Abstract
BACKGROUND
The subject of epigenetic risk of assisted reproduction treatment (ART), initiated by reports on an increase of children with the Beckwith–Wiedemann imprinting disorder, is very topical. Hence, there is a growing literature, including mouse studies.
METHODS
In order to gain information on transgenerational epigenetic inheritance and epigenetic effects induced by ART, literature databases were searched for papers on this topic using relevant keywords.
RESULTS
At the level of genomic imprinting involving CpG methylation, ART-induced epigenetic defects are convincingly observed in mice, especially for placenta, and seem more frequent than in humans. Data generally provide a warning as to the use of ovulation induction and in vitro culture. In human sperm from compromised spermatogenesis, sequence-specific DNA hypomethylation is observed repeatedly. Transmittance of sperm and oocyte DNA methylation defects is possible but, as deduced from the limited data available, largely prevented by selection of gametes for ART and/or non-viability of the resulting embryos. Some evidence indicates that subfertility itself is a risk factor for imprinting diseases. As in mouse, physiological effects from ART are observed in humans.
In the human, indications for a broader target for changes in CpG methylation than imprinted DNA sequences alone have been found. In the mouse, a broader range of CpG sequences has not yet been studied. Also, a multigeneration study of systematic ART on epigenetic parameters is lacking.
CONCLUSIONS
The field of epigenetic inheritance within the lifespan of an individual and between generations (via mitosis and meiosis, respectively) is growing, driven by the expansion of chromatin research. ART can induce epigenetic variation that might be transmitted to the next generation.
Keywords: assisted reproduction, epigenetics, human, genomic imprinting, transgenerational epigenetic inheritance
Introduction
In 2009, Katari et al. have reported that some genes from babies conceived by means of IVF show a gene expression pattern that is different from naturally conceived children (Katari et al., 2009). The observed changes were associated with the mechanism that switches genes on and off, which is heritable to forthcoming cell generations, hence under epigenetic control. In their study, Katari et al. have stated that this mechanism could put children conceived by means of assisted reproduction treatment (ART) at a greater risk of diseases, such as diabetes and obesity, later in life.
Epigenetic deregulation already received increasing attention as a possible common cause of adverse ART outcomes, since the incidence of disorders that involve imprinted genes, especially the Beckwith–Wiedemann syndrome (BWS), is often reported to be increased in the offspring (Amor and Halliday, 2008, Ceelen et al., 2008b, Manipalviratn et al., 2009). This led to an expansion of ART literature on epigenetic effects, also including mouse model studies. The subfertility of one or both parents as a causative factor has to be taken into account, which often is difficult to achieve. Moreover, mouse models in this area are almost absent. Another question is if the assumed epigenetic effects of ART can be transmitted to the next generation.
Besides classical Mendelian inheritance of information stored in the DNA sequence, other mechanisms are active in the transmission of phenotypic traits across generations too. No insight into underlying molecular mechanisms was available when the theory of non-genetic transmission was first put forward by Waddington (1953) to describe the acquired characteristics in the offspring of Drosophila exposed to heat (McLaren, 1999). In the broad sense coined by Waddington, these observations are termed transgenerational epigenetic effects (Youngson and Whitelaw, 2008). When these phenotypic alterations are caused by transfer of chromosome/chromatin modifications through the gametes, the term transgenerational epigenetic ‘inheritance’ is used (Youngson and Whitelaw, 2008; Jablonka and Raz, 2009). In this definition, the word epigenetic refers to the mechanisms involved in the mitotic and meiotic transfer of non-genetic (i.e. not DNA sequence based) information. Transgenerational epigenetic inheritance has been proved in organisms ranging from bacteria and plants to the mouse and humans (Jablonka and Raz, 2009).
Epigenetic mechanisms exist as an interplay between DNA methylation, RNA-mediated chromatin modifications, histone modifications and histone variants, but likely also less well-studied mechanisms, such as the organization of nuclear structure including chromosome replication behaviour (Jablonka and Raz, 2009; Margueron and Reinberg, 2010). In the context of this review, inheritance via DNA methylation will be mainly discussed.
The molecular epigenetic mechanisms are instrumental in the specification of cell identity and potency within generations. Epigenetic mechanisms are thought to concertedly orchestrate the spatial and temporal regulation of cell differentiation throughout development (Goldberg et al., 2007; Margueron and Reinberg, 2010; Zaidi et al., 2010). Since all cells of an organism have the same genotype, epigenetic marks are deposited to alter transcription and achieve cell-type specific gene expression patterns in different tissues. In fact, the definition of epigenetics given by Jabonka and Raz (2009) as ‘the study of the processes that underly developmental plasticity and canalization and that bring about persistent developmental effects in both prokaryotes and eukaryotes’ underlines its role in embryogenesis and cell differentiation. Sex-specific genomic imprinting and stable female X-inactivation are also under epigenetic control.
Between generations, the germ line is subjected to two distinct reprogramming events [one in the primordial germ cells (PGCs) and one in the preimplantation embryo], in order to prepare the cells for pluri- and totipotency and down-regulate the inheritance of epigenetic information between generations [reviewed in (Reik et al., 2001; Morgan et al., 2005; Feng et al., 2010)]. Both phases are pertinent to ART and especially the second one (in the preimplantation embryo) owing to the in vitro circumstances at this epigenetically crucial phase of development.
The discovery that some loci, notably imprinted ones, escape reprogramming in the early embryo, provided a first hint regarding the mechanism behind epigenetic inheritance as this brought into question the rigidity of epigenetic erasure between generations as a principle. Epigenetic marks are generally thought to be stable through rounds of somatic mitosis after initial deposition in development (Margueron and Reinberg, 2010). This careful balance between somatic maintenance of epigenetic marks and dynamic reprogramming in the germline has led epigenetic mechanisms to be put forward as a vehicle for ‘soft inheritance’ (Youngson and Whitelaw, 2008), a term first introduced to describe a more pliable system of inheritance, which would allow organisms to quickly adapt to fluctuations in nutrition, predation or disease (Mayr and Provine, 1980; Mayr, 1982).
The question we will pursue here is whether the conditions during gametogenesis and the in vitro phases intrinsic to ART could elicit epigenetic effects and, if so, whether these could be transmitted to the next generation. We will first present the processes of mitotic epigenetic inheritance. Next, we will describe the known molecular mechanisms involved in the escape of germline reprogramming and present environmental and hormonal cues that induce alterations in the epigenome to be passed on to the next generation.
In the second part of this review, observations of a molecular epigenetic nature made in mouse and human ART will be presented. Finally, we will attempt to integrate these observations, including the likelihood of transgenerational epigenetic inheritance, and designate the areas of human reproduction in the context of ART where insight is lacking most.
Methods
In order to gain information on transgenerational epigenetic inheritance and epigenetic effects induced by ART, literature databases (Pubmed, Medline) were thoroughly searched for papers on this topic by using relevant keywords.
Epigenetic inheritance and germline reprogramming
Mitotic inheritance of epigenetic marks
To address the mechanisms involved in transgenerational epigenetic inheritance, DNA must be viewed in the context of chromatin and its modifications (see Box I). The epigenetic chromatin state is tightly linked to transcription and as cells differentiate they acquire tissue-specific patterns of DNA methylation, histone modifications and other epigenetic chromatin marks (Lange and Schneider, 2010; Zaidi et al., 2010). In this context, the faithful transmission of this epigenetic signature to daughter cells is essential for lineage commitment. Mitotic transmission of epigenetic marks is observed throughout somatic cellular development (Zaidi et al., 2010) and insight into the mechanisms involved is gradually increasing (Cheng and Blumenthal, 2008; Probst et al., 2009; Kaufman and Rando, 2010; Margueron and Reinberg, 2010).
Box I: Chromatin and epigenetic mechanisms.
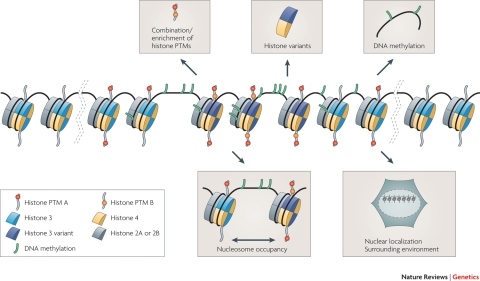
Chromatin is the structure in which DNA is packaged inside the cell nucleus. The fundamental unit of chromatin is the nucleosome, which consists of 147 base pairs of DNA wrapped around a histone octamer that contains two duplicates of histone H3, H4, H2A and H2B (Kouzarides, 2007). Nucleosomes are subsequently compacted further into higher-order chromatin structures. The degree of chromatin packing is dynamically regulated and can be either heterochromatic (densely packed chromatin, transcriptionally repressed) or euchromatic (accessible chromatin, transcriptionally active).
An intricate network of epigenetic mechanisms operates to regulate the chromatin state and thereby transcription (Fig. 1). The best-studied chromatin modifications are the methylation of DNA, which occurs at CpG sites in mammals (Doerfler, 2008), and the modification of the N-terminal histone tails (Kouzarides, 2007).
Figure 1.
Characteristics of a chromatin domain. Schematic representation of the covalent and structural features that define a certain chromatin domain. Different contributing factors are highlighted in shaded boxes. The dashed line represents a separation between two adjacent domains. Figure from Margueron and Reinberg (2010).
In DNA, the cytosine member of a CG dimer when methylated at carbon 5 acts as a fifth base. Depending on density and position, this mark is associated with gene inactivation, and the formation of heterochromatin. Once methylated, the 5methyl CpG is copied during S phase. Hence, the deposition of methyl groups to DNA in mammals is catalysed by two families of DNA methyltransferases (DNMTs) that exhibit de novo (DNMT3) and predominantly maintenance (DNMT1) activity (Cheng and Blumenthal, 2008), whereas no demethylases have been described (Lucifero et al., 2004, Popp et al., 2010). In PGCs, effects on the conversion of 5methyl CpG to CpG by ablation of the cytidine deaminase AID has been found (Popp et al., 2010) that can achieve this effect in combination with base excision repair that is indicated to be active at this stage (Hajkova et al., 2010). Another pathway may make use of the Tet1,2,3 family of enzymes that convert 5methylCpG into 5hydroxymethylCpG. This pathway is preferentially active in the male pronucleus of the zygote (Wossidlo et al., 2011) and Tet enzymes have been shown to be present in PGCs (Hajkova et al., 2010). The epigenetic effect of this conversion is currently under investigation (Szulwach et al., 2011).
Post-translational modifications of histone tails include methylation, acetylation, phosphorylation, ubiquitination and sumoylation and are thought to be of a more dynamic nature than DNA methylation as they are deposited and removed by a wide variety of enzymes (Kouzarides, 2007). As opposed to DNA methylation, which is generally involved in the repression of transcription, histone tail modifications display a wide variety of functions, both repressive and activating, and are often useful for describing the functional state at the level of a single gene to large chromatin domains (Filion et al., 2010). Histone marks exert their influence either by directly changing the structure of chromatin or via the recruitment of chromatin-binding factors like transcription factors and ATP dependent chromatin-remodelling complexes (Clapier and Cairns, 2009). Together with the incorporation of histone variants and the use of a large variety of non-coding long and small RNAs (Moazed, 2009), these modifications interact in a complex and delicate web to regulate structural aspects of chromatin domains, such as chromatin compaction, nucleosomal occupancy and the localization inside the nucleus. Insight in what constitutes these domains and how this affects transcription is slowly becoming available (Filion et al., 2010), unravelling a web of dazzling complexity. Most of the elements touched upon appear in Fig. 1.
To enable replication of DNA during the S-phase of the cell cycle, the chromatin structure is severely disrupted, which results in a partial loss of epigenetic marks. Thus, special mechanisms need to be in place to ensure the mitotic propagation of epigenetic information. Although some insight into mitotic inheritance of DNA methylation and histone modifications exists, the propagation of other epigenetic properties, such as histone occupancy and histone variants, remains largely elusive.
DNA methylation is transmitted semi-conservatively through DNA replication via the two hemimethylated DNA double helices. The DNA methyltransferase DNMT1 interacts with PCNA (proliferating cell nuclear antigen), a processivity factor of the replication machinery that forms a clamp around the DNA template (Chuang et al., 1997), and is able to specifically recognize hemimethylated DNA (Hermann et al., 2004) catalysing the addition of a methyl group to complementary CpGs of the newly synthesized strand. The error rate in base pairing is 1 in 109. The error rate in adding the CH3 to the inserted C opposite a methylated CpG is one in 40 in somatic cell (leukocyte) divisions (Fu et al., 2010).
In contrast, the propagation of the more dynamic histone marks is more complex and not completely understood. As modified histones are removed to enable passage of the replication fork, no template is available to regulate the reassembly of histones and their marks on the daughter strands. In the leading current model, newly synthesized and old histones (the latter containing their original modifications) are incorporated into daughter-chromatin randomly, thus severely diluting the modifications (Margueron and Reinberg, 2010). To prevent the gradual fading of histone marks over cell divisions, it has been proposed that these diluted modifications are able to recruit histone-modifying enzymes directly after replication, which would catalyse further deposition of the mark in a positive-feedback loop (Lange and Schneider, 2010, Margueron and Reinberg, 2010). The limited reliability of the transmission of histone marks that follows from this model combined with the fact that, in contrast to CpG methylation, histone modifications can easily be removed by a wide range of enzymes, illustrates the notion that the methylation status of DNA is more faithfully inherited over mitotic divisions than are histone modifications. Hence, CpG methylation is a more indicative tool to predict the functionality of a certain locus over forthcoming mitotic generations (Peters and Schubeler, 2005; Feng et al., 2006).
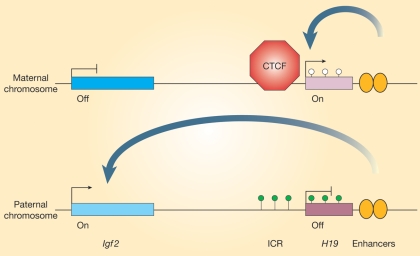
Another mechanism for the mitotic propagation of epigenetic marks is the relation between the timing of replication of a certain locus during S-phase and gene activity (Gondor and Ohlsson, 2009). Transcriptionally active genes or alleles are replicated earlier in S-phase than inactive genes or alleles. This timing pattern of DNA replication changes throughout development as cells differentiate and is intimately linked to the chromatin state. A striking example is provided by the imprinted genes (see Box II) H19 and insulin growth factor 2 (Igf2) (Fig. 4). Imprinting is maintained on the maternal allele by CpG methylation-sensitive binding of the chromatin insulator CTCF to the imprint control region (DMR) and disturbance of CTCF-binding by mutating the binding sites of the maternal DMR caused the complete maternal H19/Igf2 domain to be replicated in early, instead of late, S-phase (Bergstrom et al., 2007).
Figure 4.
Epigenetic state of the imprinted Igf2/H19 domain on the maternal and paternal genome. On the paternal chromosome, the H19 gene and the adjacent DMR are methylated preventing H19 expression and the binding of the insulator CTCF, thus allowing enhancers access to the Igf2 gene promoting its expression. In the absence of DMR methylation on the maternal chromosome, bound CTCF prevents enhancer activity reaching Igf2, effectively silencing the gene. Instead, enhancer activity is limited to the unmethylated H19 gene resulting in its expression on the maternally contributed chromosome. Figure from Reik and Murrell (2000).
Box II: Genomic imprinting.
Genomic imprinting operates to silence the maternal or paternal alleles of genes that often are organized in clusters. The gene clusters/genes subjected to genomic imprinting are regulated by an imprinting control region (ICR, also called a germline differentially methylated region or germline DMR). In a cluster, genes that are exclusively expressed from the paternal allele can co-exist with genes that are exclusively expressed from the maternal allele. However, the DMR is marked by CpG methylation in one of the two germlines (Reik and Walter, 2001). Hence methylation occurs in a sex-specific manner and is maintained throughout fertilization, embryonic and subsequent development (Fig. 2; Reik and Walter, 2001). Of the 15 human germline DMRs listed by Morison et al. (2001) (update Jan 2011) only two are methylated in the male germline. These DMRs are always located in an intergenic region, as with the DMR between H19 and IGF2 (=H19 DMR) and between DLK1 and MEG3 (=GTL2) (=IG-DMR). In mice a third intergenic paternally methylated DMR is located between Rasgrf1 and A19 (Rasgrf1 DMR) (Thorvaldsen and Bartolomei, 2007). In the female germline, the DMR is always located in a promotor region. When more than one gene is regulated by this DMR, the promotor often involves a long non-coding RNA (i.e. no protein product) that is instrumental in the allelic exclusion by inducing a local chromatin change in cis, as with the KvDMR1 in the KCNQ1OT1 (=LIT1) promotor (Kacem and Feil, 2009). For other imprinted genes like MEST (=PEG1) or PLAGL1 (=ZAC1) the imprint mechanism is unknown yet.
Figure 2.
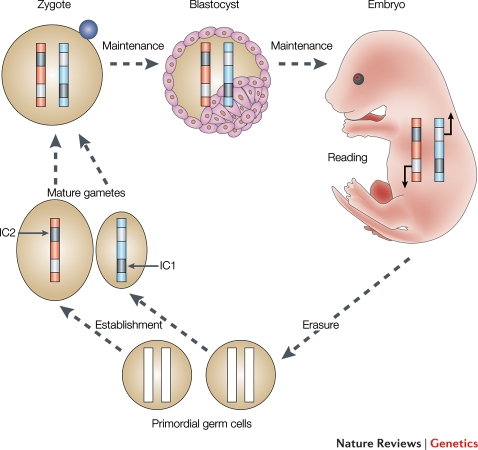
Imprinting in the germline. Erasure, establishment and maintenance of methylation imprints at imprinting control regions during germ cell and embryonic development. Imprinting control regions (IC1) and IC2 are shown as examples. Grey indicates modification and white indicates no modification at the corresponding alleles. Parental chromosomes are marked according to their sex in blue (male) or red (female). The reading in the developing embryo is indicated by arrows. Figure from Reik and Walter (2001).
At the start of germ cell development when the PGCs enter the developing gonad (Fig. 3) the imprints are erased and will be re-established later according to the sex of that germline. The deposition of new imprinting methylation marks has been associated with DNMT3a (Lucifero et al., 2004; Kaneda et al., 2010) and starts at E15 (mouse prenatal Day 15) in the male germ cells (prospermatogonia). Remethylation in the oocyte follows over a wider timeframe during post-natal follicle development. The silencing of alleles within an imprinted gene cluster requires a number of additional epigenetic changes that include the methylation of promoters, antisense transcription and the loss of permissive (Ciccone et al., 2009) and acquisition of repressive histone modifications (Terranova et al., 2008). The total number of human imprinted genes is assessed at 70 (Morison et al., 2001) and new discoveries with a profound physiological impact, such as involvement in type II diabetes and high-density lipoprotein cholesterol metabolism (Small et al., 2011) and social behaviour (Garfield et al., 2011) are currently being made.
Reprogramming the genome towards totipotency
As already touched upon in the previous paragraphs, in contrast to the mitotic transmission of epigenetic marks within the lifespan of an individual, the inheritance of epigenetic information between generations is generally actively prevented. The classical view is that to restore the germline to the totipotent state of the early preimplantation embryo, differentiating epigenetic marks are removed by two phases of epigenetic reprogramming: at the PGC stage until just after their entry in the incipient gonad and after fusion of sperm and oocyte in the zygote and during the first cleavage divisions (Morgan et al., 2005; Feng et al., 2010). Although reprogramming also entails dynamic changes in histone modifications and variants (Hajkova et al., 2008), the erasure of DNA methylation has been studied most extensively (Popp et al., 2010).
Box III: Retrotransposons in the Mouse and Human.
Almost half of the genome of the mouse (∼50%) and humans (∼40%) consists of transposable elements (TEs): mobile DNA elements which had the ability to allocate cellular transcription machinery for their replication. The majority of these fall into the category of retrotransposons, which require an RNA intermediate to duplicate themselves. Two types of retrotransposons can be discerned, based on whether or not they have long terminal repeats (LTRs) at their ends. LTR-retrotransposons that make up about 8–10% of the genome are also known as endogenous retroviruses as they are believed to be remnants of infectious retroviruses that have lost the ‘envelope’ gene necessary for capsid production. They do still possess Gag and Pol proteins, allowing them to reverse-transcribe their RNA in the cytoplasm and reintegrate into a new genomic locus. Non-LTR retrotransposons are most abundant in the mammalian genome and consist of short and long interspersed nucleotide elements (SINEs and LINEs) of which the LINE-1 (L1) subclass represents 17–20% of the human and mouse genome mass (Brouha et al., 2003). SINEs have no autonomous duplication potential but instead are mobilized in trans by the LINE encoded machinery (Goodier and Kazazian, 2008).
The presence of retrotransposons in the genome has various structural effects ranging from simple disruption of genes by insertional mutagenesis to chromosome rearrangements caused by homologous recombination (Goodier and Kazazian, 2008). Although these genomic alterations have mostly negative effects in the short-term, retrotransposons are involved in the expansion and structural evolution of the genome and are crucial in the evolution of new proteins and regulatory sequences (Volff, 2006). In mammals, retrotransposons are mostly located in euchromatic regions scattered around genes (Goodier and Kazazian, 2008). The strong, constitutive promoters of retrotransposons can disturb expression of nearby genes, either directly or via the spread of repressive epigenetic marks associated with retrotransposons.
Although most retrotransposons are rendered inactive by truncation or mutation, some are still potentially able to duplicate and reintegrate in the genome, of which the LTR-containing intracisternal A particles (IAPs) in the mouse are an excellent example. These elements are silenced throughout the germline by means of epigenetic mechanisms that include DNA methylation, chromatin modifications and RNAi pathways (Zamudio and Bourc'his, 2010).
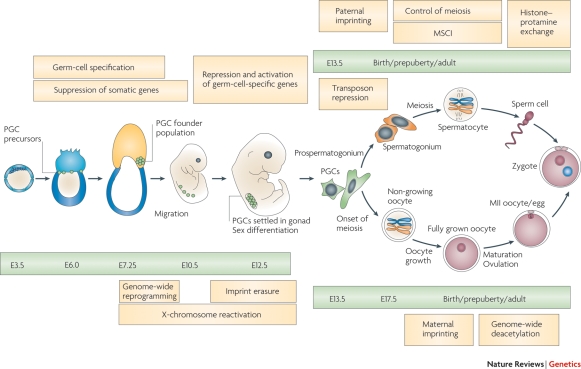
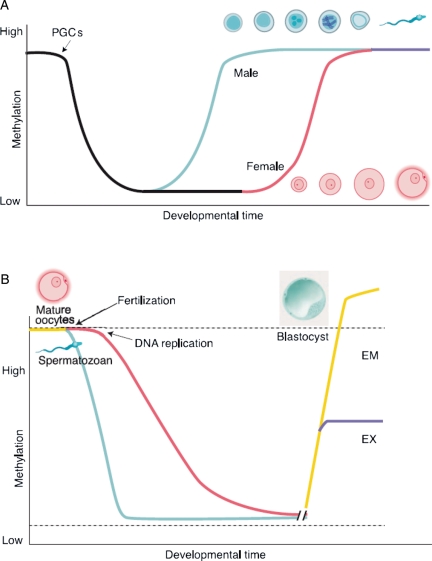
The start of germ cell development from epiblast cells around E7.25 (Figs 3 and 5A) announces the first phase of epigenetic reprogramming (demethylation) in the mouse, which continues after these PGCs have reached the gonad at E10.5 and lasts until E13.5 (Seki et al., 2005) just before female meiosis commences. At this point, <10% of CpGs retain their methylation mark, whereas 70–80% of CpG sites are methylated in embryonic stem (ES) and somatic cells indicating the magnitude of reprogramming (Popp et al., 2010). Demethylation is observed at nearly all sequence elements including promoters and genic, intergenic and transposon sequences (see Box III) and is thought to be active, as maintenance DNMT1 is present in the nucleus throughout the process (Hajkova et al., 2002; Morgan et al., 2005). The allele-specific methylated DMRs of imprinted regions are also demethylated between E10.5 and E12.5, with the precise timing individually controlled for each gene cluster (Lee et al., 2002; Hajkova et al., 2008). However, at the DMRs of the H19 and Snrpn gene clusters, the originally methylated sequences are de novo methylated at an earlier stage in subsequent spermatogenesis (H19) and oogenesis (Snrpn) than the originally unmethylated ones, suggesting that some kind of memory is conferred (Davis et al., 1999, Lucifero et al., 2004). The de novo methylase Dnmt3a in collaboration with Dnmt3L is responsible for resetting the sex specific germline DMR imprint (Shovlin et al., 2007; Kaneda et al., 2010).
Figure 3.
Chronology of mouse germ cell development and the main epigenetic events that occur. PGCs first emerge at embryonic Day 7.25 (E7.25) after which they migrate to the gonad where they enter the process of sex differentiation and eventually develop into full-grown gametes. Green bars indicate the developmental stage and yellow bars indicate the epigenetic processes occurring at these points in the germ line. MSCI stands for meiotic sex chromosome inactivation. Figure from Sasaki and Matsui (2008).
Figure 5.
(A) Methylation reprogramming in the germ line. PGCs in the mouse become demethylated early in development, between E7.5 and E13.5. Remethylation begins in prospermatogonia in male germ cells, and after birth in growing oocytes. (B) Methylation reprogramming in preimplantation embryos. The paternal genome (blue) is demethylated by an active mechanism immediately after fertilization. The maternal genome (red) is demethylated by a passive mechanism. Both are remethylated around the time of implantation to different extents in embryonic (EM) and extraembryonic (EX) lineages. Methylated imprinted genes and some repeat sequences (dashed line) do not become demethylated. Unmethylated imprinted genes (dashed line) do not become methylated. Figure from Reik et al. (2001).
The most evident exception to genome-wide demethylation in mouse PGCs known to date is formed by a class of retrotransposons called IAPs (see Box III), which show only partial demethylation (Popp et al., 2010). As PGCs proceed into gametogenesis, the acquisition of specific male and female methylation patterns (Fig. 5A) is accompanied by a range of other epigenetic changes. In spermatogenesis, considerable chromatin-remodelling occurs, especially during meiosis in the adolescent and adult male, involving the incorporation of testis-specific histone variants (Gaucher et al., 2010). Ultimately, histones are exchanged for protamines as a stress resistant packaging tool in spermatogenesis. This was long thought to limit the potential for male transgenerational epigenetic inheritance, despite the sizable portion of histones that, especially in the human, remains (Tanphaichitr et al., 1978; Gatewood et al., 1987). Recent studies provide strong indications that histone modifications of these remnant nucleosomes play a role in embryonic development (Hammoud et al., 2009; Brykczynska et al., 2010).
The chromatin changes that prepare the oocyte for driving embryonic development have hardly been characterized, likely by limitations in cell numbers that limit technical approaches and lack of theoretical concepts but molecular biology will also tackle this very interesting subject.
A second phase of reprogramming (Fig. 5B) starts early after gamete fusion with the rapid, active conversion of the five cytosine methyl group of CpG dimers into a hydroxymethyl group on mainly paternal DNA (Wossidlo et al., 2011). At this point, protamines are already exchanged for hyperacetylated histones from the maternal pool in an unknown but active process. In the zygote, the female genome also passively loses methylation related to a decrease in DNMT1 activity in the pronuclei (Cirio et al., 2008). Passive demethylation is continued until the morula stage after which the inner cell mass is differentially methylated from extra-embryonic lineages (Santos et al., 2002). Both paternally and maternally imprinted DMRs retain methylation during this phase (Howell et al., 2001; Hirasawa et al., 2008), along with IAPs (Lane et al., 2003) and regions of centromeric heterochromatin (Rougier et al., 1998). At implantation, DNMT3b catalyses de novo methylation to repress the germline expression programme and mediate the transition to terminal differentiation programmes (Borgel et al., 2010). This recent study identified a number of non-imprinted genes that inherited promoter DNA methylation from the parental gametes, suggesting that even more sequence elements are capable of escaping early embryonic reprogramming (Borgel et al., 2010). Also in this area, a more comprehensive insight of chromatin reprogramming at the onset of embryonic development is still lacking. However, there is no doubt about the concept as such, as is amply demonstrated by the poor and variable outcomes of reproductive cloning from somatic cell nuclei (Hochedlinger and Jaenisch, 2002).
One other aspect, which is pertinent in this context, is that of epigenetic asymmetry between male and female chromatin in the early embryonic phase of reprogramming originating from the grossly different chromatin states at the onset of fertilization: a protamine-dominated chromatin in sperm versus an exclusively nucleosomal chromatin organization of the female meiotic chromosomes. Male and female pronuclei at their formation both differ in histone modifications on the N-terminal tail and in histone variants, notably of H3 (van der Heijden et al., 2005, 2009). During the first cleavage divisions, paternal and maternal chromosomes will become more similar as to their gross histone modification pattern (Puschendorf et al., 2008).
Transgenerational epigenetic inheritance: escaping reprogramming in the mouse occurs in both male and female germline
When looking at transgenerational epigenetic inheritance, sequence elements that escape reprogramming are of special interest as these might retain information over multiple generations. IAPs are observed to escape demethylation in the mouse preimplantation embryo, although stable methylation of both elements serves different purposes. IAPs are among the most active retrotransposons in the mouse genome and silencing of these elements is thought to be the original evolutionary function of repressive CpG methylation and crucial for the prevention of retrotransposon induced mutations (Walsh et al., 1998; Zamudio and Bourc'his, 2010). Imprinting has only been described in mammals, plants and insects as part of the sex-specific developmental programme and is in this sense a much more recent phenomenon in evolution, indicating that imprinting mechanisms might be originally derived from those involved in retrotransposon silencing, also suggesting mechanistic similarities (Ideraabdullah et al., 2008). Indeed, the fact that both DMRs of imprinted loci and the LTRs of IAP retrotransposons behave similarly in the targeting of de novo methylation in the developing oocyte after PGC reprogramming (Lucifero et al., 2004), and in the protection from active demethylation in the zygote (Lane et al., 2003) are strong hints in this direction. The repeat-like nature of both sequence elements has been suggested to be an important factor in the acquisition of CpG methylation (Lucifero et al., 2004). In the mouse, IAPs are still the best candidate for building up epigenetic inheritance as their methylation levels are relatively high at the time of active demethylation in early PGCs, in contrast to DMRs (Popp et al., 2010).
The most striking evidence for transgenerational epigenetic inheritance in the mouse comes from experiments studying the Agouti (A) locus, a determinant of coat colour (Morgan et al., 1999). Agouti alleles carrying an upstream IAP element exist of which agouti viable yellow (AVY) has been most extensively studied. The cryptic promoter in the LTR of the IAP causes aberrant expression of the Agouti locus dependent on its level of CpG methylation. A gradient of yellow via mottled to pseudo agouti (wildtype) fur phenotypes exists caused by a similar gradient from an undermethylated LTR promoter (gene expression, yellow) to a fully methylated LTR (restricted expression, pseudo agouti) (Morgan et al., 1999). It was observed that the average coat colour of the offspring was correlated to the coat colour of the mother, excluding the possibility of a maternally contributed environment (Wolff et al., 1998; Morgan et al., 1999). Thus, this effect had to be generated by the gametic transfer of epigenetic information. Epigenetic transmission of coat colour via the father is only observed in certain genetic backgrounds (Rakyan et al., 2003). A parallel example of inheritance of epigenetic marks in the mouse is provided by the Axin-fused (AxinFu) allele, which is involved in embryonic axis formation (Rakyan et al., 2003). AxinFu also contains an inserted IAP retrotransposable element that causes aberrant expression of Axin leading to a kinked tail. Again, the allele is differentially expressed dependent on the methylation status of the LTR promoter and epigenetic transmission of the kinky tail phenotype is observed in both the male and female germline (Rakyan et al., 2003). Thus, it seems that the evolutionary conserved ability of IAPs to resist epigenetic reprogramming between generations renders their epigenetic state transgenerationally relatively stable, facilitating a mechanism of epigenetic inheritance. Typical for this mode of inheritance is the inter-individual variation that is shown in each generation. Pseudo agouti mums will always produce yellow as well as pseudo agouti offspring, the distribution of which depends on the phenotype of the mother.
The mechanism by which sequence elements like the LTRs of IAPs and the DMRs of imprinted regions are able to conserve their methylation status through reprogramming events is not yet fully clear. In the early mouse embryo, Dnmt1 can occur in a somatic form (Dnmt1s) and a more stable maternal form, inherited from the oocyte (Dnmt1o). Dnmt1o is not present in the nucleus during the first three cleavage divisions and its migration to the nucleus at the 8-cell stage has been deemed necessary for the maintenance of imprints (Howell et al., 2001). More recently, Dnmt1s has been shown to be present in the preimplantation embryo from the 2-cell stage on (Cirio et al., 2008), providing the necessary tools for selective preservation of DNA methylation in the germline. The involvement of Dnmt1o in the preservation of methylation was not only shown for a cryptic promoter such as an IAP (Gaudet et al., 2004) but also for DMRs. Loss of maternal Dnmt1o led to loss of imprinting at many loci, including paternally imprinted H19 and maternally imprinted Snrpn (Howell et al., 2001), resulting in profound phenotypic variation in the offspring expressed as a retarded development at mid gestation in 60% of the embryos owing to the deficiency in the maintenance of imprinting marks (Toppings et al., 2008). In addition to Dnmt1, various other proteins have been suggested to play a role in the prevention of demethylation. A poorly understood aspect of the rescue of certain sequence elements from demethylation is how these target sequences are recognized. Several proteins have been suggested to be involved in conferring chromatin specificity. These include the CpG binding protein Mbd3 (Reese et al., 2007) and the maternal-effect proteins Zfp57 (Li et al., 2008), Stella (Payer et al., 2003) and the RNA elongator factor Elp3 (Okada et al., 2010). These results show that the oocyte is involved in the maintenance of transgenerational epigenetic marks.
Although DNA methylation has been proved to be an important tool for silencing imprinted alleles and IAPs in the germline, other epigenetic mechanisms are almost certainly at play. Despite the observed epigenetic inheritance at the AVY and AxinFu alleles, the LTR of the IAP of these has been shown to be demethylated at the blastocyst stage (Blewitt et al., 2006; Fernandez-Gonzalez et al., 2010) thus indicating that other mechanisms might contribute to conferring memory of the repressed epigenetic state. Additional evidence for this is provided by a recent study in mouse ES cells carrying mutations for Dnmt1, in which no increase in IAP transcripts was detected despite the absence of DNA methylation at IAP loci (Hutnick et al., 2010). To explain these observations, different epigenetic mechanisms have been proposed to operate concertedly with CpG methylation in early embryonic silencing. A recent study showed active and inactive alleles of AxinFu to be differentially modified at the blastocyst stage with activating and inactivating histone marks, respectively (Fernandez-Gonzalez et al., 2010), and repressive histone modifications were found to be involved in the paternal marking of the Kcnq1ot1 imprinted cluster from the zygote stage on (Terranova et al., 2008).
In addition to these chromatin-based epigenetic marks, regulatory small RNAs have recently been implicated to be directly involved in the transfer of epigenetic variation via sperm in the mouse (Rassoulzadegan et al., 2006; Wagner et al., 2008; Grandjean et al., 2009). The observation that human sperm also contains a wide variety of RNA-species led to the speculation that similar RNA-based mechanisms play a role in epigenetic inheritance in humans (Lalancette et al., 2008; Cuzin and Rassoulzadegan, 2010). It is becoming more and more clear that RNAs play a major role in fertility (Bourc'his and Voinnet, 2010; Cuzin and Rassoulzadegan, 2010). A hint of the mechanisms by which they facilitate transgenerational epigenetic effects is provided by the observation that different species of small RNAs are involved in the suppression of retrotransposons. In the male mouse germline, piRNAs (PIWI interacting RNAs) assist in the degradation of retrotransposon RNA transcripts, thus preventing reintegration in the genome, and guide de novo methylation of partially demethylated TEs during epigenetic reprogramming in the embryo and during spermatogenesis (Aravin et al., 2008, 2009; van der Heijden et al., 2010). In the female germline, other classes of small interfering RNAs (miRNAs and siRNAs) have also been demonstrated to assist in retrotransposon silencing (Murchison et al., 2007; Watanabe et al., 2008). Experimental results (Lykke-Andersen et al., 2008) and theoretical considerations (Bourc'his and Voinnet, 2010) predict small RNAs to be involved in early embryonic development as well. The recently observed dynamic changes in populations of small RNA species from the mature secondary oocyte to the blastocyst might facilitate the necessary requirements for developmental gene regulation as well as retrotransposon silencing (Ohnishi et al., 2010).
As demonstrated by the discussed observations that RNA as well as modified histones retained in sperm likely are involved in early embryonic development, the belief that the male germline contributes no more than the paternal genome sequence to the zygote is less and less valid (Yamauchi et al., 2011). In line with this growing comprehension and in addition to the above-introduced ‘maternal-effect’ chromatin involved genes (such as Zfp57, Stella and Elp3 contributed in either RNA or protein by the oocyte), evidence for ‘paternal-effect’ genes has been presented (Chong et al., 2007). Here it was shown that ‘chromatin metabolism’ during spermatogenesis influences paternal gene expression in the next generation. Differences between female and male inheritance of epigenetic variation might be partially explained by these chromatin alterations during gametogenesis mediated by maternal- and paternal-effect genes.
In summary, both the maternal and paternal germline possess the tools necessary for the transmission of epigenetic marks. However, differences in the maternal and paternal genotype, expressed before, at and after fertilization, are likely to influence the inheritance of epigenetic variation in a sex-specific manner. The epigenetic pathways outlined here likely also have a bearing on the recently discovered phenomenon of transgenerational genetic effects (Nadeau, 2009). As an example, a paternal ancestral genotype determines food intake and body weight for multiple generations in offspring not carrying the obesity resistance conveying ancestral genetic variant (Yazbek et al., 2010). As more conditional mutants (i.e. their expression is restricted to the germline) for chromatin involved genes become available in the mouse, our insight into these very intriguing aspects of transmission biology will undoubtedly grow.
Stress, hormone and nutrition-induced transgenerational epigenetic variation
In experimental animals, such as mice and rats, a range of external influences, including irradiation stress, exposure to hormones and nutrition, has been shown to induce variation in the epigenome (Jirtle and Skinner, 2007; Kovalchuk and Baulch, 2008). Some studies have focused on investigating the possibility that this variation might be transmitted to subsequent generations. When F0 pregnant females carrying F1 embryos that already contain germ cells that will produce the F2 are treated with an inducing agent, observations have to be extended into the F3 generation in order to exclude a direct effect of the treatment on the germ cells, observed in the F2 individuals (Jablonka and Raz, 2009). When F0 adult males are treated, a transmissible epigenetic effect can be concluded at the earliest in the gametes of the F1. Evidence for the induction of heritable epigenetic variation by hormones, nutrition and stress will be briefly evaluated.
Genotoxic stress-induced transgenerational epigenetic variation
Currently known examples of stress-induced transgenerational epigenetic effects in the mouse mostly involve irradiation as the triggering event and these have recently been reviewed (de Boer et al., 2010). In addition to the direct genotoxic effects of radiation on cells, notably the nucleus, it has been recognized that radiation can induce an increased rate of DNA breaks and mutations in descendent cells and even across generations (Morgan, 2003a, b). This phenomenon is termed radiation-induced genomic instability. Germline transmission has been shown to occur exclusively after male exposure although subsequent transmission is observed via both sexes. The most widely used experimental setup to detect transmission of delayed effects of irradiation uses the mutation frequency at expanded simple tandem repeat (ESTR) loci as the readout of genomic instability (the mutation is read as a change in repeat number; Dubrova et al., 1993). The fact that non-Mendelian inheritance of an increased ESTR mutation rate is usually observed led epigenetic disturbances to be suggested as a causative factor (Barber et al., 2006). This notion was further supported by the observation that irradiation results in a quickly induced global hypomethylation of DNA (Koturbash et al., 2005; Pogribny et al., 2005; Loree et al., 2006). After testis irradiation, hypomethylation of retrotransposed interspersed repeat elements (LINEs and SINEs) was found in the offspring (Filkowski et al., 2010) next to lower levels of the methylated CpG binding protein MeCP2 and the DNA homologous recombination repair protein RAD51 in the thymus (Koturbash et al., 2006). These experiments show that genotoxic stress in the male germline can induce genetic and epigenetic variation in the offspring.
Nutrition, hormones and epigenetic variation
Epigenetic effects caused by nutrition as well as hormones have mainly been described after induction in late embryos and early fetuses, when PGCs are arising and migrate to the early gonad. Recent publications have demonstrated that the adult male is also capable of acquiring nutrition-induced epigenetic variation in the germline (Carone et al., 2010; Ng et al., 2010). The first hint was provided by the discovery that premating fasting of male mice led to altered serum glucose levels in offspring (F1) (Anderson et al., 2006). Chronic exposure of male rats to a high-fat diet was associated with pancreatic beta cell dysfunction in the offspring (Ng et al., 2010), while a low-protein diet in the male mouse affected the hepatic expression of genes involved in proliferation and cholesterol biosynthesis (Carone et al., 2010). In the mouse, the diet was found to be correlated with differential methylation of several lipid metabolism-related genes in the liver of both male and female offspring. These results show that, next to the oocyte and the developing embryo in the female, sperm is sensitive to nutrition-induced epigenetic variation. The next step in this line of research is to verify that these effects are transmitted to subsequent generations (F2) and thus elicit true long-lasting heritable epigenetic variation.
Current evidence suggests that humans are also sensitive to the effects of environmental influences on the deposition of epigenetic marks during human embryonic development (Heijmans et al., 2008). This evidence was obtained by studying the level of DNA methylation at the imprinted IGF2 gene in individuals who were prenatally exposed to famine during the Dutch Hunger Winter (1944–1945). For individuals who were conceived during the famine and thus were exposed in the earliest stages of development when epigenetic reprogramming events occur, the DMR of IGF2 was significantly hypomethylated compared with non-exposed siblings (Heijmans et al., 2008). This was not the case for individuals who were exposed at a later gestational stage. Although this study only described epigenetic effects within one generation, it shows that nutrition can also induce epigenetic variation in humans. In another study, it has been observed that the mortality rate of tested individuals related to cardiovascular disease and diabetes was significantly reduced if their grandfather experienced scarcity of food during prepuberty (Kaati et al., 2002). Subsequent analysis led to the conclusion that transgenerational epigenetic effects had to be the cause of these observations (Kaati et al., 2002; Pembrey, 2002, 2006). The study of the mechanisms behind transgenerational epigenetic inheritance in human populations is hindered by the genetic variation that exists within the population, this being the natural cause of most variation in DNA methylation (Heijmans et al., 2007). Although the use of siblings of the same sex can eliminate these genetic factors in cohort studies within one generation, this advantage is lost when looking at transgenerational epigenetic effects.
After the exposure of midgestation rats to the anti-androgenic compound vinclozolin, a decreased spermatogenic capacity and increased incidence of male infertility up to F4 was found (Anway et al., 2005) and recently evidence for persistent CpG methylation changes in selected gene promoters of F3 sperm was obtained (Guerrero-Bosagna et al., 2010). As further testimony to its epigenetic activity, vinclozolin was shown to increase and decrease DMR methylation levels of respective maternally and paternally imprinted gene clusters in the male germline (Stouder and Paoloni-Giacobino, 2010). The effect was transgenerational, as hypo- and hypermethylation were not yet completely lost in the sperm of F3 males.
The mechanistic link between estrogen and androgen hormones and epigenetic variations in chromatin architecture might be mediated by nuclear receptors (Biddie, 2011). In humans, the best-described case of hormone-induced epigenetic variation is the exposure of pregnant women to the estrogen receptor agonist diethylstilbestrol (DES), leading to developmental abnormalities in uterus structure, increased cancer risk in the daughters of pregnant mothers exposed to DES (Li et al., 2003) and an abnormal methylation of the lactoferrin promotor (Li et al., 1997). In mice, these defects in uterine development and carcinogenesis can be observed up to the F3 generation (Walker and Haven, 1997; Newbold et al., 2006), hence showing transgenerational epigenetic inheritance. For the human this has not been proven as only modest effects were found in the F2 (Blatt et al., 2003; Titus-Ernstoff et al., 2006, 2008, 2010), despite the similar nature of the phenotypic effects of DES in humans and the mouse (Ruden et al., 2005). In this context, it was suggested that the stress-induced chaperone Hsp90, of which the estrogen receptor is one of many clients, might be involved in the induction of epigenetic variation (Ruden et al., 2005).
Epigenetic effects of ART
Studies on mice designed to evaluate epigenetic and physiological aspects of ART
In all publications on epigenetic and physiological, behavioural readouts after ART in the mouse (Table I), effects have been noted. Most experiments address the effects of ovulation induction and preimplantation embryo culture on maintenance of imprinting up to mid-gestation. Imprinting status is followed using the methylation status of the DMR, in which surveys the paternally imprinted H19 is often studied. Imprinted gene expression has been determined as well. Examples of unaffected (Fauque et al., 2007) or slightly affected methylation (Market-Velker et al., 2010a, b) followed by affected expression, are given. Hence, all details of regulation by CpG methylation of imprinted gene expression are not yet available, which is a lacuna in our understanding.
Table I.
Survey of the mouse experiments aimed at testing imprinting and physiological parameters after ART.
| Reference | Genotype | Conditions |
Readouts |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| In vivo | Superov in vivo | Superov in vitro cleavage | Superov IVF + cleavage | ET | Media comparison | Blastocyst gDMR methyl | Blastocyst Impr expr | Mid gest Embryo gDMR methyl | Mid gest Embryo Impr expr | Mid gest Placenta gDMR methyl | Mid gest Placenta Impr expr | ||
| Market-Velker et al. (2010a) | B6(CAST7) | x | x | x | x | x | |||||||
| Market-Velker et al. (2010b) | B6(CAST7) | x | x | x | |||||||||
| Doherty et al. (2000) | B6(CAST7) | x | x | x | x | x | |||||||
| Mann et al. (2004) | B6(CAST7) | x | x | x | x | x | x | x | x | x | x | ||
| Khosla et al. (2001) | B6CBA F1 | x | x | x | x | ||||||||
| Fauque et al. (2007) | B6CBA F1 x F1 | x | x | x | x | x | x | x | |||||
| Fauque et al. (2010a) | B6CBA F1 x F1 | x | x | x | x | x | x | x | x | x | |||
| Fauque et al. (2010b) | B6CBA F1 x F1 | x | x | x | a | ||||||||
| Rivera et al. (2008) | B6(CAST7) | x | x | x | x | x | x | x | x | ||||
| Fortier et al. (2008) | CD1 x B6(CAST7) | x | x | x | x | x | x | x | |||||
| Li et al. (2005) | B6 x Spretus | x | x | x | |||||||||
| Adult readout | |||||||||||||
| Morgan et al. (2005) | FVB/N x B6 Avy/a | x | x | x | Phenotype epiallele | ||||||||
| Fernandez-Gonzalez et al. (2010) | B6CBA F1 x 129/Rr Axin1fu | x | x | x | Phenotype epiallele | ||||||||
| Ecker et al. (2004) | 129Sv x B6 | xb | x | x | Behavioural testing | ||||||||
| Fernandez-Gonzalez et al. (2004) | B6CBA F1 | x | x | x | Behavioural testing | ||||||||
| Watkins et al. (2007) | CBAB6 F1 x MF1 | x | x | x | Blood pressure, serum angiotensin converting enzyme, hepatic phosphoenolpyruvate carboxykinase | ||||||||
| Scott et al. (2010) | B6C3 F1 | x | xc | x | Glucose parameters | ||||||||
Superov, superovulation; ET, embryo transfer; gDMR, germline differentially methylated region (DMR); Impr expr, Imprinted expression; Mid gest, mid gestation.
aTranscriptome analysis.
bIn vivo fertilization was followed by in vivo and in vitro development.
cCulture to the 2-cell stage.
There are clear indications for both an effect of ovulation induction and of in vitro embryo culture on maintenance of DMR methylation. Also, the physiological status of the maternal tract after a ovulation induction procedure adds to deregulation of imprinting. As the only paper devoted to DMR methylation in maturing and mature oocytes after ovulation induction did not find an effect of hormonal priming (Sato et al., 2007), a maternal ‘zygotic’ effect and/or an effect of the female tract after ovulation induction, and (and should not be removed) an effect of the in vitro environment, is indicated.
In the experiments reported upon, inbred strains or hybrids between inbred strains have been predominantly used. No experiments with oocytes from random bred stocks have been reported in the literature. So principally, the oocyte inbred genotype could already confer cell biological stress (Nadeau, 2009) that exacerbates after hormonal priming and during in vitro culture.
In general the placenta is much more vulnerable than is the embryo proper, which is an illustration of the great significance of imprinting regulation for placental gene expression (Kawahara et al., 2009). One reason for the increased sensitivity of the placenta for lack of maintenance of DMR methylation might be related to the overall lower level of 5methylCpG (Monk et al., 1987; Fig. 5). In the mouse at least, effects on placental imprinted gene expression of H19, the fine regulator of prenatal growth, translate into deregulation of the imprinted gene network (Gabory et al., 2009, 2010; Fauque et al., 2010a, b) most likely also influencing gene expression among non-imprinted genes.
Effects of in vitro culture have also been observed in the Avy an Axinfu genetic systems, leading to hypomethylation of the IAP cryptic promoters. In the mouse, ART also affects physiological parameters at adult age, such as insulin sensitivity and blood pressure. Effects on adult behaviour have been reported too. For more information regarding the effects in the mouse, the reader is referred to the Supplementary Data (available online).
Epigenetic aspects of ART in human
Imprinting disorders in children born after ART
Since 2002, a number of reports have shown an association between ART and the frequency of imprinting disorders, notably BWS (Table II). Risk, expressed as the relative abundance of ART in the BWS cases compared with relative abundance of ART in the general population, was estimated at between 3.1 and 16.1 (Table II). Cases occurred irrespective of cause of infertility and are reported after IVF and ICSI, after transfer of fresh and frozen embryos, after transfer on Day 2, 3 or Day 5 and after different levels of hormonal stimulation, with in vitro culture being the common denominator (Gicquel et al., 2003; Chang et al., 2005; Sutcliffe et al., 2006; Doornbos et al., 2007). However, also after intrauterine insemination that often involves gonadotrophin stimulation, or the use of fertility drugs alone, BWS cases have been reported (Chang et al., 2005; Sutcliffe et al., 2006; Doornbos et al., 2007), which led Doornbos et al. to speculate that not ART practice but subfertility is at the heart of this increase in BWS. Indications for a link between ART and epigenetic regulation are that in the general population BWS is caused by a DMR CpG methylation error in 50-60% of the cases (Manipalviratn et al., 2009), while after ART almost all cases are related to hypomethylation of the maternal KCNQ1OT1 DMR (Table II). Also, more often other maternally methylated regions are hypomethylated in ART-BWS than in non-ART BWS children (Lim et al., 2009). To investigate whether children born after ART might have subclinical forms of BWS, Bowdin et al. (2007) analysed 1524 probands for clinical features linked to BWS. Four children had at least one of these signs, of which one was already diagnosed as having BWS. None of the other three children showed loss of methylation at KCNQ1OT1, suggesting that no milder forms of BWS have been missed in previous ART-BWS reports.
Table II.
Reports on the incidence of imprinting disorders after human IVF.
| Reference | Type of study | N cases | % IVF in cases | % IVF in ref | Estimated risk | Type of IVF | Molecular defect |
|---|---|---|---|---|---|---|---|
| Beckwith–Wiedemann syndrome | |||||||
| DeBaun et al. (2003) | Case series | 65 | 4.6 | 0.76 | 6.1 | IVF and ICSI | 5/6 LOM KCNQ1OT1 gDMRc |
| 1/6 GOM H19 DMR | |||||||
| 1/6 no imprint defect | |||||||
| Maher et al. (2003) | Case series | 149 | 4.0 | 0.997 | 4.0* | IVF (n = 3) and ICSI (n = 3) | 2/6 LOM KCNQ1OT1 gDMR |
| 4/6 not analysed | |||||||
| Gicquel et al. (2003) | Case series | 149 | 4.0 | 1.3 | 3.1* | IVF (n = 4) and ICSI (n = 2) | 6/6 LOM KCNQ1OT1 gDMR |
| Halliday et al. (2004) | Case control | 37 | 10.8 | 0.67 | 16.1* | IVF (n = 3) and ICSI (n = 1) | 3/4 LOM KCNQ1OT1 gDMR |
| 1/4 not analysed | |||||||
| Chang et al. (2005) | Case series | 341 | 5.6a | – | IVF (n = 5) and ICSI (n = 5)a | NA | |
| Sutcliffe et al. (2006) | Survey | 209 | 2.9–7.6b | 0.8 | 3.6–9.5b* | IVF (n = 1) and ICSI (n = 5) | 6/6 LOM KCNQ1OT1 gDMR |
| Doornbos et al. (2007) | Survey | 71 | 5.6 | 0.92 | 6.1* | IVF (n = 4) | 4/4 LOM KCNQ1OT1 gDMR |
| Angelman Syndrome | |||||||
| Cox et al. (2002) | Case series | 2 | – | – | – | ICSI (n = 2) | 2/2 LOM SNRPN |
| Orstavik et al. (2003) | Case report | 1 | – | – | – | ICSI (n = 1) | 1/1 LOM SNRPN |
| Ludwig et al. (2005) | Survey | 79 | 3.8 | – | – | ICSI (n = 3) | 1/3 LOM SNRPN |
| 2/3 maternal deletion 15q11 | |||||||
| Sutcliffe et al. (2006) | Survey | 75 | 0 | 0.8 | – | – | – |
| Doornbos et al. (2007) | Survey | 63 | 0 | 0.92 | – | – | – |
LOM, loss of methylation; GOM, gain of methylation; –, not analysed.
aAll 19 ART cases are included, 10 after IVF (and ICSI), 2 after hormonal stimulation and insemination and 7 for which no data on type of ART were available.
bRange takes into account the large number of lost to follow-up by assuming that all non-responders conceived naturally.
cSix ART- Beckwith–Wiedemann syndrome cases were identified in a database. Three patients were from before 2001, when use of ART was not systematically assessed. This period was excluded from the risk assessment.
*Risk is significantly increased in IVF compared with non-IVF pregnancies.
Angelman syndrome (AS) is caused by a shortage of maternal UBE3A expression in the SNRPN imprinting cluster. Less than 5% of cases are caused by an imprinting defect. Six cases of AS have been reported after ICSI (Table II; Cox et al., 2002; Orstavik et al., 2003; Ludwig et al., 2005), of which the unexpected high number of four shows a methylation defect. In two other studies covering more than 400 AS cases, none were conceived by IVF or ICSI. Instead, seven AS cases originated from ovulation induction and/or intrauterine insemination (Sutcliffe et al., 2006; Doornbos et al., 2007).
To date, five cases of Silver–Russell Syndrome (SRS) have been published in children born after IVF or ICSI (Svensson et al., 2005; Kagami et al., 2007; Galli-Tsinopoulou et al., 2008; Kallen et al., 2010b). In one, hypermethylation of the paternal MEST DMR was reported (Kagami et al., 2007). Generally, around 44% of the SRS cases is caused by H19 DMR hypomethylation and 5–10% by maternal uniparental disomy of chromosome 7. Thus far, no imprinted candidate gene on chromosome 7 could be identified (Binder et al., 2011). The number of cases involving ART is too small for a relation to be indicated.
Retinoblastoma (RB) and Prader–Willi syndrome (PWS) are two (epi)genetic disorders involving imprinting. In most cases the underlying molecular mechanism is a (point)mutation or a deletion and not an epimutation, just as in three reported PWS-ART cases (Sutcliffe et al., 2006; Doornbos et al., 2007) and two out of seven RB-ART cases (Marees et al., 2009). For the other five cases no gene defect was found and methylation was not analysed. In the PWS/AS region, methylation was normal in 92 children born after ICSI (Manning et al., 2000).
In a large follow-up study of children born after IVF in Sweden, one BWS, two SRS and four PWS patients were found (n = 31 850) (Kallen et al., 2005, 2010b). In the Danish National Cohort study (Lidegaard et al., 2005) among 6052 children there were none with a genomic imprinting disease. Recently, the French follow-up association reported 6 BWS cases (and no PWS, AS or SRS) in a cohort of 15 162 IVF children (Viot et al., 2010). With a spontaneous BWS incidence of 1 out of 13 700 (Amor and Halliday, 2008), the results of this study follow the tendency of the case series towards an increased risk after ART.
Effect of ART on epigenetic parameters in human gametes and embryos
Oocytes
Spontaneous oogenesis
In humans, studies on imprinting directed epigenetic reprogramming during oogenesis are very limited for ethical reasons. Only one study used immature oocytes from growing follicles in non-stimulated fertile patients after laparoscopy [(Sato et al., 2007) Table III]. At the primary follicle stage almost 50% of the maternally imprinted MEST, KCNQ1OT1 and PLAGL1 DMR alleles was methylated. This level gradually increased in growing preantal follicles and at the antral follicle stage, almost all alleles were methylated (Sato et al., 2007), just as in mice (Obata and Kono, 2002; Lucifero et al., 2004). The paternal imprint of the H19 DMR was partially erased at the primary follicle stage. A remnant of around 10% methylation was found at the antral follicle stage, which is different from the expectation based on mice, where in E13.5 PGCs the imprint is already removed (Hajkova et al., 2002).
Table III.
DNA CpG methylation in human oocytes at different stages of development with and without ovarian stimulation and IVM.
| Reference | gDMR | Methylation | No ovarian stimulation |
Ovarian stimulation, in vivo maturation |
Ovarian stimulation, IVM of GV |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary follicle | Pre-antral follicle | Antral follicle | GV | MI | MII | Arrested GV | Arrested MI | MII | |||
| Sato et al. (2007) | MEST | M | 50.0 | 57.0 | 91.6 | 55.6 (5/9) | 71.4 (5/7) | ||||
| PLAGL1 | M | 40.0 | 10.4 | 83.3 | |||||||
| KCNQ1OT1 | M | 55.5 | 66.6 | 87.5 | |||||||
| Geuns et al. (2007b) | KCNQ1OT1 | M | 100 (6/6) | 83.3 (5/6) | 100c (4/4) | ||||||
| Khoueiry et al. (2008) | KCNQ1OT1 | M | 67.8 (GV)a | 62.5 | 66.7 | 89.5 | 61.2 | 65.2 | 78.3 | ||
| 70.3 (MI)a | |||||||||||
| Geuns et al. (2003) | SNRPN | M | 100 (7/7) | 100 (3/3) | 100c (3/3) | ||||||
| El-Maarri et al. (2001) | SNRPN | M | 0b | ||||||||
| NDN | M | 33.3b | |||||||||
| Sato et al. (2007) | H19 | P | 28.2 | 33.4 | 12.5 | 0 (0/3) | 66.7 (2/3) | ||||
| Borghol et al. (2006) | H19 | P | 0 (0/5) | 24.3 | 13.8 | 16.7 | |||||
| Geuns et al. (2007a) | DLK1 | P | 0 (0/4) | 0 (0/6) | 0c (0/6) | ||||||
Methylation is depicted as the percentage of methylated alleles (greater-than two-third of DMR is methylated) of the total number of alleles analysed.
M, maternal; P, paternal.
If known, the number of methylated oocytes and the total number of oocytes are put in parentheses.
aGV and MI are obtained from non-stimulated patients with polycystic ovaries and after culture arrested in GV and MI.
bUnfertilized oocytes.
cMII obtained after culture of both GV and MI.
Ovulation induction
Table III gives a numerical overview of the data on genomic imprinting in oocytes from ovulation induction in women. The analysis of a possible effect of hormonal priming on imprinting can be confounded with maternal age and/or general suboptimal oogenesis. Proper control oocytes are scarce but, nevertheless, important information can be obtained.
In contrast to the almost complete methylation at the antral follicle stage of non-stimulated oocytes, after ovarian stimulation only 10 of the 16 germinal vesicle (GV)/metaphase I (MI) oocytes were methylated at MEST (Sato et al., 2007). The cause of subfertility in the couples donating the affected GV/MI primary oocytes was male factor or tuba obstruction, suggesting a genuine ovulation induction effect. The methylation status of two other maternally imprinted DMRs [SNRPN (Geuns et al., 2003) and KCNQ1OT1 (Geuns et al., 2007b)] was in most cases undisturbed. However, in one of the 12 GV/MI oocytes KCNQ1OT1 DMR was completely unmethylated (Table III). In the study of GV and MI oocytes of Khoueiry et al., around 60–70% of the alleles was methylated at KCNQ1OT1 DMR, while in metaphase II (MII) oocytes, which are mostly used for IVF treatment, the methylation level was around 90% (Khoueiry et al., 2008). This would suggest that around 10% of the MII oocytes can lead to BWS, which is not in agreement with the true incidence of BWS in ART. In this study of (Khoueiry et al., 2008), that included 52 ICSI cycles, some women suffered from polycystic ovary syndrome (PCOS, n = 11), endometriosis (n = 4) or dysovulation (n = 3). The methylation level of oocytes from women with or without PCOS was similar. Concerning SNRPN, El-Maarri et al. (2001) found a completely unmethylated DMR in a pool of 20–30 human MII oocytes obtained after ovarian stimulation. This is in agreement with neither the data from Geuns et al. (2003) where at the GV and MI stage SNRPN is already completely methylated, nor with the SNRPN methylation pattern in mouse MII oocytes (Lucifero et al., 2002). Also, the relative low incidence of AS and PWS after ART is not what would be expected with a completely unmethylated DMR.
Regarding the expected paternal DMR demethylation in oocytes, two out of six GV/MI oocytes were erroneously methylated at H19 after ovarian stimulation (Sato et al., 2007), a finding not substantiated by Borghol et al. (2006) where the paternal methylation imprint at H19 DMR was absent, as expected. Also, the paternally imprinted intergenic DMR of DLK1 and MEG3 (IG-DMR) was mainly unmethylated in superovulated oocytes at the GV and MI stages (Geuns et al., 2007a).
In vitro maturation
IVM of oocytes has been introduced to retrieve several oocytes for IVF treatment avoiding exogenous gonadotrophins, especially for patients at risk for the ovarian hyperstimulation syndrome and/or PCOS (Jurema and Nogueira, 2006). In general, small and medium-sized antral follicles are aspirated and the associated oocytes are cultured for 24–48 h before fertilization, depending on procedure at the clinic.
At the antral follicle stage, most DMR CpG methylation has been established although not completely so (see text above and Table III). Hence, in theory, IVM could interfere with imprint establishment or maintenance. To prove this point, GV and MI oocytes from women undergoing IVF treatment with hormonal stimulation were cultured in maturation medium. The normal maternal imprints of SNRPN and KCNQ1OT1 in the GV and MI oocytes were stably maintained in vitro up to the secondary oocyte stage (Geuns et al., 2003, 2007b; Table III). However, in a study comparing in vivo derived and IVM derived MII oocytes, the methylation level of KCNQ1OT1 DMR in the second group was statistically lower (Khoueiry et al., 2008). These authors point out that the maturation time (28 h when compared with ∼36 h in vivo) might be too short to finish the methylation process but this is disputed by the correct methylation pattern in oocytes cultured overnight as reported in Geuns et al. (2003, 2007b).
The paternal imprint at the IG-DMR was correctly absent after IVM of GV and MI oocytes (Geuns et al., 2007a). Borghol et al. (2006) obtained evidence that the H19 DMR is more vulnerable to the environment. After GV maturation for 24 h, two of the six pools with a maximum of three MII oocytes showed complete methylation in at least one allele, while in MI-derived MII oocytes methylation was completely absent.
Spermatozoa
In the human male germline (Table IV and Supplementary data, Table), the imprints of both maternal (MEST) and paternal (H19) DMRs are completely erased in fetal prospermatogonia (Kerjean et al., 2000). The maternally methylated MEST DMR remains unmethylated during spermatogenesis. For H19, the imprint is established during the adult spermatogonial stage or at least before the spermatocytes enter meiosis I, and is maintained thereafter (Kerjean et al., 2000), resembling the reprogramming in the mouse (Ueda et al., 2000). Hence in mature spermatozoa the paternally imprinted DMRs are completely methylated, while the maternally imprinted ones are unmethylated (see Table IV for references).
Table IV.
DNA CpG methylation studies on human spermatozoa from normal probands and subfertile patients.
| Reference | Conditions analysed |
DMR | Methylation (M/P) | Technique | ART outcome analysed | ||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Concentration | Motility | Morphology | Other | |||||
| Geuns et al. (2007a) | + | IG-DMR | P | Bisulphite sequencing | |||||
| Geuns et al. (2003) | + | SNRPN | M | Bisulphite sequencing | |||||
| Geuns et al. (2007b) | + | KCNQ1OT1 | M | Bisulphite sequencing | |||||
| Kerjean et al. (2000) | + | Spermatids, Testicular (fetal) spermatogonia and spermatocytes |
|
|
Bisulphite sequencing | ||||
| Marques et al. (2004) | + | + |
|
|
Bisulphite sequencing | ||||
| Marques et al. (2008) | + | + |
|
|
Bisulphite sequencing | ||||
| Kobayashi et al. (2007) | + | + |
|
|
Bisulphite sequencing + COBRA | + (1 child) | |||
| Sato et al. (2011) | + | + |
|
|
Bisulphite polymerase chain reaction Luminex, bisulphite sequencing and COBRA | ||||
| Hammoud et al. (2010) | + | + | Protamine replacement defect |
|
|
Bisulphite sequencing | |||
| Poplinski et al. (2010) | + | + | + | + |
|
|
Bisulphite sequencing | ||
| Benchaib et al. (2003) | + | + | + | + | Overall | global | Immunostaining | ||
| Houshdaran et al. (2007) | + | + | + | + |
|
|
|
||
| Boissonnas et al. (2010) | + | + | OAT |
|
|
Pyrosequencing | + (fertilization rate, cleavage and fragmentation) | ||
| Navarro-Costa et al. (2010) | + | OAT |
|
|
Bisulphite sequencing | ||||
| El Hajj et al. (2011) | + | OAT |
|
|
Pyrosequencing | + (Fertilization rate, pregnancy rate, live birth rate, abortion rate) | |||
| Manning et al. (2001) | + | Testicular round spermatids, testicular elongated spermatids | SNRPN | M | Methylation sensitive PCR with fragment length analysis | ||||
| Hartmann et al. (2006) | + |
|
H19 | P | Bisulphite PCR with single-strand conformation polymorphism (SSCP) analysis | ||||
| Marques et al. (2010) | Testicular spermatozoa form patients azoospermic due to ANJ, OAZI, CBAVD, HP |
|
|
Bisulphite sequencing | + (embryonic developmental arrest in HP patient) | ||||
COBRA = combined bisulphite-PCR restriction analysis, OAT = patients patients presenting with combined oligozoospermia, asthenozoospermia and teratozoospermia, ANJ = Anejaculation, OAZI = secondary inflammatory obstructive azoospermia, CBAVD = obstructive azoospermia due to congenital bilateral absence of the vas deferens, HP = secretory azoospermia due to hypospermatogenesis.
Effect of male subfertility on the epigenetic status of DMRs in spermatozoa
ART as such is unlikely to affect methylation in spermatozoa since these patterns, including the paternal imprints, are established before any manipulation occurs, the normal sperm nucleus being metabolically inert. Several studies show that a disturbed spermatogenesis itself is associated with incorrect imprinting (Table IV, Supplementary data, Table). In spermatozoa from oligozoospermic men, the occurrence of hypermethylation of several maternally imprinted DMRs or hypomethylation of the H19 and IG-DMR is increased, especially in ejaculates of <10 × 106/ml (Supplementary data, Table). The number of affected CpG sites ranges from only a few in normozoospermia to the whole DMR in severe azoospermia, only occurring in a minority of alleles sampled (Supplementary data, Table). Further evidence for an association between methylation and sperm concentration comes from a study by Boissonnas et al. (2010) who analysed the H19 DMR (CTCF6 region) in teratozoospermic (TZ) and oligo-astheno-teratozoospermic (OAT) patients. In spermatozoa from TZ patients, only 2out of 16 CpGs were significantly hypomethylated. In OAT spermatozoa, methylation was drastically reduced for all CpGs, reaching significance in subgroups with a sperm concentration of <10 × 106/ml. Sperm concentration is positively correlated with H19 methylation and negatively correlated with MEST methylation that is normally absent (Boissonnas et al., 2010, Poplinski et al., 2010). OAT spermatozoa also show reduced IG-DMR methylation (El Hajj et al., 2011).
Alteration of the protamine 1 to protamine 2 ratio, which should be around 1, generally denotes affected spermatogenesis [either as cause or as consequence (Nanassy et al., 2011)] and led to hypermethylation of several normally maternally methylated loci DMRs (KCNQ1OT1, SNRPN, MEST, PEG3, PLAGL1, IGF2) and to hypomethylation of the H19 DMR (Hammoud et al., 2010). Together, these data clearly indicate that DMR methylation defects are associated with poor spermatogenesis. Besides oligozoospermia, also other aetiologies of male subfertility are associated with epigenetic defects (Table IV, Supplementary data, Table). Azoospermia caused by anejaculation and secondary inflammatory obstruction was related to an increase in MEST methylation (Marques et al., 2010), as was a sperm motility <40, or <5% of sperm with normal morphology (Poplinski et al., 2010).
Global DNA methylation of non-imprinted repetitive sequences, such as long and short interspersed nucleotide elements (LINE1 and SINE (Alu)), did not show a decrease in spermatozoa from oligozoospermic or OAT patients (Kobayashi et al., 2007; Marques et al., 2008; Boissonnas et al., 2010; El Hajj et al., 2011), except for Alu element methylation in the study by El Hajj et al. (Supplementary data, Table). The methylation of non-imprinted genes and a repetitive sequence was also affected (Houshdaran et al., 2007), typically for sequences showing large intra- and interindividual methylation variation in spermatozoa from normozoospermic males (Flanagan et al., 2006).
Recently, it has been shown that patients with OAT had an increased level of methylation in the promotor of the normally unmethylated germline regulator gene DAZL, the autosomal substitute of Y-linked DAZ that correctly remained unmethylated (Navarro-Costa et al., 2010; Supplementary data, Table).
Effect of spermatozoa methylation defects on IVF outcome
It is not known to what extent DMR CpG methylation in both degree and prevalence can be ablated before germline transmission of this mark suffers. Kobayashi et al. (2009) compared the methylation defect that was found in trophoblastic villi from ART-miscarriages between 6-9 weeks of gestation with the imprints in the semen from the father. In 7 out of the 17 ART pregnancies with a placental H19 methylation defect, this was also found in the spermatozoa, suggesting transfer from the father. In a patient with hypospermatogenesis and with almost complete hypomethylation of the H19 DMR, the embryos obtained after ICSI all showed developmental arrest (stage unknown) (Marques et al., 2010). As in the human H19 is not expressed up to the blastocyst stage (Salpekar et al., 2001), a common paternal factor might be at stake. As no analysis of the embryos was undertaken, there is no formal proof of paternal inheritance of H19 DMR hypomethylation. In patients with OAT with a partial hypomethylation of H19, the fertilization rate after ICSI was reduced (Boissonnas et al., 2010). Developmental parameters, such as embryo quality, implantation rate, gestational age and birthweight, were similar to normally methylated paternal controls (Boissonnas et al., 2010). In another case, with spermatozoa showing both a maternal and a paternal methylation imprinting error (MEST DMR was methylated in 60% of the alleles and PLAGL1 in 20%, and H19 DMR was unmethylated in 90% of the alleles), a normal pregnancy was achieved with normal methylation (Kobayashi et al., 2007).
Methylation analysis in 19 ISCI children born small for gestational age revealed that one of them had hypermethylation in KCNQ1OT1 and MEST. As both parents had a normal methylation profile (and hypermethylation can only refer to the normally hypomethylated paternal allele), the methylation must have appeared de novo, maybe in the male germline because of the oligozoospermia of the father (Kanber et al., 2009; Table V).
Table V.
DNA CpG methylation studies of mainly DMRs in human offspring following ART.
| Reference | ART | n | Control | Sample | DMR | M/P | Results |
|---|---|---|---|---|---|---|---|
| Kanber et al. (2009) | ICSI + SGA | 19 | Normal weight children after spontaneous conception | Buccal smear |
|
|
1/19 children had hypermethylation of KCNQ1OT1 and MEST |
| Manning et al. (2000) | ICSI | 92 | – | Blood | SNRPN | M | In all 92 children the expected methylation pattern was seen |
| Gomes et al. (2009) | IVF and ICSI | 18 | Naturally conceived children and BWS patients | Peripheral blood, UCB or placenta | KCNQ1OT1 | M | 3 of 18 IVF children showed hypomethylation at KCNQ1OT1, without BWS phenotype |
| Tierling et al. (2010) | IVF and ICSI | 112 | Naturally conceived children | UCB, amnion membrane |
|
|
Only MEST was slightly hypermethylated in IVF compared with ICSI and control samples |
| Katari et al. (2009) | IVF | 10 | Naturally conceived children | UCB, placenta | 1536 CpG sites | – | 23% CpG sites differed in UCB and 16% in placenta. Imprinted genes are not extra vulnerable for deregulation. 4/11 tested genes with differential methylation also showed differential expression |
| Zechner et al. (2010) | IVF and ICSI | 42 | Abortions/stillbirths after spontaneous conception | Chorion villi |
|
|
IVF villi showed a hypomethylation (3% less) of KCNQ1OT1 |
| Turan et al. (2010) | IVF (ICSI unknown) | 45–98 | Naturally conceived children | UCB, cord, placenta | H19 | P | After IVF the intra- and inter-individual variation in methylation is higher. The expression of H19 and IGF2 in placenta and UCB was reduced in the IVF group |
M/P, maternally or paternally methylated; SGA, small for gestational age; UCB, umbilical cord blood.
There is only one case report in which part of the methylation defect of the child was also detected in leucocytes from the father. In a child with SRS conceived by IVF, eight CpGs were hypermethylated in PEG1/MEST DMR, four of which were also hypermethylated in the father (Kagami et al., 2007).
The preimplantation embryo
Loss of 5methyl CpG immunostaining in human embryos after fertilization resembles that reported in mammalian embryos (Santos et al., 2002, 2010). Active loss of immuno-recognition of paternal 5methylC also takes place in the human zygote indicating active demethylation (Beaujean et al., 2004; Fulka et al., 2004). After gamete fusion, a global passive maternal demethylation takes place, clearly visible at the 4-cell stage. At the end of the morula stage, remethylation starts (Fulka et al., 2004; Santos et al., 2010). In blastocysts, the methylation level of the trophectoderm and inner cell mass diverge, with more methylation in the inner cell mass (Santos et al., 2010). Abnormal chromatin organization, as observed via DNA-specific YOYO staining and aberrant mCpG staining, seemed to be correlated in arrested IVF embryos, suggesting proper chromatin organization for early development (Santos et al., 2010).
Transcripts of several imprinted genes like SNRPN, MEST, UBE3A and IGF2 (but not H19) are already present at the preimplantation embryonic stages (Lighten et al., 1997; Huntriss et al., 1998; Salpekar et al., 2001). The monoallelic expression starts from the 4 (SNRPN) and 8 (IGF2) cell stage, meaning that the primary imprints laid down during oogenesis and spermatogenesis are resistant to active and passive demethylation during the cleavage divisions.
Effects of IVF and embryo culture
Effects of IVF or subsequent development in culture medium alone are difficult to investigate in the human, since both are inevitably connected with each other. Moreover, the in vivo comparison cannot be made. In vitro conditions could affect maintenance of imprinting: at Day 3, 19% of human surplus embryos of low-quality (not suitable for transfer or for cryopreservation) showed hypomethylation of H19 (Chen et al., 2010). Paternal transmission was unlikely as none of the sperm samples showed hypomethylation. Similar results were obtained in a study where 8 of the 21 arrested surplus embryos showed loss of paternal methylation at H19 DMR, while the corresponding sperm samples were normal (Ibala-Romdhane et al., 2011) It is not known whether the hypomethylation (likely as a correlated response) leads to growth arrest or whether the growth arrest (induced by in vitro conditions) leads to loss of methylation. In the same study, eight arrested embryos showed methylation of the maternal allele (three of which also had hypomethylation at the paternal allele). After analysis of the unfertilized oocytes of two patients by using a distinguishing single nucleotide polymorphism, it was hypothesized that a defect in the erasure of the paternal imprint in the maternal germline led to this hypermethylation. Interestingly, five cryopreserved blastocysts that were donated for research after several years, all showed normal methylation (Ibala-Romdhane et al., 2011).
As to the mode of IVF, there is no convincing evidence that ICSI elevates the risk for epigenetic abnormalities when compared with IVF or vice versa. Nucleus structure and methylation levels (immunofluorescence) seen in arrested embryos and fully grown blastocysts did not differ between IVF and ICSI (Santos et al., 2010). An increased risk of imprinting disorders applies to pregnancies originating from both IVF and ICSI (Table II).
Recently, proof has been obtained of an effect of culture medium on offspring. IVF children derived from embryos that were cultured in two different media showed a significant difference in birthweight of almost 250 g (Dumoulin et al., 2010). This resembles the animal studies where the addition of serum to the culture medium affects the growth of the fetus (Khosla et al., 2001; Young et al., 2001; Fernandez-Gonzalez et al., 2004), although the effect in the human (not involving serum) seems less severe and a causative epigenetic variable has not been found yet.
Epigenetic effect of IVF on offspring other than imprinting diseases
Except for the described imprinting disorders, induced epigenetic variants (Table V) that do not have clear phenotypical effects might be transmitted to the offspring.
Gomes et al. (2009) analysed KCNQ1OT1, that was hypomethylated in 3 out of 18 IVF children. These were all part of a dizygotic twin, with the co-twin showing normal methylation. The methylation level was reduced from 41.5% in naturally conceived children to around 14% in these three probands without clinical symptoms. BWS patients show 1% methylation.
Another group did not find a difference in KCNQ1OT1 methylation in amnion/chorion membranes, umbilical cord blood and maternal peripheral blood of IVF and control conceptions (Tierling et al., 2010). Eight other DMRs also showed a normal methylation pattern (H19, SNRPN, GRB10, IG-DMR and 4 DMRs in the GNAS region). Only MEST was slightly hypermethylated in IVF compared with ICSI and control samples. No correlation between the methylation level of any of these genes and birthweight was found.
In chorion villus samples from spontaneous miscarriages and stillbirths, a hypomethylation of KCNQ1OT1 (significant) as well as H19 (trend) was seen in samples derived after IVF (n = 42) (Zechner et al., 2010). Five other DMRs did not show a difference compared with a control group.
The intra- and interindividual variation in methylation as assessed for H19 makes comparison in humans harder (Turan et al., 2010). The variation is higher in placental tissue compared with umbilical cord blood but also increased after IVF compared with in vivo fertilization, probably because IVF offspring result from embryos with fewer trophoblast stem cells (Turan et al., 2010). The expression of both IGF2 and H19 was reduced in placental tissue of ART pregnancies. The expression of IGF2 was not correlated with birthweight.
An extended DNA methylation analysis of more than 1500 genes, including DMRs, in placental tissue and umbilical cord blood from IVF and control pregnancies indicated that imprinted genes are not more vulnerable to methylation differences than non-imprinted genes (Katari et al., 2009). Around 16% of the analysed CpG sites showed a difference (hypo- or hypermethylation) in placental tissue and 23% in umbilical cord blood. Four out of 11 tested genes that showed a difference in methylation between the two groups also showed a difference in level of transcription.
Zhang et al. (2010) analysed global gene expression patterns in placentae from three IVF and three control pregnancies. Twenty-six genes were differentially expressed, none of them imprinted.
In all the above mentioned studies that found a methylation effect in ART children, the methylation of the single investigated CpG within the analysed tissue was never completely (100%) methylated or demethylated (0%). This suggests that the methylation defects are not transmitted from the oocyte or sperm cell. Unfortunately, the type of analyses and the presentation of the results do not allow us to accurately specify mosaicism.
Perinatal, congenital and physiological outcome of IVF children; an epigenetic response?
Three meta-analyses with similar results on perinatal outcome have been published (Helmerhorst et al., 2004; Jackson et al., 2004; McDonald et al., 2009). The studies included in the analyses were selected either on the use of an appropriate control group (Helmerhorst et al., 2004), or whether they controlled for maternal age (McDonald et al., 2009) or maternal age and parity (Jackson et al., 2004). The substantial number of included singletons ranged from 5361 (Helmerhorst et al., 2004) to more than 31 000 (McDonald et al., 2009) and all three meta-analyses showed an increased risk in the IVF group for very preterm birth [relative risk (RR) ranged from 3.0 to 3.3], preterm birth (RR 1.9–2.0), very low-birthweight (RR 2.7–3.8), low-birthweight (RR 1.4–1.8), small for gestational age (RR 1.4–1.6), Caesarean section (RR 1.5–2.1), admittance to neonatal intensive care unit (RR 1.3–1.6) and mortality (RR 1.7–2.4).
For congenital malformations, more controversy exists, also mainly because of the relative small sample sizes in comparison with the frequency of the malformation. Recently, a Swedish group analysed two consecutive cohorts, each consisting of more than 15 000 singleton IVF children (Kallen et al., 2005, 2010b). Besides esophageal atresia (OR = 5.2) and urogenital defects (OR = 2.3) (Kallen et al., 2005), in both cohorts an increased risk for limb reduction (OR = 1.7–2.0), neural tube defects (OR = 2.9–4.2) cardiovascular malformations (OR = 1.3–1.7) and syndromes associated with imprinting defects like Prader–Willi (RR 4.0) was reported. These malformations were also found in smaller cohorts [e.g. (Ericson and Kallen, 2001; Hansen et al., 2002; Klemetti et al., 2005)]. Regarding cancer, Kallen et al. found a relative risk of 1.4 in a large cohort of almost 27 000 IVF children. Pertinent to this review is the aetiology of these malformations, i.e. the balance between genetic and epigenetic aberrations.
Ceelen et al. dedicated themselves to the physical development of IVF children aged 8–18 years. They investigated systolic and diastolic blood pressure, skinfold thickness, fasting glucose/insulin levels, fat, growth velocity, bone development and endocrine status during puberty. Systolic and diastolic blood pressure, peripheral skinfold thickness, fasting glucose level, weight and height gain between 3 months and 1 year and dehydroepiandrosterone sulphate (DHEAS) and LH level in pubertal girls were all higher in the IVF group compared with a control group (consisting of naturally conceived children from subfertile couples) even after adjustment for potential confounders, such as maternal BMI (Ceelen et al., 2007, 2008a, c, 2009). The higher blood pressure was confirmed in a cohort of 4–14-year-old IVF children, together with a higher triglyceride level. DHEAS levels did not differ from the control group (Sakka et al., 2010). In a younger IVF group of around 6 years old, the IVF children were taller and had a slightly more favourable lipid profile when compared with naturally conceived children (Miles et al., 2007).
For all these parameters, under the assumption of a random participation in ART among the genotypic variance present in our population, no genetic component is indicated. It may therefore well be the result of an epigenetic adaptive response to the (preimplantation) environment.
Conclusions
It is clear that a number of questions regarding possible epigenetic effects of ART can be answered (see Fig. 6).
Figure 6.
Overview of the results of studies on the effect of ART on methylation and expression of imprinted genes. (A) Overview of mouse data. (B) Overview of human data. A/C = Amnion/Chorion, Bl = Blastocyst, E = embryo with the age, Ex = Expression which indicates either the level of expression or the allelic expression, ET = embryo transfer, GV = germinal vesicle oocyte, M = methylation, MI = oocyte in meiosis I, MII = oocyte in meiosis II, P = placenta with the age, UCB = umbilical cord blood. The numbers refer to the studies: 1. Sato et al. (2007), 2. Market-Velker et al. (2010a), 3. Fauque et al. (2007), 4. Fortier et al. (2008), 5. Rivera et al. (2008), 6. Doherty et al. (2000), 7. Mann et al. (2004), 8. Li et al. (2005), 9. Market-Velker et al. (2010b), 10. Khosla et al. (2001), 11. Fauque et al. (2010a), 12. Geuns et al. (2003), 13. Geuns et al. (2007b), 14. Khoueiry et al. (2008), 15. El-Maarri et al. (2001), 16. Borghol et al. (2006), 17. Geuns et al. (2007a) 18. Chen et al. (2010), 19. Ibala-Romdhane et al. (2011), 20. Gomes et al. (2009), 21. Tierling et al. (2010), 22. Turan et al. (2010), 23. Zechner et al. (2010), 24. Zhang et al. (2010), 25. Katari et al. (2009).
In the mouse, an effect of ART, from imprinting maintenance to physiological homeostasis to behaviour, has generally been found. The placenta stands out as much more vulnerable to the influences of ART on imprinting compared with the embryo proper. This is likely related to the underlying theoretical basis of imprinting, the male–female conflict hypothesis in mammalian reproduction (Moore and Haig, 1991).
With respect to the information that is lacking in the mouse, the clearest omissions are the absence of an OAT model and the effects of ART at increased maternal age. Also, the effect of in vitro culture without ovulation induction has not yet been studied.
The effect of ovulation induction on maintenance of imprinting can be expressed at three levels. (i) That on maintenance of imprinting at recruitment of an antral follicle, (ii) that expressed as a maternal early embryonic (‘zygotic’) cellular effect on maintenance of the imprint after gamete fusion and (iii) that expressed via the maternal tractus. Not much work has been done on the first effect, for which some indication is found in the human but none in the mouse. It should be noted that in mice maturation of multiple oocytes in one cycle is natural, while in humans it is not. That might explain why in humans ovulation induction interferes with the maintenance of imprinting in the antral follicle while in the mouse the effect is only after in vitro preimplantation development and/or through the maternal tractus. In both species, the unusual phenomenon of methylation of the H19 DMR has been observed in oocytes, the etiology of which could be different as in the human germline, erasure is indicated to be later (during first meiotic prophase) compared with the mouse (before first meiotic prophase).
Since in the mouse no ovulation induction effect in oocytes was reported (except H19), and a mosaic pattern for DMR methylation status was observed in blastocysts and the placenta, a ovulation induction effect on imprinting must be based on the maternal cellular effect in combination with the maternal environment. In the mouse, a negative effect of ovulation induction on the maternal tractus is generally accepted (de Boer et al., 1991; Van der Auwera et al., 1999; Van der Auwera and D'Hooghe, 2001). After changing this environment to normal by transferring the embryo to a non-stimulated uterus, the ovulation induction effect on imprinting lessened (Fortier et al., 2008). At mid-gestation, in vitro culture aggravated the impact of ovulation induction on imprinting maintenance (Rivera et al., 2008). In all, enough evidence has been obtained in the mouse as to an effect of hormonal intervention and the in vitro steps (fertilization and subsequent culture) on maintenance of imprinting, especially for the placenta.
In the only study that analysed superovulated in vivo matured human MII oocytes, the number of the oocytes not methylated at KCNQ1OT1 was much higher than the prevalence of BWS after ART. Most likely this suggests that the great majority of embryos derived from these oocytes are not viable. This non-viability of methylation disturbed embryos is substantiated by the hypo- and hypermethylation of H19 DMR in arrested embryos, while non-arrested embryos showed normal methylation (Ibala-Romdhane et al., 2011).
It would be of interest to know whether a milder ovarian stimulation would lead to a reduction of BWS cases (single embryo transfer is increasingly applied and therefore fewer embryos are needed).
In the human, the influence of poor spermatogenesis on maintenance of imprinting methylation and methylation of other sequences is undisputed, making sperm a potential vehicle for transmitting paternal methylation abnormalities. However, the chance of transmission to the offspring appears rather small.
The type of IVF (conventional IVF or ICSI) does not seem to make a difference regarding methylation defects. In vitro culture conditions and methylation defects might be associated since in human arrested embryos, H19 hypomethylation is reported, without methylation defects in the spermatozoa (Chen et al., 2010; Ibala-Romdhane et al., 2011). An in vivo comparison can, however, not be made.
The first array-based analysis on CpG methylation at birth (Katari et al., 2009) gives the impression of an ART-induced vulnerability for CpG methylation disturbances for a considerable frequency of sites, with more CpGs differentially methylated in umbilical cord blood than in placental tissue. When a difference was observed, in cord blood most CpGs were hypermethylated in the in vitro group while in placental tissue most CpGs were hypomethylated.
An affected 5methyl CpG maintenance can be without any effect but might bring IVF progeny closer to a threshold, making them more vulnerable to physiological reported effects at adolescence (Ceelen et al., 2007, 2008a, c, 2009) or late-onset diseases, such as cardiovascular disease, cancer (Kallen et al., 2010a) or other minor effects that have not yet been observed. Differences in gene expression of metabolism-related genes set by embryonic and/or fetal programming are assumed to underlie the relation between low-birthweight [also reported in IVF neonates (Helmerhorst et al., 2004; Jackson et al., 2004; McDonald et al., 2009)] and chronic diseases in later life, like cardiovascular diseases and diabetes mellitus (Barker, 2006). This finding could also be one explanation of the parallel observation of an effect of ART on both systolic blood pressure and glucose tolerance in mice and men (Watkins et al., 2007; Ceelen et al., 2008a; Scott et al., 2010). The recently isolated maternally imprinted gene KLF14 (Small et al., 2011) could well be the master regulator, at least for diabetes II and adipocyte-related metabolic disease risk.
In general, more evidence for an epigenetic effect of ART has been obtained in the mouse than in man. However, the definite answer to many questions in both mouse and human will await genome-wide epigenetic profiling in the different variants of ART. From the patient's perspective, those that contribute sperm from poor spermatogenesis stand out as very interesting for such an analysis as do the oocytes from older women.
Supplementary data
Supplementary data are available at http://humupd.oxfordjournals.org/.
Authors' roles
A.P.A.vM., L.H., P.dS., S.V., J.P.M.G. and P.dB. played a role in conception and design. A.P.A.vM., L.H., P.dS., S.V., J.P.M.G. and P.dB. were involved in acquisition, analysis and interpretation of the data. A.P.A.vM., L.H. and P.dB. drafted the manuscript. A.P.A.vM., L.H., P.dS., S.V., J.P.M.G. and P.dB. contributed to the revision of the manuscript and approval of the final version.
Supplementary Material
Acknowledgements
The publication of Karati et al. on CpG methylation in newly borns and their placentas was also communicated in the lay press. Thereafter the European Society of Human Reproduction and Embryology (ESHRE) was asked for expert advice. After consultation of the Special Interest Groups (SIG) Safety and Quality and Reproductive Genetics, it became clear that no document was available to which could be referred. For that reason it was decided to put together a working group consisting of members of both SIG's (J.G.; P.dS.; S.V.) and a few external experts (P.dB.; L.H.; A.P.A.vM.) to review the epigenetic state of ART as reported in the literature, supplemented by an update on epigenetic inheritance in mammals.
Funding
Funding to pay the Open Access publication charges for this article was provided by the European Society of Human Reproduction and Embryology (ESHRE). No other funding was obtained.
References
- Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23:2826–2834. doi: 10.1093/humrep/den310. doi:10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- Anderson LM, Riffle L, Wilson R, Travlos GS, Lubomirski MS, Alvord WG. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition. 2006;22:327–331. doi: 10.1016/j.nut.2005.09.006. doi:10.1016/j.nut.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and mate fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. doi:10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. doi:10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic Compartmentalization of the Fetal piRNA Pathway in Mice. Plos Genet. 2009;5:12. doi: 10.1371/journal.pgen.1000764. doi:10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber RC, Hickenbotham P, Hatch T, Kelly D, Topchiy N, Almeida GM, Jones GDD, Johnson GE, Parry JM, Rothkamm K, et al. Radiation-induced transgenerational alterations in genome stability and DNA damage. Oncogene. 2006;25:7336–7342. doi: 10.1038/sj.onc.1209723. doi:10.1038/sj.onc.1209723. [DOI] [PubMed] [Google Scholar]
- Barker DJ. Adult consequences of fetal growth restriction. Clin Obstet Gynecol. 2006;49:270–283. doi: 10.1097/00003081-200606000-00009. doi:10.1097/00003081-200606000-00009. [DOI] [PubMed] [Google Scholar]
- Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, Meehan R, Young L. Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol. 2004;14:R266–267. doi: 10.1016/j.cub.2004.03.019. doi:10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Benchaib M, Ajina M, Lornage J, Niveleau A, Durand P, Guerin JF. Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: a preliminary study. Fertil Steril. 2003;80:947–953. doi: 10.1016/s0015-0282(03)01151-8. doi:10.1016/S0015-0282(03)01151-8. [DOI] [PubMed] [Google Scholar]
- Bergstrom R, Whitehead J, Kurukuti S, Ohlsson R. CTCF regulates asynchronous replication of the imprinted H19/Igf2 domain. Cell Cycle. 2007;6:450–454. doi: 10.4161/cc.6.4.3854. doi:10.4161/cc.6.4.3854. [DOI] [PubMed] [Google Scholar]
- Biddie SC. Chromatin Architecture and the Regulation of Nuclear Receptor Inducible Transcription. J Neuroendocrinol. 2011;23:94–106. doi: 10.1111/j.1365-2826.2010.02079.x. doi:10.1111/j.1365-2826.2010.02079.x. [DOI] [PubMed] [Google Scholar]
- Binder G, Begemann M, Eggermann T, Kannenberg K. Silver–Russell syndrome. Best Pract Res Clin Endocrinol Metab. 2011;25:153–160. doi: 10.1016/j.beem.2010.06.005. doi:10.1016/j.beem.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Blatt J, Le LV, Weiner T, Sailer S. Ovarian carcinoma in an adolescent with trans generational exposure to diethylstilbestrol. J Pediatr Hematol Oncol. 2003;25:635–636. doi: 10.1097/00043426-200308000-00009. doi:10.1097/00043426-200308000-00009. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. Plos Genet. 2006;2:399–405. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, Vaiman D, Jouannet P, Tost J, Jammes H. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. doi:10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Li YF, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. doi:10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- Borghol N, Lornage J, Blachere T, Sophie Garret A, Lefevre A. Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics. 2006;87:417–426. doi: 10.1016/j.ygeno.2005.10.008. doi:10.1016/j.ygeno.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. doi:10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- Bowdin S, Allen C, Kirby G, Brueton L, Afnan M, Barratt C, Kirkman-Brown J, Harrison R, Maher ER, Reardon W. A survey of assisted reproductive technology births and imprinting disorders. Hum Reprod. 2007;22:3237–3240. doi: 10.1093/humrep/dem268. doi:10.1093/humrep/dem268. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigget S, Farley AH, Moran JV, Kazazian HH. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. doi:10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters A. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. doi:10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li RW, Bock C, Li CJ, Gu HC, Zamore PD, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. doi:10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceelen M, van Weissenbruch MM, Roos JC, Vermeiden JP, van Leeuwen FE, Delemarre-van de Waal HA. Body composition in children and adolescents born after in vitro fertilization or spontaneous conception. J Clin Endocrinol Metab. 2007;92:3417–3423. doi: 10.1210/jc.2006-2896. doi:10.1210/jc.2006-2896. [DOI] [PubMed] [Google Scholar]
- Ceelen M, van Weissenbruch MM, Vermeiden JP, van Leeuwen FE, Delemarre-van de Waal HA. Cardiometabolic differences in children born after in vitro fertilization: follow-up study. J Clin Endocrinol Metab. 2008a;93:1682–1688. doi: 10.1210/jc.2007-2432. doi:10.1210/jc.2007-2432. [DOI] [PubMed] [Google Scholar]
- Ceelen M, van Weissenbruch MM, Vermeiden JP, van Leeuwen FE, Delemarre-van de Waal HA. Growth and development of children born after in vitro fertilization. Fertil Steril. 2008b;90:1662–1673. doi: 10.1016/j.fertnstert.2007.09.005. doi:10.1016/j.fertnstert.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Ceelen M, van Weissenbruch MM, Vermeiden JP, van Leeuwen FE, Delemarre-van de Waal HA. Pubertal development in children and adolescents born after IVF and spontaneous conception. Hum Reprod. 2008c;23:2791–2798. doi: 10.1093/humrep/den309. doi:10.1093/humrep/den309. [DOI] [PubMed] [Google Scholar]
- Ceelen M, van Weissenbruch MM, Prein J, Smit JJ, Vermeiden JP, Spreeuwenberg M, van Leeuwen FE, Delemarre-van de Waal HA. Growth during infancy and early childhood in relation to blood pressure and body fat measures at age 8–18 years of IVF children and spontaneously conceived controls born to subfertile parents. Hum Reprod. 2009;24:2788–2795. doi: 10.1093/humrep/dep273. doi:10.1093/humrep/dep273. [DOI] [PubMed] [Google Scholar]
- Chang AS, Moley KH, Wangler M, Feinberg AP, Debaun MR. Association between Beckwith–Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril. 2005;83:349–354. doi: 10.1016/j.fertnstert.2004.07.964. doi:10.1016/j.fertnstert.2004.07.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Shi XY, Zheng HY, Wu FR, Luo C. Aberrant DNA methylation of imprinted H19 gene in human preimplantation embryos. Fertil Steril. 2010;94:2356–2358. doi: 10.1016/j.fertnstert.2010.01.120. 2358 e2351 doi:10.1016/j.fertnstert.2010.01.120. [DOI] [PubMed] [Google Scholar]
- Cheng XD, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. doi:10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SY, Vickaryous N, Ashe A, Zamudio N, Youngson N, Hemley S, Stopka T, Skoultchi A, Matthews J, Scott HS, et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39:614–622. doi: 10.1038/ng2031. doi:10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LSH, Ian HI, Koh TW, Ng HH, Xu GL, Li BFL. Human DNA (cytosine-5) methyltransferase PCNA complex as a target for p21(WAF1) Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. doi:10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu GL, Li E, Chen TP. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. doi:10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Ann Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. doi:10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. doi:10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzin F, Rassoulzadegan M. Non-Mendelian epigenetic heredity: gametic RNAs as epigenetic regulators and transgenerational signals. Essays Biochem. 2010;48:101–106. doi: 10.1042/bse0480101. doi:10.1042/bse0480101. [DOI] [PubMed] [Google Scholar]
- Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics. 1999;58:18–28. doi: 10.1006/geno.1999.5813. doi:10.1006/geno.1999.5813. [DOI] [PubMed] [Google Scholar]
- de Boer P, van der Hoeven FA, Wolters EM, Mattheij JA. Embryo loss, blastomere development and chromosome constitution after human chorionic gonadotropin-induced ovulation in mice and rats with regular cycles. Gynecol Obstet Invest. 1991;32:200–205. doi: 10.1159/000293031. doi:10.1159/000293031. [DOI] [PubMed] [Google Scholar]
- de Boer P, Ramos L, de Vries M, Gochhait S. Memoirs of an insult: sperm as a possible source of transgenerational epimutations and genetic instability. Mol Hum Reprod. 2010;16:48–56. doi: 10.1093/molehr/gap098. doi:10.1093/molehr/gap098. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith–Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. doi:10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. In pursuit of the first recognized epigenetic signal-DNA methylation - A 1976 to 2008 synopsis. Epigenetics. 2008;3:125–133. doi: 10.4161/epi.3.3.6249. doi:10.4161/epi.3.3.6249. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. doi:10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Doornbos ME, Maas SM, McDonnell J, Vermeiden JP, Hennekam RC. Infertility, assisted reproduction technologies and imprinting disturbances: a Dutch study. Hum Reprod. 2007;22:2476–2480. doi: 10.1093/humrep/dem172. doi:10.1093/humrep/dem172. [DOI] [PubMed] [Google Scholar]
- Dubrova YE, Jeffreys AJ, Malashenko AM. Mouse minisatellite mutations induced by ionizing radiation. Nat Genet. 1993;5:92–94. doi: 10.1038/ng0993-92. doi:10.1038/ng0993-92. [DOI] [PubMed] [Google Scholar]
- Dumoulin JC, Land JA, Van Montfoort AP, Nelissen EC, Coonen E, Derhaag JG, Schreurs IL, Dunselman GA, Kester AD, Geraedts JP, et al. Effect of in vitro culture of human embryos on birthweight of newborns. Hum Reprod. 2010;25:605–612. doi: 10.1093/humrep/dep456. doi:10.1093/humrep/dep456. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Stein P, Xu Z, Williams CJ, Kopf GS, Bilker WB, Abel T, Schultz RM. Long-term effects of culture of preimplantation mouse embryos on behavior. Proc Natl Acad Sci USA. 2004;101:1595–1600. doi: 10.1073/pnas.0306846101. doi:10.1073/pnas.0306846101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, Schorsch M, Haaf T. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–69. doi: 10.1159/000323806. doi:10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. doi:10.1038/85927. [DOI] [PubMed] [Google Scholar]
- Ericson A, Kallen B. Congenital malformations in infants born after IVF: a population-based study. Hum Reprod. 2001;16:504–509. doi: 10.1093/humrep/16.3.504. doi:10.1093/humrep/16.3.504. [DOI] [PubMed] [Google Scholar]
- Fauque P, Jouannet P, Lesaffre C, Ripoche MA, Dandolo L, Vaiman D, Jammes H. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. doi:10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauque P, Mondon F, Letourneur F, Ripoche MA, Journot L, Barbaux S, Dandolo L, Patrat C, Wolf JP, Jouannet P, et al. In vitro fertilization and embryo culture strongly impact the placental transcriptome in the mouse model. PLoS One. 2010a;5:e9218. doi: 10.1371/journal.pone.0009218. doi:10.1371/journal.pone.0009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauque P, Ripoche MA, Tost J, Journot L, Gabory A, Busato F, Le Digarcher A, Mondon F, Gut I, Jouannet P, et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum Mol Genet. 2010b;19:1779–1790. doi: 10.1093/hmg/ddq059. doi:10.1093/hmg/ddq059. [DOI] [PubMed] [Google Scholar]
- Feng SH, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. doi:10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y-Q, Desprat R, Fu H, Olivier E, Lin CM, Lobell A, Gowda SN, Aladjem MI, Bouhassira EE. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2006;2:e65. doi: 10.1371/journal.pgen.0020065. doi:10.1371/journal.pgen.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Moreira P, Bilbao A, Jimenez A, Perez-Crespo M, Ramirez MA, Rodriguez De Fonseca F, Pintado B, Gutierrez-Adan A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc Natl Acad Sci USA. 2004;101:5880–5885. doi: 10.1073/pnas.0308560101. doi:10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Ramirez MA, Pericuesta E, Calle A, Gutierrez-Adan A. Histone modifications at the blastocyst axin1(Fu) locus mark the heritability of in vitro culture-induced epigenetic alterations in mice. Biol Reprod. 2010;83:720–727. doi: 10.1095/biolreprod.110.084715. doi:10.1095/biolreprod.110.084715. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. doi:10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkowski JN, Ilnytskyy Y, Tamminga J, Koturbash I, Golubov A, Bagnyukova T, Pogribny IP, Kovalchuk O. Hypomethylation and genome instability in the germline of exposed parents and their progeny is associated with altered miRNA expression. Carcinogenesis. 2010;31:1110–1115. doi: 10.1093/carcin/bgp300. doi:10.1093/carcin/bgp300. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. doi:10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–1665. doi: 10.1093/hmg/ddn055. doi:10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- Fu AQ, Genereux DP, Stoger R, Laird CD, Stephens M. Statistical inference of transmission fidelity of dna methylation patterns over somatic cell divisions in mammals. Ann Appl Stat. 2010;4:871–892. doi: 10.1214/09-AOAS297SUPPA. doi:10.1214/09-AOAS297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka H, Mrazek M, Tepla O, Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. doi:10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. doi:10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. doi:10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- Galli-Tsinopoulou A, Emmanouilidou E, Karagianni P, Grigoriadou M, Kirkos J, Varlamis GS. A female infant with Silver Russell Syndrome, mesocardia and enlargement of the clitoris. Hormones (Athens) 2008;7:77–81. doi: 10.14310/horm.2002.1111040. [DOI] [PubMed] [Google Scholar]
- Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, Gilroy K, Baker S, Xia J, Dalley JW, Hurst LD, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469:534–538. doi: 10.1038/nature09651. doi:10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human-sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. doi:10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. doi:10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Rideout WM, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. doi:10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12:2873–2879. doi: 10.1093/hmg/ddg315. doi:10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- Geuns E, De Temmerman N, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of the intergenic differentially methylated region of DLK1-GTL2 in human. Eur J Hum Genet. 2007a;15:352–361. doi: 10.1038/sj.ejhg.5201759. doi:10.1038/sj.ejhg.5201759. [DOI] [PubMed] [Google Scholar]
- Geuns E, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of KvDMR1 in human oocytes. J Med Genet. 2007b;44:144–147. doi: 10.1136/jmg.2006.044149. doi:10.1136/jmg.2006.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith–Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–1341. doi: 10.1086/374824. doi:10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. doi:10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gomes MV, Huber J, Ferriani RA, Amaral Neto AM, Ramos ES. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol Hum Reprod. 2009;15:471–477. doi: 10.1093/molehr/gap038. doi:10.1093/molehr/gap038. [DOI] [PubMed] [Google Scholar]
- Gondor A, Ohlsson R. OPINION Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat Rev Genet. 2009;10:269–276. doi: 10.1038/nrg2555. doi:10.1038/nrg2555. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. doi:10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. doi:10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. doi:10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. doi:10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. doi:10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. doi:10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday J, Oke K, Breheny S, Algar E, Amor DJ. Beckwith–Wiedemann syndrome and IVF: a case-control study. Am J Hum Genet. 2004;75:526–528. doi: 10.1086/423902. doi:10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang HY, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. doi:10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346:725–730. doi: 10.1056/NEJMoa010035. doi:10.1056/NEJMoa010035. [DOI] [PubMed] [Google Scholar]
- Hartmann S, Bergmann M, Bohle RM, Weidner W, Steger K. Genetic imprinting during impaired spermatogenesis. Mol Hum Reprod. 2006;12:407–411. doi: 10.1093/molehr/gal040. doi:10.1093/molehr/gal040. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. doi:10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. doi:10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst FM, Perquin DA, Donker D, Keirse MJ. Perinatal outcome of singletons and twins after assisted conception: a systematic review of controlled studies. Br Med J. 2004;328:261. doi: 10.1136/bmj.37957.560278.EE. doi:10.1136/bmj.37957.560278.EE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. doi:10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. doi:10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear transplantation: lessons from frogs and mice. Curr Opin Cell Biol. 2002;14:741–748. doi: 10.1016/s0955-0674(02)00380-0. doi:10.1016/S0955-0674(02)00380-0. [DOI] [PubMed] [Google Scholar]
- Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. doi:10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. doi:10.1016/S0092-8674(01)00280-X. [DOI] [PubMed] [Google Scholar]
- Huntriss J, Daniels R, Bolton V, Monk M. Imprinted expression of SNRPN in human preimplantation embryos. Am J Hum Genet. 1998;63:1009–1014. doi: 10.1086/302039. doi:10.1086/302039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, Huang XH, Loo TC, Ma ZC, Fan GP. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. doi:10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibala-Romdhane S, Al-Khtib M, Khoueiry R, Blachere T, Guerin JF, Lefevre A. Analysis of H19 methylation in control and abnormal human embryos, sperm and oocytes. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.99. Advance Access published June 8, 2011, doi:2010.1038/ejhg.2011.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res-Fundam Mol Mech Mutagen. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. doi:10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. doi:10.1086/598822. [DOI] [PubMed] [Google Scholar]
- Jackson RA, Gibson KA, Wu YW, Croughan MS. Perinatal outcomes in singletons following in vitro fertilization: a meta-analysis. Obstet Gynecol. 2004;103:551–563. doi: 10.1097/01.AOG.0000114989.84822.51. doi:10.1097/01.AOG.0000114989.84822.51. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. doi:10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurema MW, Nogueira D. In vitro maturation of human oocytes for assisted reproduction. Fertil Steril. 2006;86:1277–1291. doi: 10.1016/j.fertnstert.2006.02.126. doi:10.1016/j.fertnstert.2006.02.126. [DOI] [PubMed] [Google Scholar]
- Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents and grandparents slow growth period. Eur J Hum Genet. 2002;10:682–688. doi: 10.1038/sj.ejhg.5200859. doi:10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- Kacem S, Feil R. Chromatin mechanisms in genomic imprinting. Mamm Genome. 2009;20:544–556. doi: 10.1007/s00335-009-9223-4. doi:10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- Kagami M, Nagai T, Fukami M, Yamazawa K, Ogata T. Silver-Russell syndrome in a girl born after in vitro fertilization: partial hypermethylation at the differentially methylated region of PEG1/MEST. J Assist Reprod Genet. 2007;24:131–136. doi: 10.1007/s10815-006-9096-3. doi:10.1007/s10815-006-9096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen B, Finnstrom O, Nygren KG, Olausson PO. In vitro fertilization (IVF) in Sweden: risk for congenital malformations after different IVF methods. Birth Defects Res A Clin Mol Teratol. 2005;73:162–169. doi: 10.1002/bdra.20107. doi:10.1002/bdra.20107. [DOI] [PubMed] [Google Scholar]
- Kallen B, Finnstrom O, Lindam A, Nilsson E, Nygren KG, Olausson PO. Cancer risk in children and young adults conceived by in vitro fertilization. Pediatrics. 2010a;126:270–276. doi: 10.1542/peds.2009-3225. doi:10.1542/peds.2009-3225. [DOI] [PubMed] [Google Scholar]
- Kallen B, Finnstrom O, Lindam A, Nilsson E, Nygren KG, Otterblad PO. Congenital malformations in infants born after in vitro fertilization in Sweden. Birth Defects Res A Clin Mol Teratol. 2010b;88:137–143. doi: 10.1002/bdra.20645. [DOI] [PubMed] [Google Scholar]
- Kanber D, Buiting K, Zeschnigk M, Ludwig M, Horsthemke B. Low frequency of imprinting defects in ICSI children born small for gestational age. Eur J Hum Genet. 2009;17:22–29. doi: 10.1038/ejhg.2008.177. doi:10.1038/ejhg.2008.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Hirasawa R, Chiba H, Okano M, Li E, Sasaki H. Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells. 2010;15:169–179. doi: 10.1111/j.1365-2443.2009.01374.x. doi:10.1111/j.1365-2443.2009.01374.x. [DOI] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. doi:10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol. 2010;22:284–290. doi: 10.1016/j.ceb.2010.02.002. doi:10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara M, Morita S, Takahashi N, Kono T. Defining contributions of paternally methylated imprinted genes at the Igf2-H19 and Dlk1-Gtl2 domains to mouse placentation by transcriptomic analysis. J Biol Chem. 2009;284:17751–17765. doi: 10.1074/jbc.M109.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, Jouannet P, Jeanpierre M. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. doi:10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. doi:10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Khoueiry R, Ibala-Rhomdane S, Mery L, Blachere T, Guerin JF, Lornage J, Lefevre A. Dynamic CpG methylation of the KCNQ1OT1 gene during maturation of human oocytes. J Med Genet. 2008;45:583–588. doi: 10.1136/jmg.2008.057943. doi:10.1136/jmg.2008.057943. [DOI] [PubMed] [Google Scholar]
- Klemetti R, Gissler M, Sevon T, Koivurova S, Ritvanen A, Hemminki E. Children born after assisted fertilization have an increased rate of major congenital anomalies. Fertil Steril. 2005;84:1300–1307. doi: 10.1016/j.fertnstert.2005.03.085. doi:10.1016/j.fertnstert.2005.03.085. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. doi:10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, Suzuki R, Suzuki F, Hayashi C, Utsunomiya T, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17:1582–1591. doi: 10.1038/ejhg.2009.68. doi:10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue—a possible mechanism contributing to radiation carcinogenesis? Biochem Biophys Res Commun. 2005;337:526–533. doi: 10.1016/j.bbrc.2005.09.084. doi:10.1016/j.bbrc.2005.09.084. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Baker M, Loree J, Kutanzi K, Hudson D, Pogribny I, Sedelnikova O, Bonner W, Kovalchuk O. Epigenetic dysregulation underlies radiation-induced transgenerational genome instability in vivo. Int J Radiat Oncol Biol Phys. 2006;66:327–330. doi: 10.1016/j.ijrobp.2006.06.012. doi:10.1016/j.ijrobp.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. doi:10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovalchuk O, Baulch JE. Epigenetic changes and nontargeted radiation effects - Is there a link? Environ Mol Mutagen. 2008;49:16–25. doi: 10.1002/em.20361. doi:10.1002/em.20361. [DOI] [PubMed] [Google Scholar]
- Lalancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: New functional insights for spermatozoal RNA. J Cell Biochem. 2008;104:1570–1579. doi: 10.1002/jcb.21756. doi:10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. doi:10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Lange UC, Schneider R. What an epigenome remembers. Bioessays. 2010;32:659–668. doi: 10.1002/bies.201000030. doi:10.1002/bies.201000030. [DOI] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- Li S, Washburn KA, Moore R, Uno T, Teng C, Newbold RR, McLachlan JA, Negishi M. Developmental exposure to diethylstilbestrol elicits demethylation of estrogen-responsive lactoferrin gene in mouse uterus. Cancer Res. 1997;57:4356–4359. [PubMed] [Google Scholar]
- Li SF, Hursting SD, Davis BJ, McLachlan JA, Barrett JC. Environmental exposure, DNA methylation, and gene regulation - Lessons from diethylstilbesterol-induced cancers. Epigenetics Cancer Prev: Early Detect Risk Assess. 2003;983:161–169. doi: 10.1111/j.1749-6632.2003.tb05971.x. [DOI] [PubMed] [Google Scholar]
- Li T, Vu TH, Ulaner GA, Littman E, Ling JQ, Chen HL, Hu JF, Behr B, Giudice L, Hoffman AR. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11:631–640. doi: 10.1093/molehr/gah230. doi:10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- Li XJ, Ito M, Zhou F, Youngson N, Zuo XP, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. doi:10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidegaard O, Pinborg A, Andersen AN. Imprinting diseases and IVF: Danish National IVF cohort study. Hum Reprod. 2005;20:950–954. doi: 10.1093/humrep/deh714. doi:10.1093/humrep/deh714. [DOI] [PubMed] [Google Scholar]
- Lighten AD, Hardy K, Winston RM, Moore GE. IGF2 is parentally imprinted in human preimplantation embryos. Nat Genet. 1997;15:122–123. doi: 10.1038/ng0297-122. doi:10.1038/ng0297-122. [DOI] [PubMed] [Google Scholar]
- Lim D, Bowdin SC, Tee L, Kirby GA, Blair E, Fryer A, Lam W, Oley C, Cole T, Brueton LA, et al. Clinical and molecular genetic features of Beckwith–Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod. 2009;24:741–747. doi: 10.1093/humrep/den406. doi:10.1093/humrep/den406. [DOI] [PubMed] [Google Scholar]
- Loree J, Koturbash I, Kutanzi K, Baker M, Pogribny I, Kovalchuk O. Radiation-induced molecular changes in rat mammary tissue: possible implications for radiation-induced carcinogenesis. Int J Radiat Biol. 2006;82:805–815. doi: 10.1080/09553000600960027. doi:10.1080/09553000600960027. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. doi:10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mann MRW, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. doi:10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42:289–291. doi: 10.1136/jmg.2004.026930. doi:10.1136/jmg.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, Zernicka-Goetz M. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol Biol Cell. 2008;19:4383–4392. doi: 10.1091/mbc.E08-02-0219. doi:10.1091/mbc.E08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, et al. Beckwith–Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003;40:62–64. doi: 10.1136/jmg.40.1.62. doi:10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91:305–315. doi: 10.1016/j.fertnstert.2009.01.002. doi:10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. doi:10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- Manning M, Lissens W, Bonduelle M, Camus M, De Rijcke M, Liebaers I, Van Steirteghem A. Study of DNA-methylation patterns at chromosome 15q11-q13 in children born after ICSI reveals no imprinting defects. Mol Hum Reprod. 2000;6:1049–1053. doi: 10.1093/molehr/6.11.1049. doi:10.1093/molehr/6.11.1049. [DOI] [PubMed] [Google Scholar]
- Manning M, Lissens W, Liebaers I, Van Steirteghem A, Weidner W. Imprinting analysis in spermatozoa prepared for intracytoplasmic sperm injection (ICSI) Int J Androl. 2001;24:87–94. doi: 10.1046/j.1365-2605.2001.00274.x. doi:10.1046/j.1365-2605.2001.00274.x. [DOI] [PubMed] [Google Scholar]
- Marees T, Dommering CJ, Imhof SM, Kors WA, Ringens PJ, van Leeuwen FE, Moll AC. Incidence of retinoblastoma in Dutch children conceived by IVF: an expanded study. Hum Reprod. 2009;24:3220–3224. doi: 10.1093/humrep/dep335. doi:10.1093/humrep/dep335. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. doi:10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod. 2010;83:938–950. doi: 10.1095/biolreprod.110.085480. doi:10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. doi:10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700–1702. doi: 10.1016/S0140-6736(04)16256-9. doi:10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. doi:10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585–594. doi: 10.1016/j.fertnstert.2009.02.051. doi:10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- Mayr E. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Cambirdge, MA/London: Belknap Press of Harvard Univ. Press; 1982. [Google Scholar]
- Mayr E, Provine WB. The Evolutionary Synthesis: Perspectives on the Unification of Biology. Cambridge, MA/London: Harvard Univ. Press; 1980. [Google Scholar]
- McDonald SD, Han Z, Mulla S, Murphy KE, Beyene J, Ohlsson A. Preterm birth and low birth weight among in vitro fertilization singletons: a systematic review and meta-analyses. Eur J Obstet Gynecol Reprod Biol. 2009;146:138–148. doi: 10.1016/j.ejogrb.2009.05.035. doi:10.1016/j.ejogrb.2009.05.035. [DOI] [PubMed] [Google Scholar]
- McLaren A. Too late for the midwife toad—stress, variability and Hsp90. Trends Genet. 1999;15:169–171. doi: 10.1016/s0168-9525(99)01732-1. doi:10.1016/S0168-9525(99)01732-1. [DOI] [PubMed] [Google Scholar]
- Miles HL, Hofman PL, Peek J, Harris M, Wilson D, Robinson EM, Gluckman PD, Cutfield WS. In vitro fertilization improves childhood growth and metabolism. J Clin Endocrinol Metab. 2007;92:3441–3445. doi: 10.1210/jc.2006-2465. [DOI] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. doi:10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: I. Radiation-induced genomic instability and bystander effects in vitro. Radiat Res. 2003a;159:567–580. doi: 10.1667/0033-7587(2003)159[0567:nadeoe]2.0.co;2. doi:10.1667/0033-7587(2003)159[0567:NADEOE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: II. Radiation-induced genomic instability and bystander effects in vivo, clastogenic factors and transgenerational effects. Radiat Res. 2003b;159:581–596. doi: 10.1667/0033-7587(2003)159[0581:nadeoe]2.0.co;2. doi:10.1667/0033-7587(2003)159[0581:NADEOE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. doi:10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):47–58. doi: 10.1093/hmg/ddi114. doi:10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29:275–276. doi: 10.1093/nar/29.1.275. doi:10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. doi:10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH. Transgenerational genetic effects on phenotypic variation and disease risk. Hum Mol Genet. 2009;18:R202–210. doi: 10.1093/hmg/ddp366. doi:10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanassy L, Liu L, Griffin J, Carrell DT. The clinical utility of the protamine 1/protamine 2 ratio in sperm. Protein Pept Lett. 2011;5:5–10. doi: 10.2174/092986611795713934. [DOI] [PubMed] [Google Scholar]
- Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, Goncalves J, Plancha CE. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod. 2010;25:2647–2654. doi: 10.1093/humrep/deq200. doi:10.1093/humrep/deq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Jefferson WN. Adverse effects of the model environmental estrogen diethylstilbestrol are transmitted to subsequent generations. Endocrinology. 2006;147:S11–S17. doi: 10.1210/en.2005-1164. doi:10.1210/en.2005-1164. [DOI] [PubMed] [Google Scholar]
- Ng SF, Lin RCY, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. doi:10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. doi:10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010;38:5141–5151. doi: 10.1093/nar/gkq229. doi:10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. doi:10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, Buiting K. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72:218–219. doi: 10.1086/346030. doi:10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JPC, Zahn D, Colledge WH, Carlton MBL, Nakano T, Surani MA. stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. doi:10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Pembrey ME. Time to take epigenetic inheritance seriously. Eur J Hum Genet. 2002;10:669–671. doi: 10.1038/sj.ejhg.5200901. doi:10.1038/sj.ejhg.5200901. [DOI] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. doi:10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- Peters A, Schubeler D. Methylation of histones: playing memory with DNA. Curr Opin Cell Biol. 2005;17:230–238. doi: 10.1016/j.ceb.2005.02.006. doi:10.1016/j.ceb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Koturbash I, Tryndyak V, Hudson D, Stevenson SML, Sedelnikova O, Bonner W, Kovalchuk O. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res. 2005;3:553–561. doi: 10.1158/1541-7786.MCR-05-0074. doi:10.1158/1541-7786.MCR-05-0074. [DOI] [PubMed] [Google Scholar]
- Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng SH, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–U1126. doi: 10.1038/nature08829. doi:10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. doi:10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao XH, Isono KI, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. doi:10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KVK, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. doi:10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. doi:10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. Plos Genet. 2007;3:1407–1417. doi: 10.1371/journal.pgen.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Murrell A. Genomic imprinting—silence across the border. Nature. 2000;405:408–409. doi: 10.1038/35013178. doi:10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. doi:10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. doi:10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17:1–14. doi: 10.1093/hmg/ddm280. doi:10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. doi:10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden DM, Xiao L, Garfinkel MD, Lu XY. Hsp90 and environmental impacts on epigenetic states: a model for the trans-generational effects of diethylstibesterol on uterine development and cancer. Hum Mol Genet. 2005;14:R149–R155. doi: 10.1093/hmg/ddi103. doi:10.1093/hmg/ddi103. [DOI] [PubMed] [Google Scholar]
- Sakka SD, Loutradis D, Kanaka-Gantenbein C, Margeli A, Papastamataki M, Papassotiriou I, Chrousos GP. Absence of insulin resistance and low-grade inflammation despite early metabolic syndrome manifestations in children born after in vitro fertilization. Fertil Steril. 2010;94:1693–1699. doi: 10.1016/j.fertnstert.2009.09.049. [DOI] [PubMed] [Google Scholar]
- Salpekar A, Huntriss J, Bolton V, Monk M. The use of amplified cDNA to investigate the expression of seven imprinted genes in human oocytes and preimplantation embryos. Mol Hum Reprod. 2001;7:839–844. doi: 10.1093/molehr/7.9.839. doi:10.1093/molehr/7.9.839. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. doi:10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Santos F, Hyslop L, Stojkovic P, Leary C, Murdoch A, Reik W, Stojkovic M, Herbert M, Dean W. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum Reprod. 2010;25:2387–2395. doi: 10.1093/humrep/deq151. doi:10.1093/humrep/deq151. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. doi:10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22:26–35. doi: 10.1093/humrep/del316. doi:10.1093/humrep/del316. [DOI] [PubMed] [Google Scholar]
- Sato A, Hiura H, Okae H, Miyauchi N, Abe Y, Utsunomiya T, Yaegashi N, Arima T. Assessing loss of imprint methylation in sperm from subfertile men using novel methylation polymerase chain reaction Luminex analysis. Fertil Steril. 2011;95:129–134. doi: 10.1016/j.fertnstert.2010.06.076. 134 e121–124 doi:10.1016/j.fertnstert.2010.06.076. [DOI] [PubMed] [Google Scholar]
- Scott KA, Yamazaki Y, Yamamoto M, Lin Y, Melhorn SJ, Krause EG, Woods SC, Yanagimachi R, Sakai RR, Tamashiro KL. Glucose parameters are altered in mouse offspring produced by assisted reproductive technologies and somatic cell nuclear transfer. Biol Reprod. 2010;83:220–227. doi: 10.1095/biolreprod.109.082826. doi:10.1095/biolreprod.109.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. doi:10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Shovlin TC, Bourc'his D, La Salle S, O'Doherty A, Trasler JM, Bestor TH, Walsh CP. Sex-specific promoters regulate Dnmt3L expression in mouse germ cells. Hum Reprod. 2007;22:457–467. doi: 10.1093/humrep/del379. doi:10.1093/humrep/del379. [DOI] [PubMed] [Google Scholar]
- Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, Thorsteindottir U, Shin SY, Richards HB, Soranzo N, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–564. doi: 10.1038/ng.833. doi:10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. doi:10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, Clayton-Smith J, Brueton LA, Bannister W, Maher ER. Assisted reproductive therapies and imprinting disorders—a preliminary British survey. Hum Reprod. 2006;21:1009–1011. doi: 10.1093/humrep/dei405. doi:10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- Svensson J, Bjornstahl A, Ivarsson SA. Increased risk of Silver-Russell syndrome after in vitro fertilization? Acta Paediatr. 2005;94:1163–1165. doi: 10.1111/j.1651-2227.2005.tb02066.x. doi:10.1080/08035250510030125. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. doi:10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp Cell Res. 1978;117:347–356. doi: 10.1016/0014-4827(78)90148-9. doi:10.1016/0014-4827(78)90148-9. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters A. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. doi:10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Bartolomei MS. SnapShot: imprinted gene clusters. Cell. 2007;130:958. doi: 10.1016/j.cell.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Tierling S, Souren NY, Gries J, Loporto C, Groth M, Lutsik P, Neitzel H, Utz-Billing I, Gillessen-Kaesbach G, Kentenich H, et al. Assisted reproductive technologies do not enhance the variability of DNA methylation imprints in human. J Med Genet. 2010;47:371–376. doi: 10.1136/jmg.2009.073189. doi:10.1136/jmg.2009.073189. [DOI] [PubMed] [Google Scholar]
- Titus-Ernstoff L, Troisi R, Hatch EE, Wisei LA, Palmer J, Hyer M, Kaufman R, Adam E, Strohsnitter W, Noller K, et al. Menstrual and reproductive characteristics of women whose mothers were exposed in utero to diethylstilbestrol (DES) Int J Epidemiol. 2006;35:862–868. doi: 10.1093/ije/dyl106. doi:10.1093/ije/dyl106. [DOI] [PubMed] [Google Scholar]
- Titus-Ernstoff L, Troisi R, Hatch EE, Hyer M, Wise LA, Palmer JR, Kaufman R, Adam E, Noller K, Herbst AL, et al. Offspring of women exposed in utero to diethylstilbestrol (DES)—a preliminary report of benign and malignant pathology in the third generation. Epidemiology. 2008;19:251–257. doi: 10.1097/EDE.0b013e318163152a. doi:10.1097/EDE.0b013e318163152a. [DOI] [PubMed] [Google Scholar]
- Titus-Ernstoff L, Troisi R, Hatch EE, Palmer JR, Hyer M, Kaufman R, Adam E, Noller K, Hoover RN. Birth defects in the sons and daughters of women who were exposed in utero to diethylstilbestrol (DES) Int J Androl. 2010;33:377–383. doi: 10.1111/j.1365-2605.2009.01010.x. doi:10.1111/j.1365-2605.2009.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppings M, Castro C, Mills PH, Reinhart B, Schatten G, Ahrens ET, Chaillet JR, Trasler JM. Profound phenotypic variation among mice deficient in the maintenance of genomic imprints. Hum Reprod. 2008;23:807–818. doi: 10.1093/humrep/den009. doi:10.1093/humrep/den009. [DOI] [PubMed] [Google Scholar]
- Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, Coutifaris C, Sapienza C. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet. 2010;6:e1001033. doi: 10.1371/journal.pgen.1001033. doi:10.1371/journal.pgen.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Abe K, Miura A, Yuzuriha M, Zubair M, Noguchi M, Niwa K, Kawase Y, Kono T, Matsuda Y, et al. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells. 2000;5:649–659. doi: 10.1046/j.1365-2443.2000.00351.x. doi:10.1046/j.1365-2443.2000.00351.x. [DOI] [PubMed] [Google Scholar]
- Van der Auwera I, D'Hooghe T. Superovulation of female mice delays embryonic and fetal development. Hum Reprod. 2001;16:1237–1243. doi: 10.1093/humrep/16.6.1237. doi:10.1093/humrep/16.6.1237. [DOI] [PubMed] [Google Scholar]
- Van der Auwera I, Pijnenborg R, Koninckx PR. The influence of in-vitro culture versus stimulated and untreated oviductal environment on mouse embryo development and implantation. Hum Reprod. 1999;14:2570–2574. doi: 10.1093/humrep/14.10.2570. doi:10.1093/humrep/14.10.2570. [DOI] [PubMed] [Google Scholar]
- Van der Heijden GW, Dieker JW, Derijck A, Muller S, Berden JHM, Braat DDM, van der Vlag J, de Boer P. Asymmetry in Histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. doi:10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Van der Heijden GW, Van den Berg IM, Baart EB, Derijck A, Martini E, De Boer P. Parental origin of chromatin in human monopronuclear zygotes revealed by asymmetric histone methylation patterns, differs between IVF and ICSI. Mol Reprod Dev. 2009;76:101–108. doi: 10.1002/mrd.20933. doi:10.1002/mrd.20933. [DOI] [PubMed] [Google Scholar]
- Van der Heijden GW, Castaneda J, Bortvin A. Bodies of evidence - compartmentalization of the piRNA pathway in mouse fetal prospermatogonia. Curr Opin Cell Biol. 2010;22:752–757. doi: 10.1016/j.ceb.2010.08.014. doi:10.1016/j.ceb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Viot G, Epelboin S, Olivennes F. Is there an increased risk of congenital malformations after ART? Results from a prospective French long-term survey of a cohort of 15 162 children. Hum Reprod. 2010;25:i54. [Google Scholar]
- Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. doi:10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. doi:10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Walker BE, Haven MI. Intensity of multigenerational carcinogenesis from diethylstilbestrol in mice. Carcinogenesis. 1997;18:791–793. doi: 10.1093/carcin/18.4.791. doi:10.1093/carcin/18.4.791. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. doi:10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. doi:10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Platt D, Papenbrock T, Wilkins A, Eckert JJ, Kwong WY, Osmond C, Hanson M, Fleming TP. Mouse embryo culture induces changes in postnatal phenotype including raised systolic blood pressure. Proc Natl Acad Sci USA. 2007;104:5449–5454. doi: 10.1073/pnas.0610317104. doi:10.1073/pnas.0610317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in A(vy)/a mice. Faseb J. 1998;12:949–957. [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. doi:10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Non-genetic contributions of the sperm nucleus to embryonic development. Asian J Androl. 2011;13:31–35. doi: 10.1038/aja.2010.75. doi:10.1038/aja.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazbek SN, Spiezio SH, Nadeau JH, Buchner DA. Ancestral paternal genotype controls body weight and food intake for multiple generations. Hum Mol Genet. 2010;19:4134–4144. doi: 10.1093/hmg/ddq332. doi:10.1093/hmg/ddq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, Broadbent PJ, Robinson JJ, Wilmut I, Sinclair KD. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–154. doi: 10.1038/84769. doi:10.1038/84769. [DOI] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. doi:10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Architectural epigenetics: mitotic retention of mammalian transcriptional regulatory information. Mol Cell Biol. 2010;30:4758–4766. doi: 10.1128/MCB.00646-10. doi:10.1128/MCB.00646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio N, Bourc'his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105:92–104. doi: 10.1038/hdy.2010.53. doi:10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- Zechner U, Pliushch G, Schneider E, El Hajj N, Tresch A, Shufaro Y, Seidmann L, Coerdt W, Muller AM, Haaf T. Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Mol Hum Reprod. 2010;16:704–713. doi: 10.1093/molehr/gap107. doi:10.1093/molehr/gap107. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cui Y, Zhou Z, Sha J, Li Y, Liu J. Altered global gene expressions of human placentae subjected to assisted reproductive technology treatments. Placenta. 2010;31:251–258. doi: 10.1016/j.placenta.2010.01.005. doi:10.1016/j.placenta.2010.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.