Abstract
Introduction
Human dental pulp stem/progenitor cells (hDPSC) can differentiate into odontoblast-like cells and express dentin sialophosphoprotein (DSPP) and osteocalcin (OCN); thus, they may be used to regenerate dentin. However, residual bacterial components in the root canal may suppress this activity.
Purpose
This study investigated the effect of a Porphyromonas gingivalis component on the expression of DSPP and OCN by stimulated hDPSCs and the influence of blockade of TLR2-mediated P. gingivalis host recognition.
Methods
Stimulated hDPSCs were exposed to varying concentrations of P. gingivalis lipopolysaccharide (LPS), and the expression of DSPP and OCN was measured. Similar groups of stimulated hDPSCs were exposed to TLR2 blocking agents before exposure to LPS.
Results
hDPSCs exposed to 5, 10, and 20 µg/mL LPS exhibited a dose-dependent reduction in the expression of DSPP (3.19 ± 0.18, 2.60 ± 0.49, and 1.15 ± 0.29, respectively) and OCN (3.51 ± 1.18, 2.60 ± 0.67 and 1.66 ± 0.89, respectively). The expression of DSPP and OCN after exposure to 20 µg/mL of LPS was significantly lower than measured for unexposed stimulated cells (analysis of variance and post hoc Tukey test, P < .05). The blockade of TLR2 using an extra- and intracellular agent affected DSPP (4.67 ± 0.97 and 5.29 ± 1.66, respectively) and OCN (5.25 ± 1.69 and 5.82 ± 2.38, respectively) expression at levels comparable to stimulated cells unexposed to 20 µg/mL LPS (6.32 ± 2.47 and 4.70 ± 1.60 for DSPP and OCN, respectively).
Conclusions
The suppressing effect of P. gingivalis on mineralized matrix formation by hDPSCs is confirmed, and this suppression can be moderated by TLR2 blockade.
Keywords: Dental pulp stem cells, dentin sialophosphoprotein, osteocalcin, Porphyromonas gingivalis, TLR2
In recent years, there has been a concerted effort to develop therapeutic methods that can regenerate a tissue with odontogenic potential in the root canals of immature permanent teeth that succumbed to pulp necrosis and infection. This development has been driven by the recognized susceptibility of immature roots to fracture when treated with conventional methods (1). To regenerate tissues with odontogenic potential, researchers have described the use of stem cells derived from the pulps of exfoliated deciduous teeth, stem cells from the pulps of human permanent teeth (hDPSCs), and stem cells from the apical papilla of human developing permanent teeth (SCAP) (2). In essence, these sources provide a mixed population of pulp cells including stem cells and odontoblast progenitor cells (OPCs), which may differentiate to form a dentin-like mineralized matrix when properly stimulated (3). This differentiation is marked by a high gene expression of hard-tissue–forming proteins such as dentin sialophosphoprotein (DSPP) (4, 5) and osteocalcin (OCN) (6). These proteins play an essential role in dentinogenesis and are present in the dentin extracellular matrix that is formed (7).
A recent in vivo demonstration of the de novo synthesis of pulp-like tissue and the formation of a continuous layer of dentin-like tissue on the existing dentin in the canal space using SCAP and DPSCs further highlights the potential of these stem/progenitor cells in dental tissue regeneration (8). This potential, however, may be impaired by the presence of microorganisms and their components that persist after disinfection of the root canal. A particular concern is lipopolysaccharide (LPS), a toxic and immunostimulatory surface molecule of gram-negative microorganisms that persists in dentin and resists elimination (9, 10). LPS derived from Porphyromonas gingivalis, a gram-negative obligate anaerobe prevalent (48%) in primary endodontic infections (11), has been shown to adversely affect DNA production in hDPSCs in a dose-dependent manner (12). Also, LPS derived from another putative endodontic pathogen, Porphyromonas endodontalis, has been shown to up-regulate the production of the proinflammatory cytokine interleukin 1β (13) as well as impede the survival of several mammalian cell lines (14, 15). Hence, for successful regeneration of a pulp-like tissue in a disinfected root canal, there is a need to neutralize the adverse effect of residual LPS.
Proinflammatory cytokines are produced in response to the recognition of pathogen-related molecular patterns such as an LPS fragment by a class of transmembrane receptors known as toll-like receptors (TLRs) located in the cell wall of specific host cells (16). Odontoblasts and OPCs express TLRs; as a result, they retain an ability to participate in the innate immune response (17). An in vivo study suggested that TLR2 was specifically required for the host response to P. gingivalis challenge (18), even though the activation of TLR2 may have been caused by heterogeneity and contamination of the isolated P. gingivalis LPS by lipoprotein (LP) (19). Blockade of TLR4s by anti-TLR4 antibodies has been shown to inhibit proinflammatory cytokine production in response to Escherichia coli derived LPS in rabbit whole blood cell culture (20). This suggests the possibility that a blockade of TLR2s may have a similar effect on OPCs exposed to a P. gingivalis component, thereby mitigating the deleterious effects associated with its presence.
The current study was designed to confirm the affect of a P. gingivalis component on the expression of genes involved in mineralized matrix formation by a population of hDPSCs and to investigate the effects of intra- and extracellular TLR2 blockade on gene expression of DSPP and OCN by hDPSCs exposed to this component. This study was undertaken to advance the understanding of the challenges facing therapeutic methods aimed at the regeneration of tissues with odontogenic potential in the infected root canals of immature permanent teeth.
Materials and Methods
Cell Culture
Human DPSCs were isolated based on our previous reports (8, 21). Briefly, teeth and/or pulp tissue was collected from the teeth of healthy patients at the University of Maryland Dental Clinics and stored in serum-free culture medium for transportation to the laboratory for processing. Sample collection conformed to the approved protocols by the Medical Institutional Review Boards at the University of Maryland (GH’s previous work location). Pulps were minced into 2 × 2 × 1 mm fragments and digested in a solution of 3 mg/mL type I collagenase and 4 mg/mL dispase for 30 to 60 minutes at 37°C (Sigma, St Louis, MO). Cell suspensions were obtained by passing the digested tissues through a 70-µm cell strainer (Becton/Dickinson, Franklin Lakes, NJ). Single-cell suspensions were seeded in 60- or 100-mm culture dishes and maintained in growth media consisting of α-minimum essential medium (α-MEM; Invitrogen, Carlsbad, CA) supplemented with 15% fetal bovine serum, 100 units/ml penicillin-G and 100 µg/mL streptomycin (Invitrogen), in a humidified atmosphere of 5% CO2 at 37°C. Colony-forming units of cells were normally observed within 1 to 2 weeks after cell seeding and were passaged at 1:3 ratio when they reached ~80% confluence. Heterogeneous populations of these hDPSCs were frozen and stored in liquid nitrogen at passages 0 to 2.
Stimulation and Exposure to LPS
Cells were thawed and expanded for experimentation at passage 3. Cells were seeded onto 12-wells plates at a density of 5,000 cells/mL. At 90% confluence, groups of cells were transferred to a dentinogenic stimulation medium consisting of original growth media supplemented with 2 mmol/L KH2PO4, 20 mmol/L HEPES (Sigma, St Louis, MO), 100 nmol/L dexamethasone prepared in α-MEM, and 5 × 10−8 mol/L 1,25-dihydroxyvitamin D3 prepared in ethanol (all from Sigma) for 7 weeks. They were exposed to 5, 10, or 20 µg/mL of P. gingivalis LPS (#05H23-SV) (InvivoGen, San Diego, CA) for 48 hours before initiating the extraction of their RNA. The negative control was comprised of unstimulated cells maintained in growth media throughout the experiment without exposure to LPS.
Blockade of TLR2 Pathway
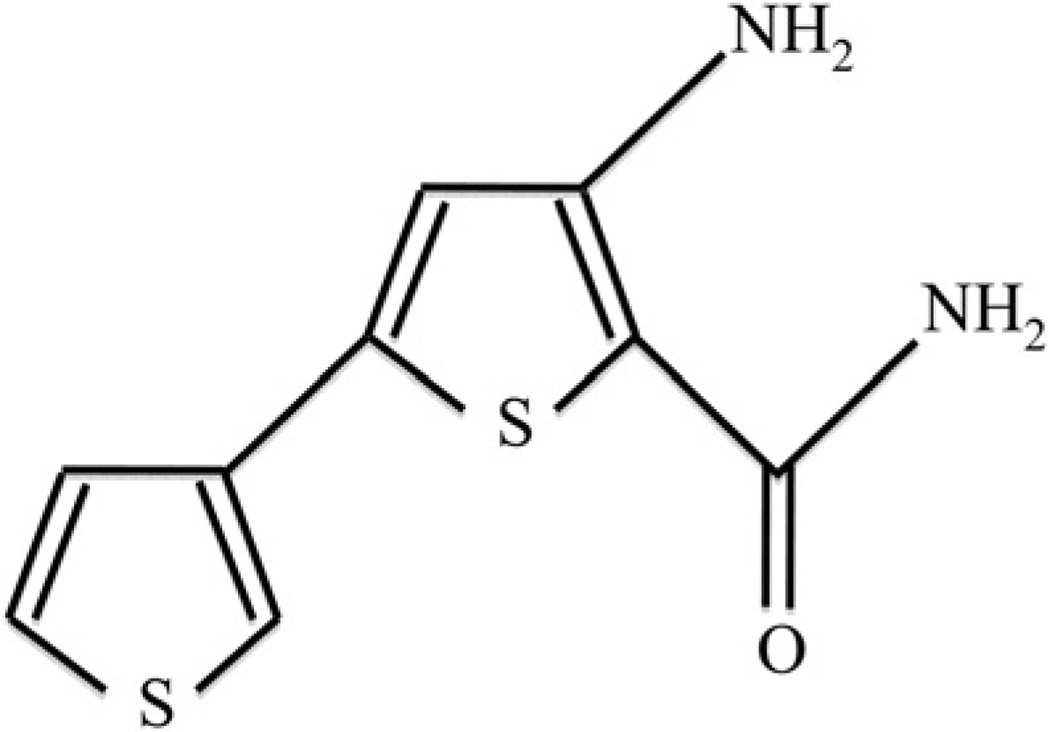
To study the effect of intra- and extracellular TLR2 blockade on DSPP and OCN gene expression before LPS exposure, cells stimulated with the dentinogenic stimulation media were incubated with 25 mg/mL of anti-TLR2 neutralizing antibody, a recognition blocker (Abcam, Cambridge, MA), or SC514 100 nmol/L prepared in 0.2% dimethyl sulfoxide (DMSO) (Cayman Chemical, Ann Arbor, MI) (Fig. 1), a pharmacological inhibitor of a downstream component (IκB kinase [IKK]) of the TLR2 recognition pathway. These groups of cells were stored in a humidified atmosphere of 5% CO2 at 37°C for 1 hour before the addition of 20 µg/mL of P. gingivalis LPS to the media in the culture well. Subsequently, they were maintained in a humidified atmosphere of 5% CO2 at 37°C for 48 hours before the extraction of RNA to assess DSPP and OCN gene expression. Cells exposed to anti-TLR2, SC514, or 0.2% DMSO but not to LPS were used as a positive control.
Figure 1.
The molecular structure of the pharmacological inhibitor SC514 used as intracellular blockade of the TLR2 pathway (MF: C9H8N2OS2). SC514 is a selective and reversible inhibitor of IKK.
RNA Extraction and Complementary DNA Preparation
Total RNA was extracted using the RNeasy Kit (Qiagen, Valencia, CA) and eluted to a final volume of 30 µL of sterile water as recommended by the manufacturer. Cells were disrupted using 1% 2-β-mercaptoenthanol in RNeasy lysis buffer with cell scrapers then vortexed for 3 × 10 seconds. Genomic DNA was eliminated using the RNase-Free DNase treatment (Qiagen). The concentration and purity of the samples were then verified with a Nanodrop 1000 Spectrophotometer (Thermo Scientific, Rockford, IL) and the samples stored at −80°C until used. Ten random samples were analyzed for RNA quality with a 2100 Bioanalyzer (Agilent Technologies Inc, Santa Clara, CA). A 1-µg aliquot of the extracted RNA was used for oligo(dT) reverse transcription to obtain complementary DNA with SuperScript II Reverse Transcriptase (Invitrogen).
Real-time Quantitative Polymerase Chain Reaction
Human primer sets for DSPP, OCN, and the control gene GAPDH were designed using a Basic Local Alignment Search Tool–assisted Internet search of a nonredundant nucleotide sequence database (National Library of Medicine, Bethesda, MD). The program MFOLD (http://mfold.bioinfo.rpi.edu/cgi-bin/dna-form1.cgi) was used to analyze the amplicon for potential secondary structures that could prevent efficient amplification. Primers were then synthesized (ACGT Corporation, Toronto, Ontario, Canada), as shown in Table 1.
TABLE 1.
Primer Sequences for the Genes Analyzed
| Gene | Sequence | Product size (bp) | |
|---|---|---|---|
| Human GAPDH (Accension no. M33197) | Forward | 5′-TTGACGCTGGGGCTGGCATT-3′ | 97 |
| Reverse | 5′-AGGTCCACCACCCTGTTGCTGT-3′ | ||
| Human DSPP (Accension no. NM_014208) | Forward | 5′-GCCAGSGCAAGTCTGGTAACGGT-3′ | 277 |
| Reverse | 5′-TGTCTCTGCSGGAGTTAGGTCTTGGT-3′ | ||
| Human OCN (Accension no. X51699.1) | Forward | 5′-TCCGGACTGTGACGACTTGGCT-3′ | 155 |
| Reverse | 5′-GCAAGGGCAAGGGGAAGAGGAA-3′ |
Each 20 µL of reaction mixture contained 5.0 µL of complementary DNA, 1 µL (10 µmol/L) of forward and reverse oligonucleotide primers, 10.0 µL of SsoFast EvaGreen Supermix (Bio-Rad, Mississauga, Ontario, Canada), and 3 µL of RNase-free water. Quantification of results from the real-time quantitative polymerase chain reaction (PCR) assays were analyzed with the CFX Manager (Bio-Rad).
To ensure that primers annealed efficiently to their targets, a range of temperatures around and at the calculated melting temperature was tried, using a Bio-Rad thermal cycler with a temperature gradient feature. The optimal temperature for annealing was determined to be 63°C. Care was taken to prevent nonspecific annealing and primerdimer formation during the test procedure. A standard curve was used to determine reaction efficiency with a twofold dilution of complementary DNA over seven points. A 1:16 complementary DNA dilution yielded optimal PCR efficiency for all primer pairs. To check reaction specificity, the PCR product was analyzed by running samples on a 2% agarose gel, which displayed a single band, and by examining the melt curve of the quantitative PCR report, which displayed a single peak.
Data Analysis
The experiments were performed in triplicate, and the means and standard error were calculated. Descriptive data and statistical analyses were performed using the SPSS 16.0 software package (SPPS Inc, Chicago, IL). Results were recorded as a ratio of normalized fold expression to GAPDH (2Ct[gene of interest]/2Ct [GAPDH]). One-way analysis of variance and the post hoc Tukey test were performed to explore a statistical difference in gene expression among groups. All statistical analyses were two tailed and interpreted at a 5% level of significance.
Results
Characterization of Isolated hDPSCs
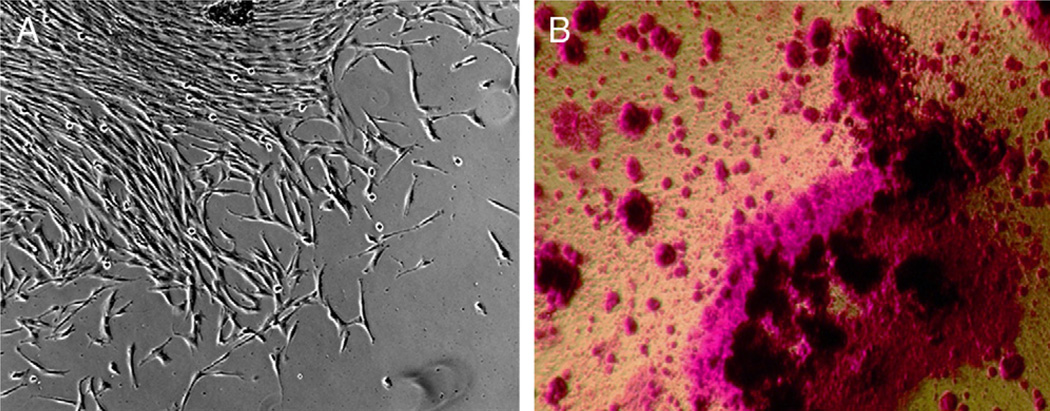
hDPSCs isolated from normal dental pulps typically formed colonies as shown in Figure 2A. After cell expansion, they were stimulated with dentinogenic differentiation medium for 7 weeks. Matrix production and mineralization of the cultures were observed by the presence of red stain (Fig. 2B), indicating their potency in differentiating into dentinogenic cell lineages upon stimulation. These hDPSCs were used in our studies to determine the effects of the P. gingivalis component on their dentinogenic gene expression after differentiation.
Figure 2.
hDPSCs grown and analyzed in cultures. (A) Typical clonogenic DPSCs forming a colony in the culture dish after being isolated from human pulp tissues using enzyme digestion. (B) A representative image showing cultured hDPSCs that underwent dentinogenic differentiation after being stimulated by media containing dexamethasone and 1,25-dihydroxyvitamin D3 for 7 to 8 weeks. Mineralized particles were stained red by alizarin red S.
Effect of P. gingivalis LPS on DSPP and OCN Gene Expression
To determine whether the P. gingivalis component affects the activity of hDPSCs differentiated into dentinogenic lineage, cells were first allowed to grow under the dentinogenic differentiation condition for 7 weeks. In pilot studies, dentinogenic gene markers DSPP and OCN were up-regulated significantly after stimulation for this time period. To observe the effect of P. gingivalis on the expression of DSPP and OCN, the LPS was added to the cells at 7 weeks after dentinogenic stimulation.
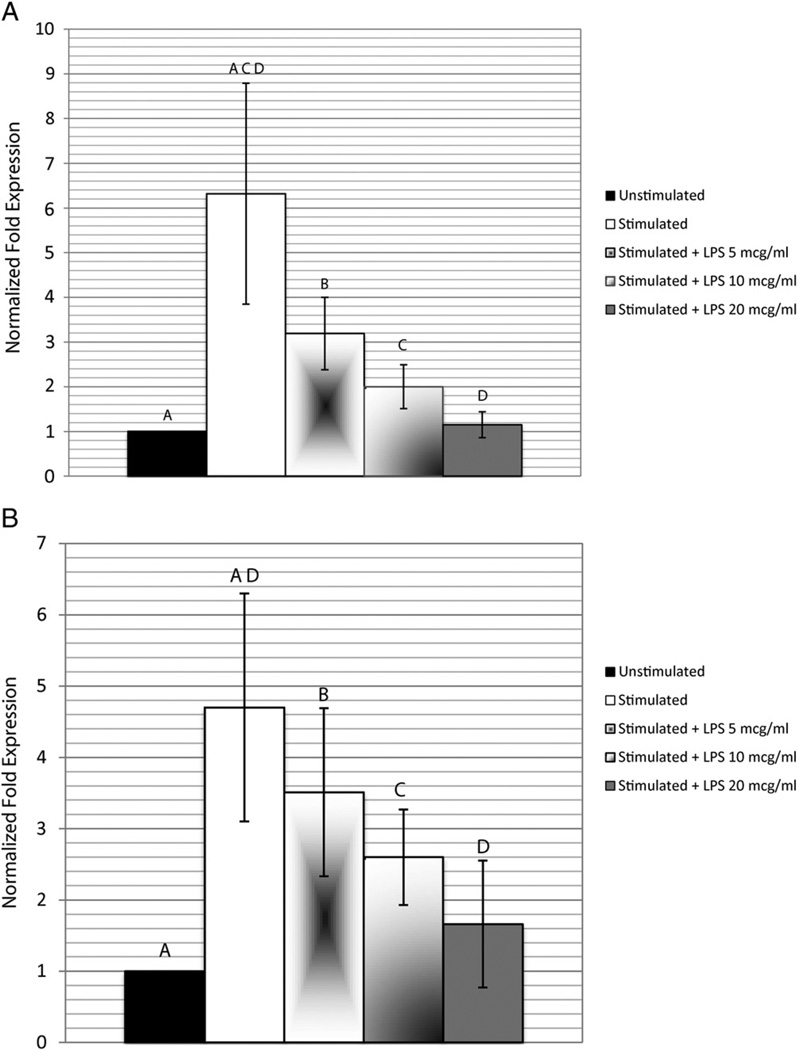
The expression of DSPP (6.32 ± 2.47) and OCN (4.70 ± 1.60) of the stimulated hDPSCs was significantly higher (P < .05) than that of the nonstimulated cells (Fig. 3). Exposure of stimulated cells to 5, 10, and 20 µg/mL LPS produced a dose-dependent decrease in DSPP and OCN gene expression (Fig. 3). When compared with the control gene GAPDH, DSPP expression decreased to 3.19 ± 0.18, 2.60 ± 0.49, and 1.15 ± 0.29, respectively, and OCN expression decreased to 3.51 ± 1.18, 2.60 ± 0.67, and 1.66 ± 0.89, respectively, compared with the stimulated cells normalized against GAPDH. Only after exposure of 20 µg/mL LPS, the expression of DSPP (1.15 ± 0.29) and of OCN (1.66 ± 0.89) was significantly lower (P < .05) than that of the unexposed stimulated cells and nonsignificantly different from that of the nonstimulated cells.
Figure 3.
The effect of the P. gingivalis component on DSPP and OCN gene expressions. Cells stimulated with dentinogenic media were exposed to P. gingivalis LPS at final concentrations of 5, 10, or 20 µg/mL for 48 hours. Quantitative real-time PCR was used to detect messenger RNA expression levels of (A) DSPP and (B) OCN normalized to GAPDH. Experiment was performed in triplicate, and error bars represent the standard error of the mean. Statistical significance is denoted by the same letter designation (ANOVA and the post hoc Tukey test, P < .05).
Effect of TLR2 Blockade on the Suppression of DSPP and OCN Gene Expression
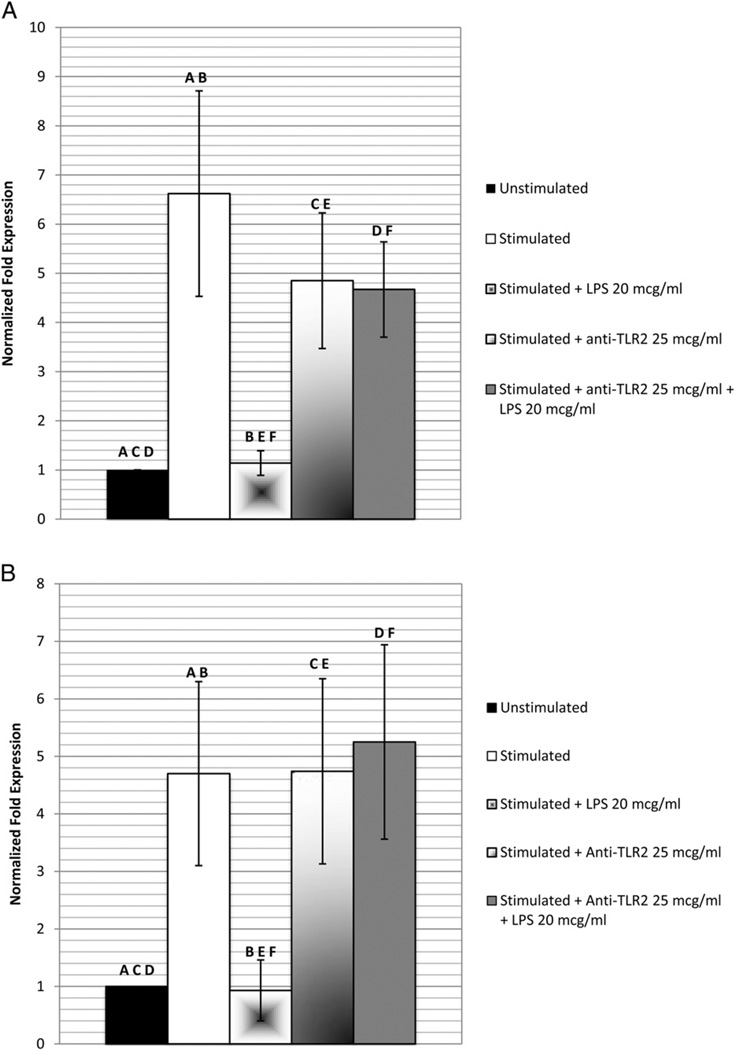
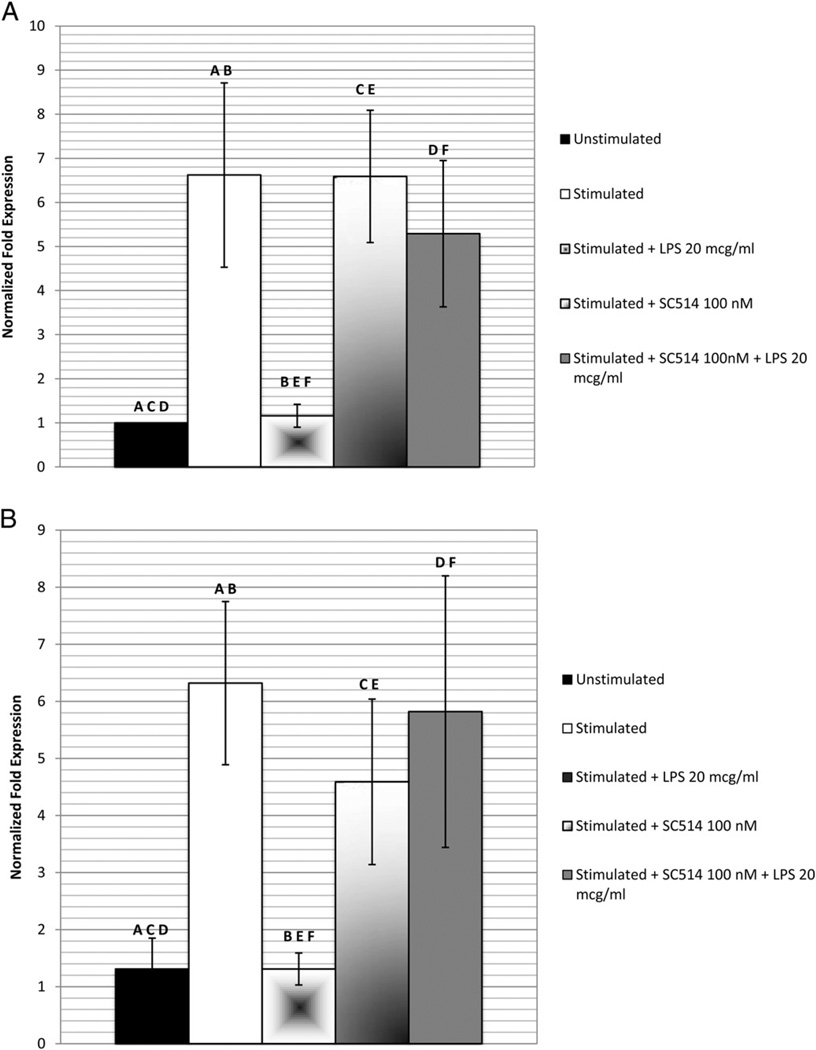
DSPP and OCN gene expression of cells exposed to anti-TLR2, SC514, or 02% DMSO alone (results not shown) did not differ significantly from the levels recorded for the stimulated cells not exposed to LPS (Figs. 4 and 5). Using the blockade of the TLR2 pathway by anti-TLR2 neutralizing antibodies (Fig. 4) and the pharmacological inhibitor SC514 (Fig. 5), the gene expression of DSPP (4.67 ± 0.97 and 5.29 ± 1.66, respectively) and OCN (5.25 ± 1.69 and 5.82 ± 2.38, respectively) after exposure to 20 µg/mL LPS was significantly (P < .05) higher than that of the stimulated, LPS-exposed cells without the blockade (1.16 ± 0.26). Both gene expression levels did not differ significantly from those of the stimulated cells not exposed to the P. gingivalis component.
Figure 4.
The effect of anti-TLR2 on P. gingivalis suppression of DSPP and OCN gene expression. Cells stimulated with dentinogenic media were incubated with anti-TLR2 before exposure to 20 µg/mL of P. gingivalis LPS for 48 hours. Quantitative real-time PCR was used to detect messenger RNA expression levels of (A) DSPP and (B) OCN normalized to GAPDH. Experiment was performed in triplicate, and error bars represent standard error of the mean. Statistical significance is denoted by the same letter designation (ANOVA and the post hoc Tukey test, P < .05).
Figure 5.
The effect of pharmacologic inhibitor SC514 on P. gingivalis suppression of DSPP and OCN gene expression. Cells stimulated with dentinogenic media were incubated with SC514 before exposure to 20 µg/mL of P. gingivalis LPS for 48 hours. Quantitative real-time PCR was used to detect messenger RNA expression levels of (A) DSPP and (B) OCN normalized to GAPDH. The experiment was performed in triplicate, and the error bars represent the standard error of the mean. Statistical significance is denoted by the same letter designation (ANOVA and the post hoc Tukey test, P < .05).
Discussion
In recent years, interest in “regenerative endodontics” as an approach to the treatment of infected pulps in partially developed teeth has been rekindled (8, 22). This interest has been fueled by the limitations of current methods of treatment to provide a favorable long-term outcome and by encouraging advances reported in stem cell research in related fields. Research is currently underway to develop a means by which the pulp space can be successfully repopulated with cells that, when stimulated, proliferate, differentiate, and reinitiate the dentinogenesis arrested when infection occurred (2, 23, 24).
Normal pulp is a soft tissue of ectomesenchymal origin composed of stem cells, immune cells, ectomesenchymal cells, fibroblasts, preodontoblasts (25), and odontoblasts supported by a vasculature and a neural network in a gelatinous extracellular matrix (8). The odontoblast is a highly specialized end cell that forms the dentin. Other pulp cells appear to have the potential to ultimately differentiate into odontoblast-like cells and secrete hard-tissue–forming proteins if properly stimulated (2, 23, 24). It has been estimated that approximately 1% of pulp cells retain this ability (26), and it is these cells that reportedly compose the cellular pool for de novo dentin formation. Collectively, cells with this dentinogenic potential have been referred to as OPCs, and although the sites in the pulp from which OPCs can be recruited have not been identified, it is known that they can, upon stimulation, express a number of dentinogenesis-associated genes (5). Two of these genes are OCN and DSPP (27, 28). It was for this reason that the expression of these genes by hDPSCs was selected in this study as a measure of dentinogenic potential, whereas suppressed expression was a measure of LPS toxicity. Therefore, variances in the expression levels of these genes during the different phases of the study would reflect the positive and negative effects of the procedures undertaken.
One of the factors identified as an impediment to the successful clinical application of regenerative pulp procedures is the persistence of microorganisms and their components in the infected root canal after attempts have been made to clean and disinfect it (9). Although current root canal disinfection regimens are relatively effective in eliminating microorganisms (10), they are less effective in eliminating microbial LPS (9). LPS has been found in the root canal, in apical tissues, and in dentin subsequent to endodontic infection (29, 30). This molecule is potentially harmful to host cells as a toxin and as an immune stimulant (9). The P. gingivalis component was used in this study because this microorganism has been identified as a putative endodontic pathogen prevalent in endodontic infections (11) and because the impact of its immunostimulatory effects on hDPSCs has yet to be investigated. Previous studies had shown that P. gingivalis LPS adversely affected cell function and survival in other cell lines (15). The unique lipid A component of naturally extracted P. gingivalis LPS is bioactive and had been classified as an agonist for host cell TLR2 and not TLR4/MD2 (31), as is characteristic of LPS derived from enteric bacteria. This behavior has been attributed to LP, a bioactive contaminant present in naturally extracted P. gingivalis lipid A, but not in its synthetic forms (31). Recent studies have also shown that the purified or synthesized molecule of the lipid A is an agonist for TLR4/MD2, but it is relatively weak as an immunostimulant (32). It is now apparent that naturally derived P. gingivalis LPS as used in this study is recognized by both TLR2 and TLR4/MD2 (33). By blocking the TLR2 recognition of LP, the overall harmful effect of P. gingivalis could be prevented.
As expected, exposure of hDPSCs to the P. gingivalis component induced a dose-dependent suppression of DSPP and OCN gene expression. This pattern was similar to the previously reported one in the suppression of DSPP and Runx2 gene expression in a rat DPSC line exposed to Aggregatibacter actinomycetemcomitans LPS (5). These findings underline the potential deleterious effect of microbial products known to remain in the root canal after disinfection procedures (10) on the regenerative potential of hDPSCs even when properly stimulated. In the current study, a blockade of TLR2 at its receptor site inhibited the suppression of DSPP and OCN gene expression by a P. gingivalis component whose recognition is mediated by TLR2. Blocking IKK, a protein component in the TLR2 downstream recognition pathway, had a similar effect. This appeared to support the premise that the immune potential of P. gingivalis might be more of an impediment to the function of hDPSCs than to its survival. Because the blockade was successful, it became apparent that P. gingivalis LPS at a concentration of 20 µg/mL was not toxic to the cell population. It also was apparent that the effect could be moderated by a TLR2 blockade.
These study results should not be interpreted as simplifying the obstacles posed by microorganisms and their byproducts in the development of effective “regenerative endodontic” strategies. As many other in vitro studies, this study addressed a single isolated factor of a complex clinical problem. The microbiota of the infected root canal is highly diverse (34) as are the types and concentrations of bacterial components that may persist in the disinfected root canal. Because the need to overcome the obstacles posed by microorganisms and their byproducts when performing regenerative pulp therapy was highlighted, future studies are warranted that will address additional aspects of this multifaceted biological challenge.
In conclusion, this study confirmed the suppression of genes involved in mineralized matrix formation by hDPSCs upon exposure to a P. gingivalis component. A dose-dependent reduction of DSPP and OCN gene expression was observed; however, only at a LPS concentration of 20 µg/mL were both gene expressions significantly lower than that of stimulated hDPSCs. In addition, an extra- and intracellular blockade of TLR2 pathway were able to inhibit the gene suppression by P. gingivalis. This study highlighted the need to address the infection of the root canal space as an integral part of regenerative procedures.
Acknowledgments
This study was supported in parts by grants from the National Institutes of Health (NIH) R01 DE019156-01 (G.T.-J.H.), the Canadian Academy of Endodontics Endowment Fund, the Alpha Omega Foundation of Canada, the Dental Research Institute of the University of Toronto, and the Canadian Institutes of Health Research (CIHR) (M.G.).
The authors thank Dr. Gajanan V. Kulkarni for providing biostatistical consultation.
Footnotes
The authors deny any conflicts of interest related to this study.
References
- 1.Cvek M. Prognosis of luxated non-vital maxillary incisors treated with calcium hydroxide and filled with gutta-percha. A retrospective clinical study. Endod Dent Traumatol. 1992;8:45–55. doi: 10.1111/j.1600-9657.1992.tb00228.x. [DOI] [PubMed] [Google Scholar]
- 2.Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitagawa M, Ueda H, Iizuka S, et al. Immortalization and characterization of human dental pulp cells with odontoblastic differentiation. Arch Oral Biol. 2007;52:727–731. doi: 10.1016/j.archoralbio.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Bleicher F, Couble ML, Buchaille R, Farges JC, Magloire H. New genes involved in odontoblast differentiation. Adv Dent Res. 2001;15:30–33. doi: 10.1177/08959374010150010701. [DOI] [PubMed] [Google Scholar]
- 5.Nomiyama K, Kitamura C, Tsujisawa T, et al. Effects of lipopolysaccharide on newly established rat dental pulp-derived cell line with odontoblastic properties. J Endod. 2007;33:1187–1191. doi: 10.1016/j.joen.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Butler WT, Ritchie H. The nature and functional significance of dentin extracellular matrix proteins. Int J Dev Biol. 1995;39:169–179. [PubMed] [Google Scholar]
- 7.Butler WT. Dentin matrix proteins. Eur J Oral Sci. 1998;106 suppl 1:204–210. doi: 10.1111/j.1600-0722.1998.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang GT, Yamaza T, Shea LD, et al. Stem/Progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng Part A. 2010;16:605–615. doi: 10.1089/ten.tea.2009.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vianna ME, Horz HP, Conrads G, Zaia AA, Souza-Filho FJ, Gomes BP. Effect of root canal procedures on endotoxins and endodontic pathogens. Oral Microbiol Immunol. 2007;22:411–418. doi: 10.1111/j.1399-302X.2007.00379.x. [DOI] [PubMed] [Google Scholar]
- 10.Paquette L, Legner M, Fillery ED, Friedman S. Antibacterial efficacy of chlorhexidine gluconate intracanal medication in vivo. J Endod. 2007;33:788–795. doi: 10.1016/j.joen.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Siqueira JF, Rocas IN, Silva MG. Prevalence and clonal analysis of Porphyromonas gingivalis in primary endodontic infections. J Endod. 2008;34:1332–1336. doi: 10.1016/j.joen.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Nakane A, Yoshida T, Nakata K, Horiba N, Nakamura H. Effects of lipopolysaccharides on human dental pulp cells. J Endod. 1995;21:128–130. doi: 10.1016/s0099-2399(06)80437-1. [DOI] [PubMed] [Google Scholar]
- 13.Hosoya S, Matsushima K. Stimulation of interleukin-1 beta production of human dental pulp cells by Porphyromonas endodontalis lipopolysaccharide. J Endod. 1997;23:39–42. doi: 10.1016/S0099-2399(97)80205-1. [DOI] [PubMed] [Google Scholar]
- 14.Miura M, Zhu H, Rotello R, Hartwieg EA, Yuan J. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 15.Yamaji Y, Kubota T, Sasaguri K, et al. Inflammatory cytokine gene expression in human periodontal ligament fibroblasts stimulated with bacterial lipopolysaccharides. Infect Immun. 1995;63:3576–3581. doi: 10.1128/iai.63.9.3576-3581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin Immunol. 2007;19:3–10. doi: 10.1016/j.smim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Staquet MJ, Durand SH, Colomb E, et al. Different roles of odontoblasts and fibroblasts in immunity. J Dent Res. 2008;87:256–261. doi: 10.1177/154405910808700304. [DOI] [PubMed] [Google Scholar]
- 18.Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting edge: TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa T, Asai Y, Makimura Y, Tamai R. Chemical structure and immunobiological activity of Porphyromonas gingivalis lipid A. Front Biosc. 2007;12:3795–3812. doi: 10.2741/2353. [DOI] [PubMed] [Google Scholar]
- 20.Smith LS, Kajikawa O, Elson G, et al. Effect of Toll-like receptor 4 blockade on pulmonary inflammation caused by mechanical ventilation and bacterial endotoxin. Exp Lung Res. 2008;34:225–243. doi: 10.1080/01902140802022492. [DOI] [PubMed] [Google Scholar]
- 21.Huang GT, Shagramanova K, Chan SW. Formation of odontoblast-like cells from cultured human dental pulp cells on dentin in vitro. J Endod. 2006;32:1066–1073. doi: 10.1016/j.joen.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Huang GT. Dental pulp and dentin tissue engineering and regeneration: advancement and challenge. Front Biosci (Elite Ed) 2011;3:788–800. doi: 10.2741/e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang GT, Sonoyama W, Liu Y, Liu H, Wang S, Shi S. The hidden treasure in apical papilla: the potential role in pulp-dentin regeneration and bioroot engineering. J Endod. 2008;34:645–651. doi: 10.1016/j.joen.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang GT. Pulp and dentin tissue engineering and regeneration: current progress. Regen Med. 2009;4:69770–69777. doi: 10.2217/rme.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruch JV, Lesot H, Begue-Kirn C. Odontoblast differentiation. Int J Dev Biol. 1995;39:51–68. [PubMed] [Google Scholar]
- 26.Smith AJ, Patel M, Graham L, Sloan AJ, Couper PR. Dentine regeneration: the role of stem cells and molecular signaling. O Biosci Med 2. 2005;2:127–132. [Google Scholar]
- 27.Wei X, Ling J, Wu L, Liu L, Xiao Y. Expression of mineralization markers in dental pulp cells. J Endod. 2007;33:703–708. doi: 10.1016/j.joen.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Gluhak-Heinrich J, Wang YH, et al. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horiba N, Maekawa Y, Matsumoto T, Nakamura H. A study of the distribution of endotoxin in the dentinal wall of infected root canals. J Endod. 1990;16:331–334. doi: 10.1016/S0099-2399(06)81944-8. [DOI] [PubMed] [Google Scholar]
- 30.Barthel CR, Levin LG, Reisner HM, Trope M. TNF-alpha release in monocytes after exposure to calcium hydroxide treated Escherichia coli LPS. Int Endod J. 1997;30:155–159. doi: 10.1046/j.1365-2591.1997.00066.x. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto M, Asai Y, Ogawa T. Separation and structural analysis of lipoprotein in a lipopolysaccharide preparation from Porphyromonas gingivalis. Int Immunol. 2004;16:1431–1437. doi: 10.1093/intimm/dxh146. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa T, Hamada S. Hemagglutinating and chemotactic properties of synthetic peptide segments of fimbrial protein from Porphyromonas gingivalis. Infect Immun. 1994;62:3305–3310. doi: 10.1128/iai.62.8.3305-3310.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa T, Yagi T. Bioactive mechanism of Porphyromonas gingivalis lipid A. Periodontol 2000. 2010;54:71–77. doi: 10.1111/j.1600-0757.2009.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Machado de Oliveira JC, Siqueira JF, Rocas IN, et al. Bacterial community profiles of endodontic abscesses from Brazilian and USA subjects as compared by denaturing gradient gel electrophoresis analysis. Oral Microbiol Immunol. 2007;22:14–18. doi: 10.1111/j.1399-302X.2007.00311.x. [DOI] [PubMed] [Google Scholar]