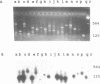
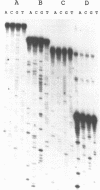
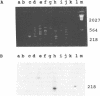
Abstract
We describe a modification of a polymerase chain reaction method called 'targeted gene walking' that can be used for the amplification of unknown DNA sequences adjacent to a short stretch of known sequence by using the combination of a single, targeted sequence specific PCR primer with a second, nonspecific 'walking' primer. This technique can replace conventional cloning and screening methods with a single step PCR protocol to greatly expedite the isolation of sequences either upstream or downstream from a known sequence. A number of potential applications are discussed, including its utility as an alternative to cloning and screening for new genes or cDNAs, as a method for searching for polymorphic sites, restriction endonuclease or regulatory regions, and its adaptation to rapidly sequence DNA of lengthy unknown regions that are contiguous to known genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Burmer G. C., Rabinovitch P. S., Loeb L. A. Analysis of c-Ki-ras mutations in human colon carcinoma by cell sorting, polymerase chain reaction, and DNA sequencing. Cancer Res. 1989 Apr 15;49(8):2141–2146. [PubMed] [Google Scholar]
- Copley C. G., Boot C., Bundell K., McPheat W. L. Unknown sequence amplification: application to in vitro genome walking in Chlamydia trachomatis L2. Biotechnology (N Y) 1991 Jan;9(1):74–79. doi: 10.1038/nbt0191-74. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Sukumar S., Barbacid M. Activation of ras oncogenes preceding the onset of neoplasia. Science. 1990 Jun 1;248(4959):1101–1104. doi: 10.1126/science.2188364. [DOI] [PubMed] [Google Scholar]
- Li H. H., Gyllensten U. B., Cui X. F., Saiki R. K., Erlich H. A., Arnheim N. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature. 1988 Sep 29;335(6189):414–417. doi: 10.1038/335414a0. [DOI] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988 Nov;120(3):621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Curiel D. T., Brantly M. L., Holmes M. D., Crystal R. G. Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J Lab Clin Med. 1989 Aug;114(2):105–113. [PubMed] [Google Scholar]
- Parker J. D., Burmer G. C. The oligomer extension "hot blot": a rapid alternative to Southern blots for analyzing polymerase chain reaction products. Biotechniques. 1991 Jan;10(1):94–101. [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Syvänen A. C., Bengtström M., Tenhunen J., Söderlund H. Quantification of polymerase chain reaction products by affinity-based hybrid collection. Nucleic Acids Res. 1988 Dec 9;16(23):11327–11338. doi: 10.1093/nar/16.23.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T., Peterson M. G., Kemp D. J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988 Aug 25;16(16):8186–8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]