Abstract
Background
To study the methylation status of genes that play a role in the p53-Bax mitochondrial apoptosis pathway and its clinical significance in cholangiocarcinoma.
Patients and Methods
Out of 36 cases cholangiocarcinoma patients from April 2000 to May 2005 were collected.Promoter hypermethylation of DAPK, p14ARF, and ASC were detected by methylation-specific PCR on cholangiocarcinoma and normal adjacent tissues samples. Mutation of the p53 gene was examined by automated sequencing. Correlation between methylation of these genes and/or p53 mutation status with clinical characteristics of patients was investigated by statistical analysis.
Results
We found 66.7% of 36 cholangiocarcinoma patients had methylation of at least one of the tumor suppressor genes analyzed. p53 gene mutation was found in 22 of 36 patients (61.1%). Combined p53 mutation and DAPK, p14ARF, and/or ASC methylation was detected in 14 cases (38.9%). There were statistically significant differences in the extent of pathologic biology, differentiation, and invasion between patients with combined p53 mutation and DAPK, p14ARF, and/or ASC methylation compared to those without (P < 0.05). The survival rate of patients with combined DAPK, p14ARF, and ASC methylation and p53 mutation was poorer than other patients (P < 0.05).
Conclusion
Our study indicates that methylation of DAPK, p14ARF, and ASC in cholangiocarcinoma is a common event. Furthermore, p53 mutation combined with DAPK, p14ARF, and/or ASC methylation correlates with malignancy and poor prognosis.
Keywords: cholangiocarcinoma, methylation-specific PCR, p53-Bax mitochondrial apoptosis pathway
Background
Apoptosis or programmed cell death is a normal physiological control mechanism that is critical in maintaining homeostasis. Apoptotic inhibition can lead to abnormal cell survival and carcinogenesis. The p53-Bax mitochondrial apoptosis pathway plays an important role in inducing cell death after DNA damage or under conditions of cellular stress [1]. However, some genes in the pathway have latent methylation sites, such as p14ARF, DAPK, and ASC/TMS1, which can be methylated and inactivated resulting in cancer development [2]. Studies investigating methylation of gene that play a role in activation of the p53-Bax mitochondrial apoptosis pathway has not been well studied in cholangiocarcinoma to date. Thus, investigation of the methylation status of the genes in this pathway may give insight into the mechanism of cholangiocarcinoma development, resulting in enhanced diagnostic and treatment capabilities. We determined the methylation status of p14ARF, DAPK, and ASC/TMS1 as well as p53 mutation status in cancerous and normal adjacent tissue for 36 cholangiocarcinoma patients. Correlation between p14ARF, DAPK, and ASC/TMS1 methylation and/or p53 mutation with the biological behavior of cholangiocarcinoma and clinical outcome was examined.
Methods
1. Patients
Cholangiocarcinoma and normal adjacent tissues samples were randomly selected from specimens obtained and immediately frozen from patients treated by surgical resection from April 2000 to May 2005 in the Department of General Surgery of the First Affiliated Hospital of China Medical University and Hepatobiliary Surgery of the Affiliated Yantai Yuhuangding Hospital of Qingdao University Medical College. Informed consent was obtained from all the patients. Samples included 18 tubular adenocarcinomas, 9 papillary adenocarcinomas, 4 mucoid carcinomas, and 5 non-differentiated carcinomas. Twenty-five samples were well-moderately differentiated and 11 samples were poorly differentiated. According to the UICC standard, 16 cases were T1, 10 cases were T2, and 10 cases were T3. Lymph node metastasis was found in 33 cases. Non-lymph node metastasis was found in the 3 cases. This group contained 23 males and 13 females, with ages ranging from 36 to 71 years (median age of 61.2 years). A11 of the samples were frozen at -80°C until DNA extraction and histological diagnosis by a pathologist.
2. DNA Extraction
DNA was obtained from samples using the QIAamp DNA Micro kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). Briefly, tissue sample weighing less than 10 mg was injected into a 1.5 ml microcentrifuge tube, 180 μl Buffer ATL and 20 μl proteinase k was immediately added followed by pulse-vortexing for 15 s. Samples were incubated overnight at 56°C. Buffer AL (200 μl) and 100% ethanol (200 μl) were added and samples incubated for 5 min at room temperature. Lysate was then added to a QIAamp MinElute Column, 500 μl Buffer AW1 and Buffer AW2 added, and samples were centrifuged at 15000 rpm. Buffer AE (100 μl) was added and DNA obtained. DNA was quantified by spectrophotometric absorption and the A260/280 ratio.
3. Methylation specific PCR
We examined the methylation status of genes by using the methylation specific polymerase chain reaction (MSP) method. Briefly, 1 mg of tissue DNA was urea/bisulfite-treated to render unmethylated cytosines to uracil according to the method previously described [3] to render unmethylated cytosines to uracil. The modified DNA was resuspended in 20 μl of TE Buffer and immediately subjected to PCR or stored at -20°C. Table 1 shows primer sequences used for PCR. Primers were synthesized by Hokkaido Bioscience Company (Ohtsu, Japan).
Table 1.
MSP primers sequences
| gene | primer sequences | product (bp) |
|---|---|---|
| p14ARF | M: GTG TTA AAG GGCTGGCCGTAGGC | 122 |
| AAA ACC CTC ACT CGC GAC GA | ||
| U: TTT TTG GTG TTA AAG GGT GGT GTA GT | 132 | |
| CAC AAA AAC CCT CAC TCA CAA CAA | ||
| DAPK | M: GGA TAG TCG GAT CGA GTT AAC GTC | 98 |
| CCC TCC CAA ACG CCG A | ||
| U: GGA GGA TAG TTG GAT TGA GTT AAT GTT | 106 | |
| CAA ATC CCT CCC AAA CAC CAA | ||
| TMS1/ASC | M: TTG TAG CGG GGT GAG CGG C | 191 |
| AAC GTC CAT AAA CAA CGC G | ||
| U: GGT TGT AGT GGG GTG AGT GGT | 196 | |
| CAA AAC ATC CAT AAA CAA CAA CAC A |
M: Methylated specific PCR primers. U: Unmethylated specific PCR primers.
PCR reactions were performed with a 20 μl reaction volume containing 1 U of Hot start EX Taq DNA polymerase (Takara Biochemical, Japan), 2 μl of 10X EXtaq Buffer, 2 μl of dNTP mixture, forward and reverse primers (8 pmol each), and 1 μl of DNA template. The conditions for the first PCR reaction were as follows: 96°C for 3 min, 40 cycles: 96°C for 30 s, 60°C for 30 s, 72°C for 30 s; 72°C for 4 min. Intergen (IVD) was used as positive control for methylation. DNA from normal lymphocytes was used as a negative control for methylation, and water was used as a negative control. PCR product (5 μl) was resolved by 1.0% TBE gel electrophoresis. Each MSP reaction was repeated at least three times.
4. PCR amplification and DNA sequencing
Primers for amplification of p53 exon 5-8 were synthesized by Hokkaido Bioscience Company(Japan)[4]. PCR used a 20 μL reaction volume containing 1 unit of Hot start EXTaq DNA polymerase (Takara, Biochemical, Japan), 2 μL of 10 × EXTaq Buffer,2 μL of dNTP mixture and each primers (8 pmol each for reaction)and 1 μL of DNA template. The condition of the first PCR was as follows: 96°C 3 min for denaturation, 40 cycles of PCR,(96°C,30s,60°C,30s,72°C,30s) with a final elongation step of 4 min at 72°C.Water was used as negative control. PCR product (5 μl) was resolved by 10 g/L TBE gel electrophoresis. Each PCR reaction was repeated three times.
All of the PCR products were purified using Auto seq TMG-50 (Amersham Biochemical Company, Piscataway, NJ). BigDye Terminator Cycle sequencing Ready Reaction (Perkin Elmer, Waltham, MA) was performed to sequence DNA. The sequencing primers used were the same as the PCR primers, but, the concentration was 1/10th of that used for the PCR reaction. The sequencing reaction condition was as follows: 95°C, 4 min; 40 cycles: 95°C for 30 s, 55°C for 30 s, 72°C for 30 s; 72°C for 4 min. Both sense and antisense products were analyzed on an ABI prism 310 Genetic Analyzer (Perkin Elmer). Each reaction was repeated three times.
5. Follow-up data
A follow-up visit at the hospital was offered every three months for the first year after surgery, every six months for the second year, and every six to nine months for the third year. All follow-up data was recorded.
6. Statistical analysis
All data were analyzed by Chi-square test or Spearman rank correlation analysis. The survival rate was calculated using the Kaplan-Meier method and compared with long-rank test using SPSS 13.0 software (IBM, Armonk, NY; size of test α = 0.05).
Results
1.p14ARF, DAPK, and ASC/TMS1 methylation status in cholangiocarcinoma
The methylation status of tumor suppressor genes that play a role in the p53-Bax mitochondrial apoptosis pathway (p14ARF, DAPK, and TMS1/ASC) was investigated in cholangiocarcinoma samples. It was found that 66.7 of 36 cases had methylation of at least one of these genes. The frequency of methylation in cholangiocarcinoma tissue was: p14ARF (24.0%), DAPK (30.6%), and TMS1/ASC (36.1%). The frequency of methylation in normal adjacent tissue was low: DAPK (5.6%) and TMS1/ASC (8.3%) (Figure 1). Methylation of all tumor suppressor genes analyzed was significantly increased in cholangiocarcinoma samples compared to normal adjacent tissue (P < 0.001).
Figure 1.
Methylation specific PCR analysis of p14ARF, TMS1/ASC, and DAPK in cholangiocarcinoma and normal adjacent tissue. L: 100 bp DNA ladder; U: unmethylated primers; M: methylated primers;. a-e:cholangiocarcinoma samples; IVD: positive control; N: negative control.
2. p53 gene mutations in cholangiocarcinoma tissue
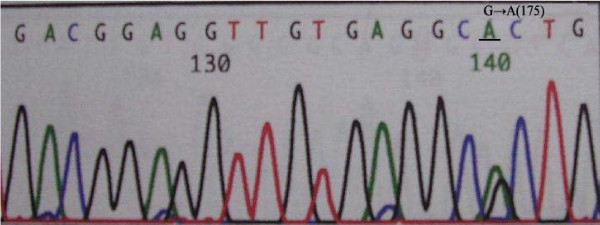
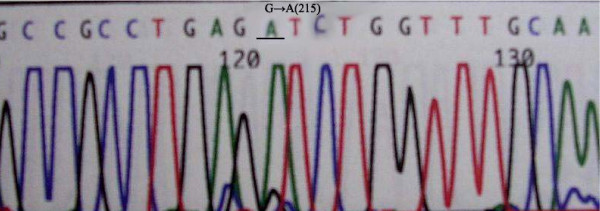
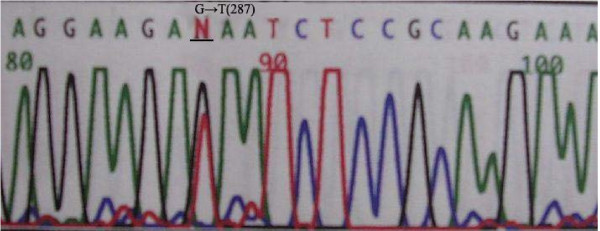
Exon 5-8 of p53 was amplified by PCR in all samples; gene mutations were detected by DNA sequencing. Mutations in p53 were detected in 22 of 36 patients (61.1%). There were 7 cases of exon 5 mutations, located on codons 161, 175 and 196 codon. All these mutations were transitions (G:C/A:T). Exon 6 mutations were detected in 6 cases, located on codons 209, 213, and 215; 4 mutations were transitions (G:C/A:T) and 2 mutations were tranversions (G-T). Exon 7 mutations were found in 3 cases, located on codons 248 and 252. Exon 8 mutations were detected in 6 cases, located on codons 282, 287, 289, and 306; 4 mutations were transitions (G:C/A:T) and 2 mutations were tranversions (G-T) (Figure 2, 3, and 4).
Figure 2.
Representative transition of p53 exon 5 (G: C/A: T) found in cholangiocarcinoma.
Figure 3.
Representative transition of p53 exon 6 (G: C/A: T) found in cholangiocarcinoma.
Figure 4.
Representative tranversion of p53 exon 8 (G → T) found in cholangiocarcinoma.
3. Methylation patterns of DAPK, p14ARF, and ASC in cholangiocarcinoma
Promoter region methylation of p14ARF, DAPK, and TMS1/ASC in cholangiocarcinoma is common. There were 15 cases (41.1%) with methylation of one of these genes, 9 cases (25.0%) with methylation of two of these genes, and no cases where all three genes were methylated. In some cases (13.9%) of one of these genes was found to be methylated in normal adjacent tissue, however, there were no cases where two of these genes were found methylated in normal tissue. None of these genes was found to be methylated at a significantly higher level in tumor tissue than the other genes (P > 0.05). The methylation profile for p14ARF, DAPK, and TMS1/ASC in all 36 cases of cholangiocarcinoma examined is summarized in Figure 5.
Figure 5.
Methylation profile of p14ARF, TMS1/ASC, and DAPK and p53 mutation in cholangiocarcinoma. Open boxes denote no detectable methylation or mutation. Shaded boxes represent positive methylation or mutation.
4. Correlation between p14ARF, DAPK, and TMS1/ASC methylation, p53 mutation, and pathologic biology in cholangiocarcinoma
There were no statistically significant differences in age or gender in the methylation of p14ARF, DAPK, and TMS1/ASC or p53 gene mutation on p53-Bax mitochondrial apoptosis pathway. Combined p53 mutation and p14ARF, DAPK, and/or TMS1/ASC methylation was found in 14 out of 36 cases (38.9%). Interestingly, p53 mutation and p14ARF, DAPK, and/or TMS1/ASC methylation was often found in mucoid carcinomas and non-differentiated carcinomas, these tumors tended to be poorly differentiated and had a deeper extent of invasion. There was a correlation between the occurrence of p53 mutations and tumor pathology, differentiation, and invasion (P < 0.05) (Table 2). Although combined p53 gene mutation and p14ARF, DAPK, and/or TMS1/ASC methylation was often found in patients with lymph node metastasis, this observation was not statistically significant (P > 0.05).
Table 2.
Correlation between combined p53 mutation and p14ARF, DAPK, and/or ASC/TMS1 methylation with pathologic biology in cholangiocarcinoma
| combined p53 mutation and p14ARF, DAPK, and/or ASC/TMS1 methylation | ||||
|---|---|---|---|---|
| Pathologic character | cases | positive | positive ratio (%) | P-value |
| Pathologic type | ||||
| Tubular carcinoma | 18 | 5 | 27.8 | |
| Papillary carcinoma | 9 | 1 | 11.1 | |
| Mucoid carcinoma | 4 | 4 | 100.0 | |
| Non-differentiated | 5 | 4 | 80.0 | < 0.05 |
| carcinoma | ||||
| Differentiation | ||||
| Well -Moderate | 25 | 6 | 24.0 | |
| Poor | 11 | 8 | 72.7 | < 0.05 |
| Invasion | ||||
| T1 | 16 | 2 | 12.5 | |
| T2 | 10 | 4 | 40.0 | |
| T3 | 10 | 8 | 80.0 | < 0.05 |
| Lymph node metastasis | ||||
| N0 | 3 | 0 | 0 | |
| N1 | 33 | 14 | 42.4 | > 0.05 |
5. Correlation of combined p14ARF, DAPK, and TMS1/ASC methylation and p53 mutation with prognosis in cholangiocarcinoma
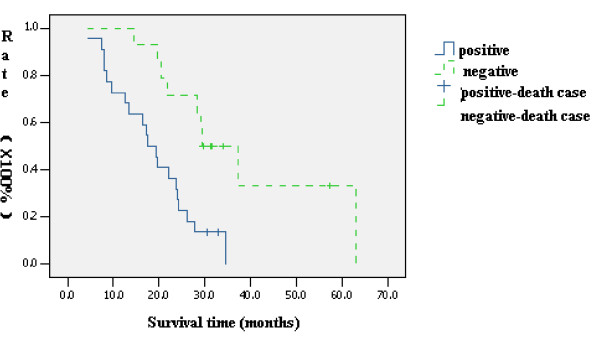
Complete follow-up information was available for all the 36 patients enrolled in this study with a mean follow-up time period of 23.78 months, in which 29 patients died and 7 survived. The one-, two-, and three-year survival rates were 44.0%, 21.0%, and 14.0%, respectively, with a median survival time of 22.29 months. The one-, two-, and three-year survival rates of patients with combined methylation of p14ARF, DAPK, and/or TMS1/ASC and p53 mutation was 28.0%, 5.0%, and 0.0%, respectively, with a median survival time of 18.6 months. The one-, two-, and three-year survival rates of patients without combined methylation of the tumor suppressor genes analyzed was 70.0%, 43.0%, and 28.0%, with a median survival time of 33.1 months. The survival rate between the two groups was significantly different (X2 = 9.06, P < 0.05) (Figure 6). Combined p53 mutation and p14ARF, DAPK, and/or TMS1/ASC methylation correlated with poor prognosis.
Figure 6.
Kaplan-Meier curve of p14ARF, TMS1/ASC, and DAPK methylation and p53 mutation on p53-Bax mitochondrial apoptosis pathway and prognosis in cholangiocarcinoma. Positive: patients with combined p53 mutation and p14ARF, TMS1/ASC, and/or DAPK; negative: patients without combined p53 mutation and p14ARF, TMS1/ASC, and/or DAPK; + = a patient death.
Discussion
Cholangiocarcinoma is a malignant tumor of the epithelial bile duct, and accounts for 3.0% of malignant tumors of the gastrointestinal tract [5,6]. In recent years, the incidence of cholangiocarcinoma and the mortality rate due to this disease have been increasing [7,8]. The mean age of patients with cholangiocarcinoma is 50 years, and the incidence in males is 1.5 times greater than that in females [9-11]. Without special clinical manifestations and tumor markers, early diagnosis and treatment of cholangiocarcinoma is very difficult.
Mammalian cells possess the capacity to epigenetically modify their genomes via the covalent addition of a methyl group to the 5-position of the cytosine ring within the context of the CpG dinucleotide. Certain regions of the genome, which are often clustered at the 5'-ends of genes possess the expected CpG frequency and have been termed CpG islands. CpG island methylation has been shown to be essential for normal development, X-chromosome inactivation, imprinting, and the suppression of parasitic DNA sequences. However, hypermethylation of CpG islands can cause inactivation of genes [12,13].
In this study, promoter hypermethylation of DAPK, p14ARF, and ASC was detected by methylation-specific PCR, and mutation of the p53 gene status at exons 5-8 was examined by automated sequencing in 36 cases of cholangiocarcinoma. We found that 66.7% of patients had methylation of at least one of the tumor suppressor genes analyzed. The frequency of methylation in cholangiocarcinoma was: p14ARF (24.0%), DAPK (30.6%), and TMS1/ASC (36.1%). The frequency of methylation in normal adjacent tissues was: DAPK (5.6%) and TMS1/ASC (8.3%). p53 gene mutations were found in 22 of 36 patients (61.1%). Combined p53 mutation and DAPK, p14ARF, and/or ASC methylation was detected in 14 cases (38.9%).
The p53-Bax mitochondrial apoptosis pathway plays an important anti-tumor role. When cells suffer DNA damage or certain types of stress, p53 is activated and sends a death signal to cells. p53 activity is regulated by MDM2, which induces the degradation of p53. MDM2 levels are regulated by p14ARF. DNA damage or cellular stress induces expression of p14ARF, resulting in degradation of MDM2 and stabilization of p53. Stress signals induce p14ARF expression, in part, through death-associated protein kinase, DAPK. Activated p53 promotes apoptosis through Bax, a molecule that stimulates the release of cytochrome-c from mitochondria, activating procaspase-9 and resulting in cell death. TMS1/ASC plays a role in caspase-9-mediated apoptosis. Expression of regulators of the p53-Bax mitochondrial apoptosis pathway, including p14ARF, DAPK, and ASC/TMS1, can become inactivated by promoter methylation and lead to the destruction of the pathway [14,15]. Our study indicates that methylation of regulators of the p53-Bax mitochondrial apoptosis pathway is a common epigenetic event in cholangiocarcinoma.
Futhermore, we found p53 gene mutation combined with DAPK, p14ARF, and/or ASC methylation in 14 cases (38.9%), which correlated with extent of pathologic biology, differentiation, and invasion of the tumors (P < 0.05). Patients with combined p53 mutation and methylation of tumor suppressor genes tended to have more malignant tumors. Moreover, the one-, two-, and three-year survival rate of patients with combined tumor suppressor gene methylation and p53 mutation was 28.0%, 5.0%, and 0.0%, respectively, with a median survival time of 18.6 months. Those patients without combined p53 mutation and tumor suppressor methylation was 70.0%, 43.0%, and 28.0%, with a median survival of 33.1 months. Therefore, combined p53 mutation and DAPK, p14ARF, and/or ASC methylation correlates with poor prognosis in cholangiocarcinoma patients.
Although surgery can relieve obstruction of the biliary tract, the distinct pathobiology of cholangiocarcinoma makes it difficult to obtain a radical surgical cure. Thus, chemotherapeutics are an important part of combined therapy. However, at present, it is unclear how effective chemotherapeutics are in treating cholangiocarcinoma patients. Our study indicates the methylation status of regulators of the p53-Bax mitochondrial apoptosis pathway could indicate the biological behavior of cholangiocarcinoma and predict prognosis of these patients, providing a basis for postoperative adjunctive treatment [16]. Further research is needed to investigate the methylation status of p53-Bax mitochondrial apoptosis pathway genes in cholangiocarcinoma patients and how this correlates with response to drug therapy.
Conclusion
In conclusion, 66.7% of 36 cholangiocarcinoma cases had methylation of at least one of the tumor suppressor genes analyzed. These data indicate that methylation of genes that regulate the p53-Bax mitochondrial apoptosis pathway is a common event cholangiocarcinoma. Mutation of the p53 gene was found in 61.1% of cases. Combined p53 mutation and DAPK, p14ARF, and/or ASC methylation was found in 38.9% of cases, and these patients had a poorer pathologic biology and prognosis than the other patients. Future studies are required to assess DAPK, p14ARF, and ASC methylation status and to the possibility of using DNA demethylating drugs to treat cholangiocarcinoma patients.
Funding
The study was supported by a grant from Natural Science Foundation of Shandong Province, China (No. Y2008C82).
Competing interests
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
Authors' contributions
LXF proposed the study and wrote the first draft of the manuscript. TK analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. LXF is the guarantor. All authors read and approved the final manuscript'
Contributor Information
Liu Xiaofang, Email: Liu634@263.net.
Tang Kun, Email: tangk@yahoo.com.
Yu Shaoping, Email: ysp@163.com.
Wang Zaiqiu, Email: wzq@263.net.
Su Hailong, Email: shl1976@163.com.
Acknowledgements
This work was supported by a grant from Natural Science Foundation of Shandong Province, China (No. Y2008C82). We thank Dr Zhang H and Dr Wang YD (China Medical University) for encouragement and technical assistance during this study.
References
- Schuler M, Green DR. Mechanisms of p53-dependent apoptosis. Biochemical Society Transactions. 2001;29(6):684–688. doi: 10.1042/BST0290684. [DOI] [PubMed] [Google Scholar]
- Jones PA. Death and methylation. Nature. 2001;409:141–143. doi: 10.1038/35051677. [DOI] [PubMed] [Google Scholar]
- Liu XF, Zhu SG, Zhang H. The methylation status of the TMS1/ASC gene in cholangiocarcinoma and its clinical significance. HBPT INT. 2006. pp. 449–453. [PubMed]
- Liu XF, Zhang H, Zhu SG. Corrlation of p53 gene mutation and expression in cholangiocarcinoma. World J Gastroenterology. 2006. pp. 4609–4772. [DOI] [PMC free article] [PubMed]
- Mosconi S, Beretta GD, Labianca R, Zampino MG, Gatta G, Heinemann V. Cholangiocarcinoma. Crit Rev Oncol Hematol. 2009;69:259–270. doi: 10.1016/j.critrevonc.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcino -ma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- Paik WH, Park YS, Hwang JH, Lee SH, Yoon CJ, Kang SG. Palliative treatment with self-expandable metallic stents in patients with advanced type III or IV hilar cholangiocarcinoma: a percutaneous versus endoscopic approach. Gastrointest Endosc. 2009;69:55–62. doi: 10.1016/j.gie.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Aljiffry M, Walsh MJ, Molinari M. Advances in diagnosis, treatment and palliation of cholangiocarcinoma: 1990-2009. World J Gastroenterol. 2009;15:4240–4262. doi: 10.3748/wjg.15.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaib YH, El-Serag HB, Nooka AK, Thomas M, Brown TD, Patt YZ, Hassan MM. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: a hospital-based case-control study. Am J Gastroenterol. 2007;102:1016–1021. doi: 10.1111/j.1572-0241.2007.01104.x. [DOI] [PubMed] [Google Scholar]
- Welzel TM, Graubard BI, El-Serag HB, Shaib YH, Hsing AW, Davila JA, McGlynn KA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: a population-based case-control study. Clin Gastroenterol Hepatol. 2007;5:1221–1228. doi: 10.1016/j.cgh.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaib Y, El-serav HB. The epidemiology of cholangiocarcinoma. Sem in Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Uriol E, Diego LB. High performance capillary electrophoretic method for the quantification of 5-methyl-2'-deoxycytidine in genomic DNA:application to plant, animal and human cancer tissues. Electrophoresis. 2002. pp. 1677–1681. [DOI] [PubMed]
- Baylin SB, Esteller M, Rountree MR. Aberrant patterns of DNA methylation, chromation and gene expression in cancer. Hum Mol Genet. 2001. pp. 687–692. [DOI] [PubMed]
- Ohtsuka T, Ryu H, Minamishima YA, Macip S. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nature Cell Biology. 2004;6(2):121–128. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Liu XF, Koga Y. Methylation-induced silencing of ASC and the effect of expressed ASC on p53-mediated chemosensitivity in colorectal cancer. Oncogene. 2006. pp. 1807–1811. [DOI] [PubMed]
- Yuasa Y. DNA methylation in cancer and aging. Mechanisms of aging and development. 2002;123:1649–1654. doi: 10.1016/S0047-6374(02)00100-8. [DOI] [PubMed] [Google Scholar]