Abstract
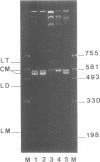
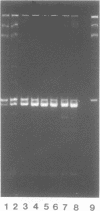
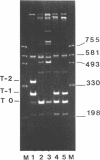
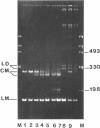
We have investigated the effect on the DNA structure of protein MC1, a basic and small polypeptide (Mr 10700) representing the major chromosomal protein in Methanosarcinaceae. The ability of protein MC1 to strongly favour cyclization upon polymerization of short DNA fragments by T4 DNA ligase indicates that protein MC1 mediates DNA bending. Several negatively supercoiled topoisomers of minicircles were obtained with DNA fragments of 203 and 146 bp, their distribution depends upon the amount of protein MC1 complexed with DNA. In addition, protein MC1 can induce a compaction of a nicked plasmid.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Chartier F., Laine B., Belaïche D., Touzel J. P., Sautière P. Primary structure of the chromosomal protein MC1 from the archaebacterium Methanosarcina sp. CHTI 55. Biochim Biophys Acta. 1989 Aug 14;1008(3):309–314. doi: 10.1016/0167-4781(89)90021-3. [DOI] [PubMed] [Google Scholar]
- Chartier F., Laine B., Bélaïche D., Sautière P. Primary structure of the chromosomal proteins MC1a, MC1b, and MC1c from the archaebacterium Methanothrix soehngenii. J Biol Chem. 1989 Oct 15;264(29):17006–17015. [PubMed] [Google Scholar]
- Chartier F., Laine B., Sautiere P. Characterization of the chromosomal protein MC1 from the thermophilic archaebacterium Methanosarcina sp. CHTI 55 and its effect on the thermal stability of DNA. Biochim Biophys Acta. 1988 Nov 10;951(1):149–156. doi: 10.1016/0167-4781(88)90035-8. [DOI] [PubMed] [Google Scholar]
- Dripps D., Wartell R. M. DNA bending induced by the catabolite activator protein allows ring formation of a 144 bp DNA. J Biomol Struct Dyn. 1987 Aug;5(1):1–13. doi: 10.1080/07391102.1987.10506370. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges-Garcia Y., Hagerman P. J., Pettijohn D. E. DNA ring closure mediated by protein HU. J Biol Chem. 1989 Sep 5;264(25):14621–14623. [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Imbert M., Laine B., Helbecque N., Mornon J. P., Hénichart J. P., Sautière P. Conformational study of the chromosomal protein MC1 from the archaebacterium Methanosarcina barkeri. Biochim Biophys Acta. 1990 May 8;1038(3):346–354. doi: 10.1016/0167-4838(90)90247-d. [DOI] [PubMed] [Google Scholar]
- Kosturko L. D., Daub E., Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989 Jan 11;17(1):317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz D., Fritsch A., Buc H. Variations of intramolecular ligation rates allow the detection of protein-induced bends in DNA. EMBO J. 1986 Apr;5(4):799–803. doi: 10.1002/j.1460-2075.1986.tb04284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine B., Belaiche D., Sautiere P., Biserte G. Characterization and structural study of the DNA-binding protein HRm From Rhizobium meliloti. Biochem Biophys Res Commun. 1982 May 14;106(1):101–107. doi: 10.1016/0006-291x(82)92063-0. [DOI] [PubMed] [Google Scholar]
- Laine B., Chartier F., Imbert M., Lewis R., Sautiere P. Primary structure of the chromosomal protein HMb from the archaebacteria Methanosarcina barkeri. Eur J Biochem. 1986 Dec 15;161(3):681–687. doi: 10.1111/j.1432-1033.1986.tb10493.x. [DOI] [PubMed] [Google Scholar]
- Mignotte B., Delain E., Rickwood D., Barat-Gueride M. The Xenopus laevis mitochondrial protein mtDBP-C cooperatively folds the DNA in vitro. EMBO J. 1988 Dec 1;7(12):3873–3879. doi: 10.1002/j.1460-2075.1988.tb03273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Shore D., Langowski J., Baldwin R. L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Comparative gel electrophoresis measurement of the DNA bend angle induced by the catabolite activator protein. Biopolymers. 1990 Jan;29(1):29–38. doi: 10.1002/bip.360290106. [DOI] [PubMed] [Google Scholar]