic Analysis
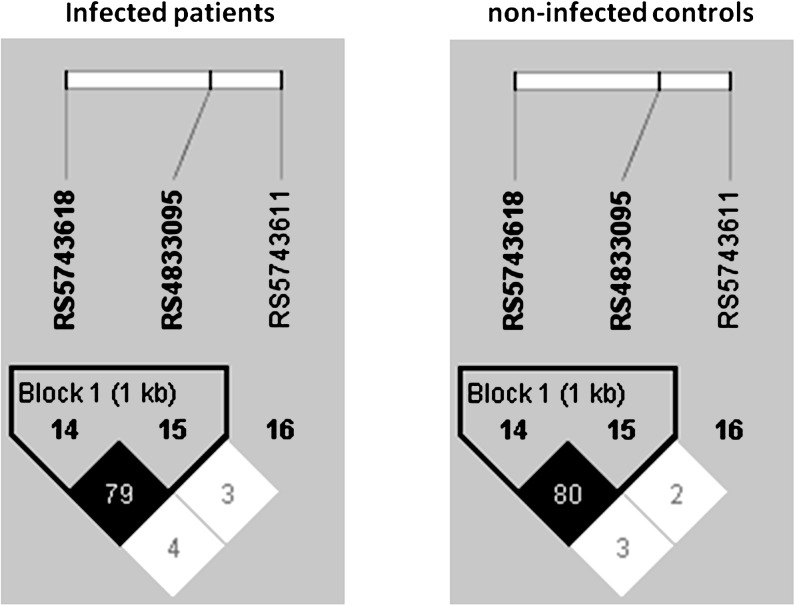
Data from adult patients (n = 338) and controls (n = 351) were included in the analysis for genetic association with susceptibility to infection. The intergroup comparison between the Dutch RUNMC and white DUMC noninfected controls and between the infected subjects recruited at DUMC and RUNMC revealed a similar genetic distribution of the genotyped SNPs, which allowed the groups to be merged into 1 group of noninfected controls and 1 group of infected subjects (data not shown). A post hoc analysis of the interaction between SNPs and race supported our stratification of this analysis by race. Genotyping of candidemia patients and noninfected controls for polymorphisms in genes encoding TLRs and their adaptor molecules MyD88 and Mal/TIRAP revealed a significantly different distribution of 3 TLR1 polymorphisms when comparing infected patients and noninfected controls in white adults (Table 4). All genotyped SNPs were in Hardy-Weinberg equilibrium (data not shown). Linkage disequilibrium analysis revealed that alleles at rs4833095 (S248N) and rs5743618 (I602S) were strongly in LD (r2 = 0.8) with each other but not with alleles at rs5743611 (R80T; r2 < 0.03) (Figure 1). No associations were observed with polymorphisms in TLR2, TLR4, TLR6, TLR9, MyD88, and TIRAP and susceptibility to candidemia in whites (data not shown).
Table 4.
Distribution of TLR1 Genotypes in the White (245 Adult Candidemia Patients, 263 Noninfected Adult Controls) and African American (93 Adult Candidemia Patients, 88 Noninfected Adult Controls) Cohorts
| Whites (245 Infected, 263 Noninfected) MAF | P Value | OR (95% CI)b | ||||
| TLR1 Ra80T | GGa | GC | CC | .02 | 1.82 (1.12–2.98) | |
| Noninfected | 88.2% | 9.9% | 1.91% | 0.068 | ||
| Infected | 80.2% | 18.9% | 0.86% | 0.103 | ||
| TLR1 S248Na | GG | GA | AAa | .04 | 0.68 (.47–.96) | |
| Noninfected | 5.3% | 41.8% | 52.9% | 0.262 | ||
| Infected | 7.2% | 30.1% | 62.7% | 0.223 | ||
| TLR1 I602Sa | TT | TG | GGa | .02 | 0.69 (.49–.80) | |
| Noninfected | 7.6% | 44.7% | 47.7% | 0.300 | ||
| Infected | 10.2% | 33.1% | 56.8% | 0.268 | ||
| African Americans (93 infected, 88 noninfected) MAF | P value | OR (95% CI)c | ||||
| TLR1 Ra80T | GGa | GC | CC | .62 | 2.92 (.23–155.70) | |
| Noninfected | 98.9% | 1.1% | 0% | 0.006 | ||
| Infected | 96.8% | 3.2% | 0% | 0.016 | ||
| TLR1 Sa248N | GG | GA | AAa | .09 | 3.86 (.71–39.15) | |
| Noninfected | 63.6% | 28.4% | 7.95% | 0.222 | ||
| Infected | 69.9% | 27.9% | 2.15% | 0.161 | ||
| TLR1 Ia602S | TT | TG | GGa | .60 | 3.12 (.25–166.70) | |
| Noninfected | 76.1% | 20.5% | 3.4% | 0.137 | ||
| Infected | 80.2% | 18.7% | 1.1% | 0.105 |
Abbreviations: CI, confidence interval; MAF, minor allele frequency; OR, odds ratio; TLR, Toll-like receptor.
Reference (equals major) allele in entire study population.
Logistic regression, dominant model.
Exact logistic regression.
Figure 1.
Linkage disequilibrium (LD) analysis of TLR1 single nucleotide polymorphisms R80T (rs5743611), S248N (rs4833095), and I602S (rs5743618) in white individuals. Pairwise LD was assessed using the correlation coefficient (r2). Percentage correlation between the 2 markers is noted in each box.
No significant associations with susceptibility to candidemia were observed in the African American cohort, neither with SNPs in TLR1 (Table 4) nor in the other analyzed genes (data not shown).
Association of Polymorphisms With Clinical Outcome
Among the 298 DUMC adult infected subjects, 18% (54) developed disseminated disease and 15% (45) experienced persistent fungemia; the mortality rate was 28% (83) and 39% (116) at 30 and 90 days after initial blood culture, respectively. The most common type of infection complicating the bloodstream was abdominal abscess/peritonitis (55%) followed by hepatosplenic candidiasis (13%) and endocarditis/vascular sites (11%). Persistence occurred at similar rate among those with and without central catheters (13% vs 15%; P = .67) and with and without central catheter removal by day 5 (19% vs 25%; P = .33). Median (interquartile range) time to antifungal therapy in those with and without persistence (2 [1.75–3] days vs 2 [1–3] days; P = .36) was also similar in those with and without persistence. Univariate and subsequent multivariate analysis assessing baseline patient characteristics and polymorphisms in TLR1, TLR2, TLR4, TLR6, TLR9, MyD88, and TIRAP (data not shown) indicated that there was no association between presence of polymorphisms at these loci and outcomes such as dissemination, persistence, and mortality.
In Vitro Stimulation Assays
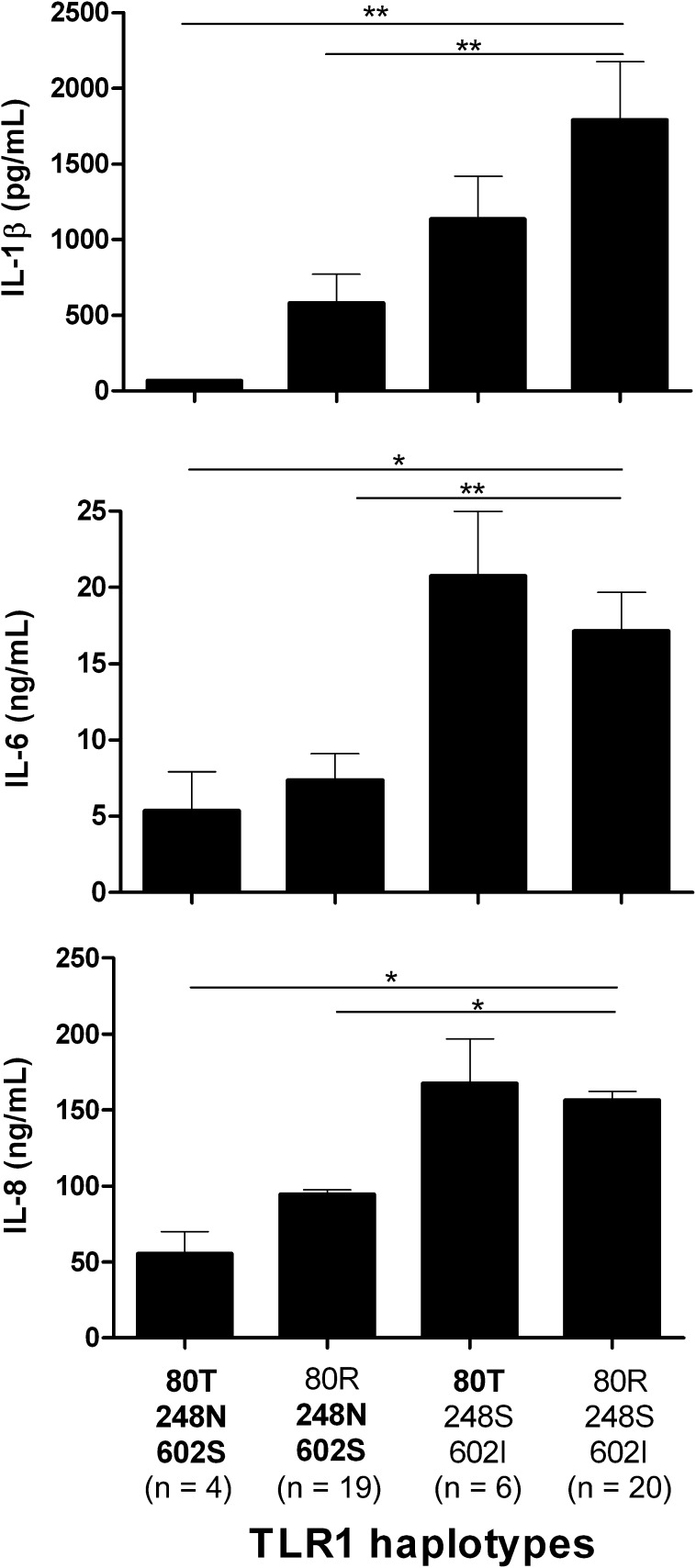
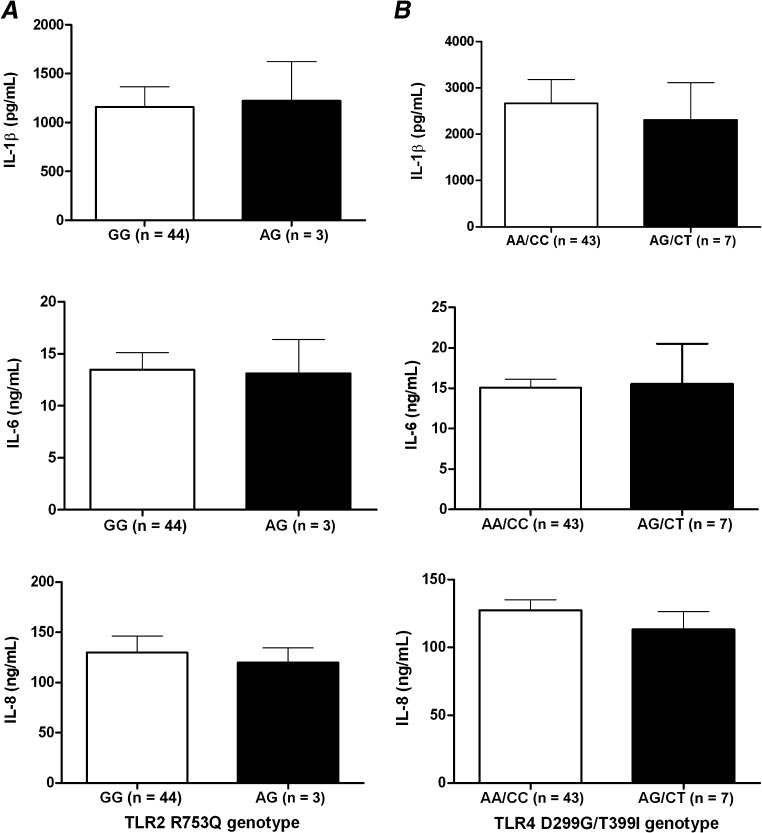
Functional consequences of the 3 TLR1 polymorphisms were studied by stimulating PBMCs obtained from 49 white, healthy volunteers bearing different TLR1 haplotypes with Pam3Cys, a specific agonist of the TLR1/TLR2 heterodimer. Cytokine production capacity of IL-1β, IL-6, and IL-8 was measured after 24 hours of stimulation (Figure 2). Unstimulated cells produced no detectable cytokine concentrations (data not shown). Dramatically decreased cytokine production was observed in cells bearing 1, 2, or 3 multilocus genotypes in TLR1 that are associated with higher susceptibility to candidemia compared with haplotypes of wild-type alleles. Cytokine responses in relation to previously reported associations of TLR2 (R753Q; Pam3Cys stimulation) and TLR4 (D299G/T399I; LPS stimulation) polymorphisms with candidemia were also evaluated but revealed no significant differences (Figure 3).
Figure 2.
Correlation of TLR1 haplotypes with cytokine measurements of interleukin 1 β (IL-1β), interleukin 6 (IL-6), and interleukin 8 (IL-8) 24 h after in vitro stimulation of peripheral blood mononuclear cells from healthy volunteers with the Toll-like receptor 1 (TLR1)/TLR2 agonist Pam3Cys (10 μg/mL). On the x-axis the different TLR1 haplotypes are depicted with protective genotypes (not bold) and risk genotypes (bold). Data are presented as means ± SEM (Mann–Whitney U test, *P ≤ .05, **P ≤ .005).
Figure 3.
Correlation of TLR2 R753Q (A) and TLR4 D299G/T399I (B) genotypes with cytokine measurements of interleukin 1 β (IL-1β), interleukin 6 (IL-6), and interleukin 8 (IL-8) 24 h after in vitro stimulation of peripheral blood mononuclear cells from healthy volunteers with either Pam3Cys (A, 10 μg/mL) or lipopolysaccharide (B, 10 ng/mL). Data are presented as means ± SEM. Abbreviation: TLR, Toll-like receptor.
DISCUSSION AND CONCLUSIONS
The present study demonstrates the association of 3 TLR1 polymorphisms with an increased susceptibility for candidemia in white patients. In addition to the genetic association between TLR1 polymorphisms and candidemia, cells isolated from individuals bearing the TLR1 variants that predispose to candidemia have shown functional consequences with decreased cytokine responses upon in vitro stimulation with specific TLR1/TLR2 ligands. In contrast to TLR1, no influence of TLR2, TLR4, TLR6, TLR9, MyD88, or TIRAP SNPs on the susceptibility to candidemia was observed.
Several TLRs have been reported to be involved in recognition of C. albicans, among which TLR2 mediates recognition of phospholipomannan [28], TLR4 of O-bound mannan [29], and TLR9 of fungal DNA [30]. Toll-like receptor 6 has also been demonstrated to contribute to Candida-mediated cytokine responses [31], whereas TLR1 forms functional heterodimers with TLR2 [32]. Because identified genetic variants of TLRs have recently been reported to influence function of these proteins and susceptibility to infections [33], we assessed the role of SNPs in these receptors and their adaptor molecules MyD88 and Mal/TIRAP for potential insights into the susceptibility to Candida bloodstream infections.
Among the genes and SNPs tested, we found that 3 polymorphisms in TLR1 were associated with increased susceptibility to development of candidemia in hospitalized patients at risk for this fungal disease. Although 2 of these polymorphisms (S248N and I602S) were strongly (80%) linked, the third (R80T) was not linked to the first 2 polymorphisms. When the 2 groups of subjects recruited at DUMC and RUNMC were considered separately, this trend of association could be observed also in the smaller RUNMC group. Due to the common Western European ancestry of the 2 groups, they were analyzed together. Importantly, frequencies of the TLR1 polymorphisms in the white noninfected controls were fairly equal compared with the general population, as assessed by genotyping 152 white healthy controls for these SNPs (data not shown), indicating that TLR1 polymorphisms confer a higher risk for developing candidaemia, but not for underlying predisposing factors. The observation that all 3 SNPs influence susceptibility to candidemia strengthens the conclusion that there is an important role for TLR1 in influencing the susceptibility to the infection. Moreover, the genetic data were complemented by functional studies demonstrating that TLR1 genotypes predisposing to candidemia are associated with decreased proinflammatory cytokine release upon stimulation.
One study in mice did not identify a nonredundant role of TLR1 in the host defense against disseminated candidiasis [31]. Hence, our data suggest that important differences exist between the role of TLR1 in mice and humans. Furthermore, the high inocula mouse model of disseminated candidiasis may very well differ from the clinical course of infection in humans with their heterogeneous and dynamic risk factors.
Toll-like receptor 1 is known to form heterodimers with TLR2, which has been shown to be involved in Candida recognition. For one of the TLR1 polymorphisms, I602S, it was demonstrated that the TLR1 protein bearing 602S is associated with impaired cell surface trafficking, resulting in a decreased availability of TLR1 on the cell membrane [34]. Consequently, lower availability of TLR1 could skew the configuration of TLR2 from heterodimers attached to TLR1 toward predominantly TLR2 homodimers or TLR2/TLR6 heterodimers. This, in turn, could alter intracellular signaling and cytokine production induced after Candida species encounter innate immune cells, which might result in deleterious effects during systemic fungal infections.
Toll-like receptor 1 could also exert its effect on antifungal host defense through beta-defensin-3, which activates immune cells through TLR1/TLR2 [35]. Beta-defensin-3 has an important lytic activity against C. albicans, and its expression is induced in mucosal layers after fungal recognition [36]. As a consequence, diminished TLR1 function could impair mucosal antifungal defense, enabling Candida to invade the tissue defenses and reach the bloodstream. Of note, the main route of Candida species to invade the bloodstream and cause invasive infection is indeed through crossing the epithelial layers of the skin and the intestine [37]. Furthermore, it cannot be excluded that other still unidentified endogenous danger-associated molecular patterns, which are possibly elicited in the context of candidemia, are able to induce TLR1 signaling and contribute to activation of antifungal mechanisms.
In contrast to the effects of TLR1 SNPs in white patients, no significant association of TLR1 polymorphisms with susceptibility to candidemia was detected in patients of African American origin. This is likely due to the smaller sample size of African Americans in our study population and consequently lower statistical power as well as a different distribution of these polymorphisms compared with those observed among whites. It has been previously noted that there are large differences in the genetic distribution of these TLR1 polymorphisms between African American and white populations [38]. In particular, the alleles encoding S248N and I602S polymorphisms are distributed very differently throughout the populations, and this may provide a partial explanation for the absence of association of TLR1 polymorphisms with candidemia in our smaller African American cohort. On the other hand, no substantial differences were observed between white and African American patients with regard to basic clinical characteristics.
Clinical parameters of infection comprising dissemination, persistent fungemia, and 30-day mortality were not associated with any of the polymorphisms analyzed in this study, including polymorphisms in TLR1. This may imply that TLR1 is mainly involved in the early host response, thereby preventing bloodstream infection with Candida rather than determining the outcome of established candidemia. Of note, previous studies with mouse models of disseminated candidiasis in TLR1 knockout mice have only addressed the latter condition [31], which may provide an additional explanation for the differences observed in the role of TLR1 in candidemia between mice and humans.
Our results demonstrating important effects of TLR1 polymorphisms on Candida bloodstream infections are strengthened by studies reporting similar effects on genetic susceptibility to aspergillosis [19], bacterial sepsis [39], or mycobacterial infections [40]. Interestingly, a recent study on the evolutionary genetics of TLRs reported a high variation in TLR1, with important evolutionary pressures exerted by infections on this receptor [38]. We must, however, concede that these specific pressures were most likely exerted by bacterial infections and not by fungal infections that are primarily caused by modern medical circumstances (eg, chemotherapy, stem cell transplantation, and invasive procedures).
Among DUMC whites and African Americans, a number of baseline risk factors were significantly more common among infected subjects on univariate analysis (data not shown). Although these differences were unavoidable due to the nature of the pathologies associated with candidemia, none of these variables were significantly associated with presence of the TLR1 polymorphisms in either infected patients or noninfected controls. Therefore it is highly unlikely that they are confounders of the association between TLR1 polymorphisms and candidemia (data not shown).
Recent studies have demonstrated that TLR2, TLR4, TLR6, and TLR9 are also implicated in recognition of components of Candida species. Polymorphisms in TLR2 have been reported as a risk factor for candidemia in a small number of patients [21], and we have reported the association of TLR4 D299G/T399I polymorphisms in a small cohort of patients [20]. These data could not be confirmed in the present study that investigated by far the largest cohort of patients with candidemia, further emphasizing the importance of recruiting cohorts of sufficient size and comparing them to controls at similar risk of infection. In contrast to TLR1, no effects of TLR6, TLR9, TIRAP, or MyD88 polymorphisms on susceptibility to Candida bloodstream infections were apparent.
Our study is subject to several limitations. Although we had a relatively large sample for a study of genetic association of an infectious disease, we had a small sample of African American patients, which limited our ability to detect differences in this segment of the population, except large differences in allelic frequency. In addition, the associations we report for TLR1 polymorphisms among whites were not significant enough to withstand adjustment for multiple comparisons. However, we demonstrated that genetic variants of TLR1 associated with higher susceptibility result in a suppressed function of TLR1, as revealed by impaired induction of cytokine responses upon activation of innate immune cells through TLR1. Together, these complementary genetic and immunologic data support a role of this receptor in antifungal host defense against candidemia.
In conclusion, the present study demonstrates the association between TLR1 polymorphisms and susceptibility to candidemia. We also found that these same TLR1 polymorphisms are associated with impaired function of TLR1 and decreased cytokine responses. Although this correlation between genetic and functional data strengthens the conclusion of the study, further research is warranted to elucidate the exact role of TLR1 signaling in host defense against Candida species in humans and how knowledge on polymorphisms in TLR1 could be used in risk assessment for strategies of prophylactic, empiric, or preemptive therapies.
Notes
Acknowledgments.
T. S. P., E. v. d. V., D. K., and M. O. performed the experiments; E. v. d. V. and D. K. selected SNPs and designed the genotyping assays; M. D. J., P. B. S., B. D. A., J. C. Y., G. M. L., J. R. P., B. J. K., and M. G. N. managed the collection of the cohort; M. D. J., D. R. V. E., and W. K. S. performed the clinical statistical analysis; T. S. P., M. D. J., J. R. P., T. W., and M. G. N. designed the study and wrote the manuscript. All authors read and approved the final manuscript. All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Financial support.
This work was supported by a Vici grant from the Netherlands Organization for Scientific Research to M. G. N. and by the National Institutes of Health (AI-51537 to M. D. J.) and (AI-73896 to J. R. P.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Gudlaugsson O, Gillespie S, Lee K, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37:1172–7. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 3.Pappas PG, Rex JH, Lee J, et al. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis. 2003;37:634–43. doi: 10.1086/376906. [DOI] [PubMed] [Google Scholar]
- 4.Tortorano AM, Peman J, Bernhardt H, et al. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur J Clin Microbiol Infect Dis. 2004;23:317–22. doi: 10.1007/s10096-004-1103-y. [DOI] [PubMed] [Google Scholar]
- 5.Viscoli C, Girmenia C, Marinus A, et al. Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC) Clin Infect Dis. 1999;28:1071–9. doi: 10.1086/514731. [DOI] [PubMed] [Google Scholar]
- 6.Wingard JR, Merz WG, Rinaldi MG, Johnson TR, Karp JE, Saral R. Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N Engl J Med. 1991;325:1274–7. doi: 10.1056/NEJM199110313251803. [DOI] [PubMed] [Google Scholar]
- 7.Trick WE, Fridkin SK, Edwards JR, Hajjeh RA, Gaynes RP. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989–1999. Clin Infect Dis. 2002;35:627–30. doi: 10.1086/342300. [DOI] [PubMed] [Google Scholar]
- 8.Marr KA, Seidel K, White TC, Bowden RA. Candidemia in allogeneic blood and marrow transplant recipients: evolution of risk factors after the adoption of prophylactic fluconazole. J Infect Dis. 2000;181:309–16. doi: 10.1086/315193. [DOI] [PubMed] [Google Scholar]
- 9.Pfaller MA, Diekema DJ, Gibbs DL, et al. Results from the ARTEMIS DISK global antifungal surveillance study, 1997 to 2005: an 8.5-year analysis of susceptibilities of Candida species and other yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J Clin Microbiol. 2007;45:1735–45. doi: 10.1128/JCM.00409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruciani M, de Lalla F, Mengoli C. Prophylaxis of Candida infections in adult trauma and surgical intensive care patients: a systematic review and meta-analysis. Intensive Care Med. 2005;31:1479–87. doi: 10.1007/s00134-005-2794-y. [DOI] [PubMed] [Google Scholar]
- 11.Vardakas KZ, Samonis G, Michalopoulos A, Soteriades ES, Falagas ME. Antifungal prophylaxis with azoles in high-risk, surgical intensive care unit patients: a meta-analysis of randomized, placebo-controlled trials. Crit Care Med. 2006;34:1216–24. doi: 10.1097/01.CCM.0000208357.05675.C3. [DOI] [PubMed] [Google Scholar]
- 12.Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 13.Agnese DM, Calvano JE, Hahm SJ, et al. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–5. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 14.Khor CC, Chapman SJ, Vannberg FO, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–8. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–32. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 16.Sutherland AM, Walley KR, Russell JA. Polymorphisms in CD14, mannose-binding lectin, and Toll-like receptor-2 are associated with increased prevalence of infection in critically ill adults. Crit Care Med. 2005;33:638–44. doi: 10.1097/01.ccm.0000156242.44356.c5. [DOI] [PubMed] [Google Scholar]
- 17.von Bernuth H, Picard C, Jin Z, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–6. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochud PY, Chien JW, Marr KA, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med. 2008;359:1766–77. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesh S, Mensah NY, Peterlongo P, et al. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. Ann N Y Acad Sci. 2005;1062:95–103. doi: 10.1196/annals.1358.012. [DOI] [PubMed] [Google Scholar]
- 20.Van der Graaf CA, Netea MG, Morre SA, et al. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. Eur Cytokine Netw. 2006;17:29–34. [PubMed] [Google Scholar]
- 21.Woehrle T, Du W, Goetz A, et al. Pathogen specific cytokine release reveals an effect of TLR2 Arg753Gln during Candida sepsis in humans. Cytokine. 2008;41:322–9. doi: 10.1016/j.cyto.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 23.Ferwerda G, Meyer-Wentrup F, Kullberg BJ, Netea MG, Adema GJ. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol. 2008;10:2058–66. doi: 10.1111/j.1462-5822.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 26.Delaney NL, Esquenazi V, Lucas DP, Zachary AA, Leffell MS. TNF-alpha, TGF-beta, IL-10, IL-6, and INF-gamma alleles among African Americans and Cuban Americans. Report of the ASHI Minority Workshops: Part IV. Hum Immunol. 2004;65:1413–19. doi: 10.1016/j.humimm.2004.07.240. [DOI] [PubMed] [Google Scholar]
- 27.Hassan MI, Aschner Y, Manning CH, Xu J, Aschner JL. Racial differences in selected cytokine allelic and genotypic frequencies among healthy, pregnant women in North Carolina. Cytokine. 2003;21:10–16. doi: 10.1016/s1043-4666(02)00489-1. [DOI] [PubMed] [Google Scholar]
- 28.Jouault T, Ibata-Ombetta S, Takeuchi O, et al. Candida albicans phospholipomannan is sensed through toll-like receptors. J Infect Dis. 2003;188:165–72. doi: 10.1086/375784. [DOI] [PubMed] [Google Scholar]
- 29.Netea MG, Gow NA, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116:1642–50. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyazato A, Nakamura K, Yamamoto N, et al. Toll-like receptor 9-dependent activation of myeloid dendritic cells by deoxynucleic acids from Candida albicans. Infect Immun. 2009;77:3056–64. doi: 10.1128/IAI.00840-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Netea MG, van de Veerdonk F, Verschueren I, Van der Meer JW, Kullberg BJ. Role of TLR1 and TLR6 in the host defense against disseminated candidiasis. FEMS Immunol Med Microbiol. 2008;52:118–23. doi: 10.1111/j.1574-695X.2007.00353.x. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi O, Sato S, Horiuchi T, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 33.Schroder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis. 2005;5:156–64. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 34.Johnson CM, Lyle EA, Omueti KO, et al. Cutting edge: a common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol. 2007;178:7520–4. doi: 10.4049/jimmunol.178.12.7520. [DOI] [PubMed] [Google Scholar]
- 35.Funderburg N, Lederman MM, Feng Z, et al. Human-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc Natl Acad Sci U S A. 2007;104:18631–5. doi: 10.1073/PNAS.0702130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoover DM, Wu Z, Tucker K, Lu W, Lubkowski J. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob Agents Chemother. 2003;47:2804–9. doi: 10.1128/AAC.47.9.2804-2809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nucci M, Anaissie E. Revisiting the source of candidemia: skin or gut? Clin Infect Dis. 2001;33:1959–67. doi: 10.1086/323759. [DOI] [PubMed] [Google Scholar]
- 38.Barreiro LB, Ben-Ali M, Quach H, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5:e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wurfel MM, Gordon AC, Holden TD, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med. 2008;178:710–20. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misch EA, Macdonald M, Ranjit C, et al. Human TLR1 deficiency is associated with impaired mycobacterial signaling and protection from leprosy reversal reaction. PLoS Negl Trop Dis. 2008;2:e231. doi: 10.1371/journal.pntd.0000231. [DOI] [PMC free article] [PubMed] [Google Scholar]